Abstract
Mitochondria govern many metabolic processes. In addition, mitochondria sense the status of metabolism and change their functions to regulate energy production, cell death, and thermogenesis. Recent studies have revealed that mitochondrial structural remodeling through division and fusion is critical to the organelle’s function. It has also become clear that abnormalities in mitochondrial division and fusion are linked to the pathophysiology of metabolic diseases such as diabetes and obesity. Here, we discuss the current understanding of the mechanisms of mitochondrial dynamics and their role in cellular and organismal metabolism.
Introduction
Popularly known as the “powerhouse of the cell”, the mitochondrion is a double membrane-bound organelle and the source of cellular energy, ATP [1,2]. Cellular metabolism relies on mitochondria to provide energy from oxidative phosphorylation. Mitochondria are highly dynamic organelles that constantly fuse and divide to maintain normal cellular functions (Figure 1) [3-8]. When this delicate balance between division and fusion is lost, mitochondrial function, metabolism, and signaling are altered. A range of pathological conditions, including cancer, aging, neurodegeneration, and metabolic disorders have been associated with altering the balance between fusion and division [9-12]. Although many studies have sought to understand the dynamic nature of this process over the past several decades, the complete molecular mechanisms, physiological function, and connection to human diseases remain unclear.
Figure 1. Mitochondrial morphology is regulated by division and fusion.
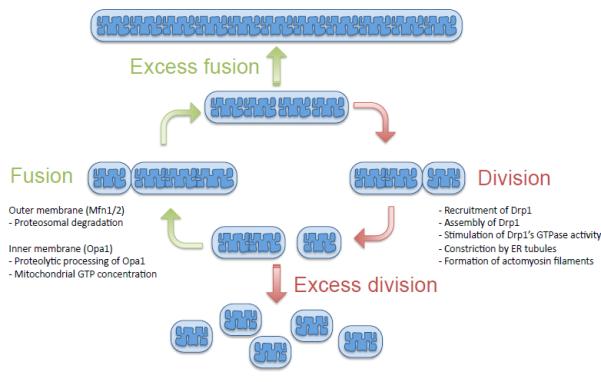
Mitochondria continuously divide and fuse and control their morphology. Mitochondrial division is initiated by recruitment of cytosolic Drp1 to the mitochondrial outer membrane by Drp1 receptors. On mitochondria, Drp1 assembles into helical filaments, wrapping around mitochondrial tubules. Drp1 filaments constrict and divide mitochondria, working together with ER tubules and actomyosin filaments. Mitochondrial fusion consists of outer membrane fusion and inner membrane fusion. Outer membrane fusion is mediated by mitofusin while inner membrane fusion is mediated by Opa1. Mitochondrial fusion is regulated by proteosomal degradation of mitofusins, proteolytic processing of Opa1 and production of GTP.
Mitochondrial dynamics refer to the perpetual process of fusion, division, movement, and morphological changes which take place in response to the ever-changing physiological demands of cells [13,14]. There is dedicated protein machinery that controls the mitochondrial dynamics in the cell (Table 1) [6,15,16]. In this review, we focus on mitochondrial division and fusion. Division is crucial for maintaining the number of mitochondria in growing cells, regulating cell death pathways, and eliminating damaged mitochondria as part of quality control through mitophagy [7,17]. In contrast, fusion is important for mixing of mitochondrial contents and maintaining electrical conductivity throughout the mitochondria [17]. These two opposing forces ensure that at any given time, the cell has a healthy mitochondrial population. Defects in the core components of these systems, three dynamin-related GTPases, give rise to several disease conditions, including neonatal death with severe neural defects (defects in outer membrane protein Drp1, which mediates division), Charcot-Marie-Tooth neuropathy type 2A, a neurodegenerative disease of peripheral neurons, (defects in outer membrane protein Mfn2 which mediates fusion), and inherited forms of dominant optic atrophy (defects in inner membrane protein Opa1, which mediates fusion) [9,10,18].
Table 1.
