Figure 2.