Key proteins involved in mitochondrial dynamics and associated disease. Main components of the mitochondrial fusion and fission machineries are indicated in model organisms from algae to mammals. Their location, functions and related diseases are shown.
| Key Mitochondrial dynamics proteins |
Algae | Yeast | Worms | Fly | Mammals | Function | Disease |
|---|---|---|---|---|---|---|---|
|
Dynamin related
protein1 (Drp1) |
Dnm1 | Dnm1 | DRP-1 | Drp1 | Drp1 | OM Fission | Post neonatal death with neurodevelopme ntal defects |
|
Mitochondrial
fission factor (Mff) |
Unknown | Unknown | Mff-1 | Mff-1 | Mff | Drp1 receptor | Unknown |
| MiD49/MiD51 | Unknown | Unknown | Unknown | Unknown | Mid49/Mid51 | Drp1 receptor | Unknown |
|
Fission Protein 1
(Fis1) |
Fis1 | Fis1 | Fis-1 | Fis1 | Fis1 | Drp1 receptor | Unknown |
|
Mdv1/Caf4/Num1/
Mdm36 |
Mda1/-/-/- | Mdv1/Caf4/Num 1/Mdm36 |
Unknown | Unknown | Unknown | Dnm1 adaptor protein |
Unknown |
|
Ganglioside induced
differentiation protein (GDAP1) |
Unknown | Unknown | Unknown | CG4623? | GDAP1 | Unknown | Charcot-Marie- Tooth type 4A |
|
Optic atrophy 1
(Opa1) |
Mgm1 | Mgm1 | EAT-3 | Opa1 | Opa1 | IM Fusion | Autosomal dominant Optic atrophy type 1 |
|
Mitofusin1/2
(Mfn1/2) |
Fzo1 | Fzo1 | Fzo-1 | Fzo1, Marf, dMfn |
Mfn1/Mfn2 | OM fusion | Charcot-Marie- Tooth type 2A |
| Ugo1 | Ugo1 | Ugo1 | Unknown | Unknown | Unknown | OM fusion Links Fzo1 and Mgm1 |
Unknown |
It is well known that mitochondrial energy production controls cellular and organismal metabolism. Studies have shown that mitochondrial division and fusion regulate these metabolic processes, and changes in metabolism affect the dynamics of mitochondria. Therefore, the sensing and adjusting of metabolism by mitochondria create physiological circuits that consist of negative and positive feedback loops to establish robust metabolic responses. In this review, we discuss recent findings linking mitochondrial dynamics to metabolism.
Components of Mitochondrial Dynamics
Mitochondrial division
The dynamic nature of mitochondria was first observed during the early twentieth century [19,20]. Identification of the molecular components has been determined in approximately the last 15 years (Table 1) [6,15]. Model organisms have been instrumental in identifying the core components of mitochondrial division and fusion. Dnm1/Drp1 is the main component of mitochondrial division. It is a cytosolic dynamin-related GTPase, which moves to the mitochondrial outer membrane where it self assembles via GTP binding. Loss of Drp1 results in long interconnected mitochondrial networks. Mitochondrial receptors with transmembrane domains are involved in targeting Drp1 to the outer mitochondrial membrane. Yeast genetics was used to identify the central division component, Dnm1 [21,22], and its receptor Fis1 [23], and adaptors Mdv1 [24-26], and Caf4 [27]. Num1 and Mdm36 are unique, separate components with dual functions, connecting Dnm1 to the actin cortex and regulating both mitochondrial division and positioning within the cell [28-31]. Analyses of dynamin homologs in Caenorhabditis elegans and in mammalian cells have identified Drp1 (a homolog of Dnm1), and shown that Dnm1/Drp1 are evolutionarily conserved division factors [32,33]. A mammalian homolog of Fis1 has been identified [34]. However, Fis1 appears to recruit Drp1 in a subset of cell types, and/or under specific physiological conditions such as mitochondrial stress [35-37]. Steady state recruitment of Drp1 likely depends on other receptors such as Mff (mitochondrial fission factor) and Mid49/51 (MIEF1/2). Mff was discovered in siRNA screens, using cultured Drosophila DS2R+ cells [38,39], while Mid49/51 was found through analyses of mitochondrial proteomes [40,41]. The crystal structure and biochemical characteristics of Mid51 suggested that it binds to ADP and GDP [42,43]. Purified Mid51 stimulated the GTPase activity of Drp1 in the presence of ADP, suggesting that Mid51 sensed the metabolic status of cells and regulated mitochondrial division [42,43]. Proteins that are involved in inner membrane fission are yet to be identified. However, it is likely that outer and inner membrane fissions are independent events which may be coordinated [18,32,44].
In addition to these mitochondrial components, the endoplasmic reticulum (ER) and actin cytoskeleton are also involved in mitochondrial division. ER tubules appear to encircle and constrict mitochondrial tubules prior to the recruitment of Drp1 to mitochondria [45]. At the inter-organelle interface, ER-associated formin, INF2, facilitates polymerization of actin to generate small patches of the actin cytoskeleton [46]. Other actin regulatory proteins such as cortactin, cofilin, and Arp2/3 complexes associate with mitochondria and regulate their division [47]. Myosin II is also assembled into filaments at the cytoskeleton and may form contractile networks to constrict mitochondrial tubules [48]. Interestingly, Drp1 receptors are located at the ER-mitochondria contact site, but in different manners. While a fraction of Mff is stably located at the organelle contact independently of Drp1, Mid proteins co-assemble with Drp1 [43,45]. Furthermore, Fis1 may be recruited to the contacts and form protein complexes with the ER proteins upon mitochondrial stress, possibly leading to mitophagy [36,37]. It would be important to understand how the actin cytoskeleton and Drp1 receptors interplay to coordinate Drp1 recruitment, assembly, and activation.
Mitochondrial fusion
Drosophila genetics discovered the first conserved fusion component, fuzzy onions, leading to subsequent identification of yeast (Fzo1) and mammalian homologs (Mfn1 and 2) (Table 1) [49-51]. Drosophila fuzzy onions is a developmentally regulated gene, and a homologous protein, Marf/Dmfn, was later found to be ubiquitously expressed [52]. Similar to Dnm1/Drp1, these proteins are dynamin-related GTPases. But unlike the division proteins, they have two transmembrane domains, which are embedded in the outer membrane and mediate fusion of the outer membrane. In addition to its role in membrane fusion, Mfn2 has also been shown to mediate membrane tethering of mitochondria with other organelles such as the ER [53] and melanosomes [54]. An inner membrane fusion component, Mgm1, was first identified in yeast, and later its mammalian homolog, Opa1, was characterized [55-59]. Mgm1 and Opa1 are also dynamin-related GTPases which are present in the inner membrane and the intermembrane space [60-64]. Interestingly, Mgm1/Opa1 has a role in the morphology of inner membrane cristae, which is important for the maintenance of the pro-apoptotic factor, cytochrome c, and the assembly of the electron transport chain complexes [65,66]. Studies of all these different organisms, along with the information from human diseases, have greatly helped elucidate the mechanism and function of the division and fusion machineries.
Metabolic control of mitochondrial dynamics
Mitochondria play a critical role in cellular bioenergetics by virtue of being the principal ATP generator in the cell. Cells sense changes in nutrient conditions and regulate mitochondrial ATP production to sustain normal physiology under stressful conditions such as starvation. Starvation induces autophagy, and mitochondria rapidly elongate in order to escape from being engulfed by autophagosomes, resulting in elimination by this quality control mechanism of the cell [67,68]. This elongation likely allows mitochondria to maintain ATP production and is mediated by post-translational modifications of Drp1 through modulating its phosphorylation status [67,68]. When mitochondria become dysfunctional and are targeted for degradation by mitophagy, mitochondrial division generates small organelles to facilitate their movements and engulfment of mitochondria by autophagosomes [69-76].
In addition to Drp1, the function of Opa1 is regulated by the mitochondrial energetic status. In healthy cells, Opa1 produces two major forms (a membrane anchored form and a soluble form in the intermembrane space) by proteolytic cleavage of its transmembrane domain by the inner membrane located ATP-dependent proteases (m-AAA and i-AAA) [77,78]. However, when mitochondria decrease their function and membrane potential, Opa1 is excessively processed by a metalloproteinase (Oma1), releasing the majority of the protein from its transmembrane anchor, and becoming incompetent for fusion [79-82]. It is likely that combinations of increased division and decreased fusion may effectively produce small mitochondria, and separate them from the rest of the population. This proteolytic processing of Opa1 is further regulated by the level of oxidative phosphorylation. Oxidative phosphorylation stimulated Opa1-mediated inner membrane fusion [83]. Surprisingly, this oxidative phosphorylation stimulated fusion required the cleavage of Opa1 by Oma1 or Yme1L [83]. Uncleaved forms of Opa1 may undergo changes in the status of assembly or conformation upon proteolytic cleavage, which directly participate at a specific step in the pathway of mitochondrial fusion. Interestingly, when both proteases were lost in Oma1 or Yme1L knockout cells, Opa1 maintained the ability to fuse the inner membrane without proteolytic processing [84]. Therefore, its cleavage may not be an essential step, and may function in a specific type of fusion such as oxidative phosphorylation stimulated fusion. These new findings suggest elegant regulation of Opa1-mediated inner membrane fusion, which is a very active area of current research in the field of mitochondrial dynamics.
GTP is a major source for the energy that drives mitochondrial division and fusion, and Drp1, Mfn, and Opa1 are all dynamin-related GTPases. This suggests that the production of GTP is coupled to membrane remodeling. Nucleoside diphosphate kinases, which generate GTP from GDP and ATP, are important for dynamin-mediated membrane remodeling in endocytosis and mitochondrial fusion [85]. Two cytoplasmic nucleoside diphosphate kinases (NM23-H1/2) provide endocytic dynamin with GTP, while a mitochondrial counterpart (NM23-H4) supplies GTP for mitochondrial fusion. Because NM23-H4 interacts with Opa1, production of GTP may be spatially coupled with biding to Opa1 [86]. It is currently unknown whether NM23-H4 supplies GTP to other mitochondrial dynamin GTPases, Mfn1/2, and Drp1.
Diabetes and obesity
Diabetes mellitus is a chronic disease which affects ~400 million people worldwide. It is a disease where the body is either unable to produce enough insulin to deal with elevated blood glucose levels or is unable to respond to insulin which is produced and secreted by pancreatic β cells [5]. Pancreatic β cells normally control excess amounts of glucose by increasing their mass and producing and releasing increased insulin [5]. It has long been known that the most significant source of oxidative stress in a cell is mitochondria [87,88]. Recent studies have indicated abnormal mitochondrial dynamics in diabetic individuals along with increased reactive oxygen species (ROS) production [87,88]. Hyperglycemia induces excessive production of ROS, which strongly impacts mitochondrial morphology [88]. A study in diabetic mice showed increased mitochondrial fragmentation along with elevated Drp1 and decreased Opa1 levels [89]. This phenotype could be rescued with superoxide anion scavengers, indicating that ROS played important roles in failure of cellular bioenergetic control during hyperglycemia through regulation of mitochondrial division and fusion [89]. In diabetics, high glucose induced ROS generation can also cause endothelial dysfunction, where abnormal mitochondrial division is the cause [90]. Hyperglycemia also stimulated mitochondrial division in podocytes in the kidney by activating Drp1 under diabetic conditions [91]. The loss of mitochondrial fusion by Opa1 deficiency in pancreatic β cells also caused hyperglycemia, without inducing apoptosis or a loss of mtDNA in mice [92]. Therefore, mitochondrial dynamics might shift the balance to excess division under hyperglycemic conditions. Readjusting the balance of division and fusion might be beneficial, and inhibition of the division machinery partially ameliorated muscle insulin insensitivity and signaling in genetically obese mice [93]. These findings highlight the importance of mitochondrial division and fusion in glucose homeostasis, and the pathological consequences from imbalanced homeostasis caused by hyperglycemia.
Obesity has been linked to dysregulation of appetite which is controlled by leptin [94]. Leptin is secreted from adipocytes and signals neurons in the hypothalamus. Leptin resistance is a key component of obesity. In the hypothalamus, there are at least two types of neurons with opposite functions; orexigenic agouti-related protein (Agrp) neurons and anorexigenic pro-opiomelanocortin precursor (POMC) neurons. In orexigenic Agrp neurons of mice, mitochondria fragment upon starvation. In contrast, when mice are fed with high fat diets, mitochondria elongate in a manner dependent on Mfns [95]. When Mfns were deleted, Agrp neurons decreased their firing frequency in response to high fat diets, perhaps due to decreased intracellular ATP levels, as the addition of exogenous ATP restored the defects in mitofusin-depleted Agrp neurons [95]. These changes in mitochondrial dynamics are important for the physiological control of metabolism and body weight. Although the mechanisms remain unclear, the mitochondrial fusion and division machinery may function to control appetite in leptin-mediated and diet-regulated signaling pathways in neurons. It is also possible that changes in mitochondrial dynamics affect respiratory functions and modulate intracellular ATP levels which, in turn, affect neuronal activities. In POMC neurons, abundance of Mfn2 decreased in diet-induced obese mice, leading to fragmentation of mitochondria, decreases in ER-mitochondria contacts, and ER stress [96]. These mice became resistant to leptin and increased their body weight. Interestingly, modest overexpression of Mfn2 decreased food intake and body weight in mice fed with high fat diets. Knockout of Mfn2 in POMC neurons led to phenotypes similar to those found in diet-induced obese mice, suggesting the pathological mechanisms underlying diet induced obesity involved changes in Mfn2, mitochondrial fusion, and mitochondria-ER interactions [96].
In addition to these neurons, mitochondria in adipocytes also changed their dynamics, and therefore their functions in thermogenesis. In brown adipocytes which control body temperature, the sympathetic neurotransmitter norepinephrine induced fragmentation of mitochondria by activating Drp1 and inactivating Opa1 [97]. This fragmentation promoted energy expenditure by inhibiting oxidative phosphorylation and enhancing heat production. It is possible that mitochondrial division and fusion in different parts of the body sense nutrient availability and respond to hormones to physiologically regulate metabolism.
Conclusions and Future Directions
Mitochondrial dynamics play an integral role in cellular and physiological remodeling of mitochondrial structure and function in response to metabolic changes. Dysregulation of these membrane remodeling processes can result in pathological conditions, including metabolic disorders. We anticipate that the list of diseases related to metabolic alterations controlled by mitochondrial division and fusion will expand as increased knowledge of the mechanistic links of these processes to diseases becomes available. For example, changes in mitochondrial morphology and respiration are associated with different types of cancers. However, it is unclear how mitochondrial division and fusion are involved in tumorigenesis. Mitochondria also remodel their structural organization and functional capacity during development of the cardiac, immune, and nervous systems, suggesting that abnormalities in mitochondrial division and fusion can contribute to developmental disorders in multiple tissues. Finally, understanding the mechanisms underlying age-related declines in mitochondrial function and disorganization in their structures will be an exciting topic for future studies.
Acknowledgements
We are grateful to many scientists who have contributed to our understanding of mitochondrial division and fusion, and apologize that we were not able to cite all of the relevant publications due to space limitation. This work was supported by NIH grants GM084015 (MI), AG042178 (PHR), AG047812 (PHR), GM089853 (HS) and NS084154 (HS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yun J, Finkel T. Mitohormesis. Cell metabolism. 2014 doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace DC. Bioenergetics in human evolution and disease: implications for the origins of biological complexity and the missing genetic variation of common diseases. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2013;368:20120267. doi: 10.1098/rstb.2012.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sesaki H, Adachi Y, Kageyama Y, Itoh K, Iijima M. In vivo functions of Drp1: Lessons learned from yeast genetics and mouse knockouts. Biochimica et biophysica acta. 2013;842:1179–1185. doi: 10.1016/j.bbadis.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lackner LL. Determining the shape and cellular distribution of mitochondria: the integration of multiple activities. Current opinion in cell biology. 2013;25:471–476. doi: 10.1016/j.ceb.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell metabolism. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamoto K, Shaw JM. Mitochondrial Morphology and Dynamics in Yeast and Multicellular Eukaryotes. Annu Rev Genet. 2005 doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 7.Elgass K, Pakay J, Ryan MT, Palmer CS. Recent advances into the understanding of mitochondrial fission. Biochimica et biophysica acta. 2013;1833:150–161. doi: 10.1016/j.bbamcr.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends in Cell Biology. 2013;23:64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy PH, Tripathi R, Troung Q, Tirumala K, Reddy TP, Anekonda V, Shirendeb UP, Calkins MJ, Reddy AP, Mao P, et al. Abnormal mitochondrial dynamics and synaptic degeneration as early events in Alzheimer's disease: Implications to mitochondria-targeted antioxidant therapeutics. Biochimica et biophysica acta. 2012;1822:639–649. doi: 10.1016/j.bbadis.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho DH, Nakamura T, Lipton SA. Mitochondrial dynamics in cell death and neurodegeneration. Cell Mol Life Sci. 2010;67:3435–3447. doi: 10.1007/s00018-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ong SB, Hall AR, Hausenloy DJ. Mitochondrial dynamics in cardiovascular health and disease. Antioxidants & redox signaling. 2013;19:400–414. doi: 10.1089/ars.2012.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shutt TE, McBride HM. Staying cool in difficult times: Mitochondrial dynamics, quality control and the stress response. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbamcr.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 15.Tamura Y, Itoh K, Sesaki H. SnapShot: Mitochondrial dynamics. Cell. 2011;145:1158. doi: 10.1016/j.cell.2011.06.018. 1158 e1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bui HT, Shaw JM. Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Current biology : CB. 2013;23:R891–899. doi: 10.1016/j.cub.2013.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoppins S. The regulation of mitochondrial dynamics. Current opinion in cell biology. 2014;29C:46–52. doi: 10.1016/j.ceb.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annual review of genetics. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 19.Lewis MR, Lewis WH. Mitochondria (and other cytoplasmic structures) in tissue cultures. Amer. J. Anat. 1915;17:339–401. [Google Scholar]
- 20.Lewis MR, Lewis WH. Mitochondria in Tissue Culture. Science. 1914;39:330–333. doi: 10.1126/science.39.1000.330. [DOI] [PubMed] [Google Scholar]
- 21.Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated Mitochondrial Fission Is a Multi-step Process Requiring the Novel Integral Membrane Component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerveny KL, McCaffery JM, Jensen RE. Division of mitochondria requires a noverl DNM1-interacting protien, Net2p. Molecular Biology of the Cell. 2001 doi: 10.1091/mbc.12.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tieu Q, Nunnari J. Mdv1p Is a WD Repeat Protein that Interacts with the Dynamin-related GTPase, Dnm1p, to Trigger Mitochondrial Division. J Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fekkes P, Shepard KA, Yaffe MP. Gag3p, an Outer Membrane Protein Required for Fission of Mitochondrial Tubules. J Cell Biol. 2000;151:333–340. doi: 10.1083/jcb.151.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffin EE, Graumann J, Chan DC. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerveny KL, Studer SL, Jensen RE, Sesaki H. Yeast mitochondrial division and distribution require the cortical num1 protein. Developmental Cell. 2007;12:363–375. doi: 10.1016/j.devcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 29.Hammermeister M, Schodel K, Westermann B. Mdm36 is a mitochondrial fission-promoting protein in Saccharomyces cerevisiae. Molecular biology of the cell. 2010;21:2443–2452. doi: 10.1091/mbc.E10-02-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Klecker T, Scholz D, Fortsch J, Westermann B. The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. Journal of cell science. 2013;126:2924–2930. doi: 10.1242/jcs.126045. The two studies (30 and 31) show that attachement of mitochondria to the cell cortex is important for mitochondrial division. Furthremore, the ER plays an important roles in Num1-mediated mitochondria-cell cortex association. [DOI] [PubMed] [Google Scholar]
- *31.Lackner LL, Ping H, Graef M, Murley A, Nunnari J. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E458–467. doi: 10.1073/pnas.1215232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 33.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *35.Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49 and MiD51 mediate Drp1 recruitment in mitochondrial fission. Molecular biology of the cell. 2013 doi: 10.1091/mbc.E12-10-0721. This article defines funtional contributions of the four Drp1 receptor proteins, Fis1, Mff, MiD49 and MiD51 to mitochondrial division. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *36.Shen Q, Yamano K, Head BP, Kawajiri S, Cheung JT, Wang C, Cho JH, Hattori N, Youle RJ, van der Bliek AM. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Molecular biology of the cell. 2014;25:145–159. doi: 10.1091/mbc.E13-09-0525. The authors reveal a specific role of the Drp1 receptor Fis1 in mitochondrial division in response to cellular stress. Fis1 and Drp1 promote mitochondrial division after Mff and Drp1 mediate mitochondrial division during stress induced mitophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamano K, Fogel AI, Wang C, van der Bliek AM, Youle RJ. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife. 2014;3:e01612. doi: 10.7554/eLife.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlen P, Tomilin N, Shupliakov O, Lendahl U, Nister M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. The EMBO journal. 2011;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO reports. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *42.Loson OC, Liu R, Rome ME, Meng S, Kaiser JT, Shan SO, Chan DC. The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor. Structure. 2014;22:367–377. doi: 10.1016/j.str.2014.01.001. These two studies (42 and 43) solve the crystal structure of the cytoplasmic domain of Mid51. ADP binds to MiD51 and regulates its functional interactions with Drp1. MiD51 also coassmbles together Drp1 during mitochonddrial division. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *43.Richter V, Palmer CS, Osellame LD, Singh AP, Elgass K, Stroud DA, Sesaki H, Kvansakul M, Ryan MT. Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission. The Journal of cell biology. 2014;204:477–486. doi: 10.1083/jcb.201311014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujioka H, Tandler B, Hoppel CL. Mitochondrial division in rat cardiomyocytes: an electron microscope study. Anatomical record. 2012;295:1455–1461. doi: 10.1002/ar.22523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *46.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. This paper shows that an ER-located actin regulator, INF2, promotes Drp1 recruitment and mitochondrial divisoin at ER-mitochodnria contact sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *47.Li S, Xu S, Roelofs BA, Boyman L, Lederer WJ, Sesaki H, Karbowski M. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. The Journal of cell biology. 2015;208:109–123. doi: 10.1083/jcb.201404050. This study reveals that Drp1 control the dynamics of actin depolymerization on mitochodnria during organelle division. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korobova F, Gauvin TJ, Higgs HN. A role for myosin II in mammalian mitochondrial fission. Current biology : CB. 2014;24:409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 50.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 51.Rapaport D, Brunner M, Neupert W, Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem. 1998;273:20150–20455. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- 52.Deng H, Dodson MW, Huang H, Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Brito OM, Scorrano L. Mitofusin 2: a mitochondria-shaping protein with signaling roles beyond fusion. Antioxid Redox Signal. 2008;10:621–633. doi: 10.1089/ars.2007.1934. [DOI] [PubMed] [Google Scholar]
- 54.Daniele T, Hurbain I, Vago R, Casari G, Raposo G, Tacchetti C, Schiaffino MV. Mitochondria and melanosomes establish physical contacts modulated by Mfn2 and involved in organelle biogenesis. Current biology : CB. 2014;24:393–403. doi: 10.1016/j.cub.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 55.Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 56.Griparic L, van der Wel NN, Orozco IJ, Peters PJ, van der Bliek AM. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J Biol Chem. 2004;279:18792–18798. doi: 10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- 57.Shepard KA, Yaffe MP. The yeast dynamin-like protein, mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance [In Process Citation] J Cell Biol. 1999;144:711–720. doi: 10.1083/jcb.144.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong ED, Wagner JA, Gorsich SW, McCaffery JM, Shaw JM, Nunnari J. The Dynamin-related GTPase, Mgm1p, Is an Intermembrane Space Protein Required for Maintenance of Fusion Competent Mitochondria. J Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sesaki H, Southard SM, Yaffe MP, Jensen RE. Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol Biol Cell. 2003;14:2342–2356. doi: 10.1091/mbc.E02-12-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sesaki H, Southard SM, Hobbs AE, Jensen RE. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem Biophys Res Commun. 2003;308:276–283. doi: 10.1016/s0006-291x(03)01348-2. [DOI] [PubMed] [Google Scholar]
- 61.McQuibban GA, Saurya S, Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- 62.Herlan M, Vogel F, Bornhovd C, Neupert W, Reichert AS. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- 63.Sesaki H, Dunn CD, Iijima M, Shepard KA, Yaffe MP, Machamer CE, Jensen RE. Ups1p, a conserved intermembrane space protein, regulates mitochondrial shape and alternative topogenesis of Mgm1p. The Journal of cell biology. 2006;173:651–658. doi: 10.1083/jcb.200603092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamura Y, Endo T, Iijima M, Sesaki H. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J Cell Biol. 2009;185:1029–1045. doi: 10.1083/jcb.200812018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- **66.Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC, et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155:160–171. doi: 10.1016/j.cell.2013.08.032. This study shows that Opa1-dependent crista morphology is required for the maintenance of electron transport chain supercomplexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **71.Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. The Journal of cell biology. 2012;197:535–551. doi: 10.1083/jcb.201110034. These studies (71 and 72) show that Drp1 mediates mitophagy in neurons, cardiomyocytes and fibroblasts in vivo and in vitro, in dependently of the ubiquitin E3 ligase parkin. The loss of Drp1-mediated mitophagy leads to neurodegneration and heart failure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **72.Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi S, Chen W, Höke A, Dawson VL, Dawson TM, et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. Embo J. 2014 doi: 10.15252/embj.201488658. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berthet A, Margolis EB, Zhang J, Hsieh I, Hnasko TS, Ahmad J, Edwards RH, Sesaki H, Huang EJ, Nakamura K. Loss of mitochondrial fission depletes axonal mitochondria in midbrain dopamine neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:14304–14317. doi: 10.1523/JNEUROSCI.0930-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Udagawa O, Ishihara T, Maeda M, Matsunaga Y, Tsukamoto S, Kawano N, Miyado K, Shitara H, Yokota S, Nomura M, et al. Mitochondrial Fission Factor Drp1 Maintains Oocyte Quality via Dynamic Rearrangement of Multiple Organelles. Current biology : CB. 2014 doi: 10.1016/j.cub.2014.08.060. [DOI] [PubMed] [Google Scholar]
- **76.Ishihara T, Ban-Ishihara R, Maeda M, Matsunaga Y, Ichimura A, Kyogoku S, Aoki H, Katada S, Nakada K, Nomura M, et al. Dynamics of mtDNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Molecular and cellular biology. 2014 doi: 10.1128/MCB.01054-14. This study shows that mitochondrial division helps segregate mitochondrial DNA in the heart. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ishihara N, Fujita Y, Oka T, Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. Embo J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baker MJ, Lampe PA, Stojanovski D, Korwitz A, Anand R, Tatsuta T, Langer T. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. The EMBO journal. 2014;33:578–593. doi: 10.1002/embj.201386474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. The Journal of cell biology. 2009;187:959–966. doi: 10.1083/jcb.200906083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, Langer T. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol. 2009;187:1023–1036. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *82.Quiros PM, Ramsay AJ, Sala D, Fernandez-Vizarra E, Rodriguez F, Peinado JR, Fernandez-Garcia MS, Vega JA, Enriquez JA, Zorzano A, et al. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. The EMBO journal. 2012;31:2117–2133. doi: 10.1038/emboj.2012.70. This report shows that mice lacking a protease which cleaves Opa1 exhibit altered balance between mitochondrial fusion and division and defects in enegy metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **83.Mishra P, Carelli V, Manfredi G, Chan DC. Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell metabolism. 2014;19:630–641. doi: 10.1016/j.cmet.2014.03.011. This paper descirbe a new form of mitochondrial fusion that is stimulated by oxydatieve phosphorylation. Interestingly, this fusion mechanism requies clevage of Opa1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E, Langer T. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol. 2014;204:919–929. doi: 10.1083/jcb.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **85.Boissan M, Montagnac G, Shen Q, Griparic L, Guitton J, Romao M, Sauvonnet N, Lagache T, Lascu I, Raposo G, et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science. 2014;344:1510–1515. doi: 10.1126/science.1253768. The two studies (85 and 86) show the production of GTP occur near Opa1 and stimulates Opa1-mediated mitochondrial fusion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **86.Schlattner U, Tokarska-Schlattner M, Ramirez S, Tyurina YY, Amoscato AA, Mohammadyani D, Huang Z, Jiang J, Yanamala N, Seffouh A, et al. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. The Journal of biological chemistry. 2013;288:111–121. doi: 10.1074/jbc.M112.408633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jezek P, Hlavata L. Mitochondria in homeostasis of reactive oxygen species in cell, tissues, and organism. Int J Biochem Cell Biol. 2005;37:2478–2503. doi: 10.1016/j.biocel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 88.Kowaltowski AJ, de Souza-Pinto NC, Castilho RF, Vercesi AE. Mitochondria and reactive oxygen species. Free Radic Biol Med. 2009;47:333–343. doi: 10.1016/j.freeradbiomed.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 89.Makino A, Scott BT, Dillmann WH. Mitochondrial fragmentation and superoxide anion production in coronary endothelial cells from a mouse model of type 1 diabetes. Diabetologia. 2010;53:1783–1794. doi: 10.1007/s00125-010-1770-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shenouda SM, Widlansky ME, Chen K, Xu G, Holbrook M, Tabit CE, Hamburg NM, Frame AA, Caiano TL, Kluge MA, et al. Altered mitochondrial dynamics contributes to endothelial dysfunction in diabetes mellitus. Circulation. 2011;124:444–453. doi: 10.1161/CIRCULATIONAHA.110.014506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang W, Wang Y, Long J, Wang J, Haudek SB, Overbeek P, Chang BH, Schumacker PT, Danesh FR. Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell metabolism. 2012;15:186–200. doi: 10.1016/j.cmet.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Z, Wakabayashi N, Wakabayashi J, Tamura Y, Song WJ, Sereda S, Clerc P, Polster BM, Aja SM, Pletnikov MV, et al. The dynamin-related GTPase Opa1 is required for glucose-stimulated ATP production in pancreatic beta cells. Molecular biology of the cell. 2011;22:2235–2245. doi: 10.1091/mbc.E10-12-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jheng HF, Tsai PJ, Guo SM, Kuo LH, Chang CS, Su IJ, Chang CR, Tsai YS. Mitochondrial fission contributes to mitochondrial dysfunction and insulin resistance in skeletal muscle. Mol Cell Biol. 2012;32:309–319. doi: 10.1128/MCB.05603-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haslam DW, James WP. Obesity. Lancet. 2005;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- **95.Dietrich MO, Liu ZW, Horvath TL. Mitochondrial dynamics controlled by mitofusins regulate Agrp neuronal activity and diet-induced obesity. Cell. 2013;155:188–199. doi: 10.1016/j.cell.2013.09.004. The two article (95 and 96) report that mitochondrial fusion mediated by mitofusins regulates two neurons with two opposing functions in the hypothalamus and controls animal metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **96.Schneeberger M, Dietrich MO, Sebastian D, Imbernon M, Castano C, Garcia A, Esteban Y, Gonzalez-Franquesa A, Rodriguez IC, Bortolozzi A, et al. Mitofusin 2 in POMC neurons connects ER stress with leptin resistance and energy imbalance. Cell. 2013;155:172–187. doi: 10.1016/j.cell.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **97.Wikstrom JD, Mahdaviani K, Liesa M, Sereda SB, Si Y, Las G, Twig G, Petrovic N, Zingaretti C, Graham A, et al. Hormone-induced mitochondrial fission is utilized by brown adipocytes as an amplification pathway for energy expenditure. The EMBO journal. 2014;33:418–436. doi: 10.1002/embj.201385014. This study shows that shifting a balance of mitochondrial dynamics toward increased division promotes hormone-induced energy expenditure in brown adipocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]


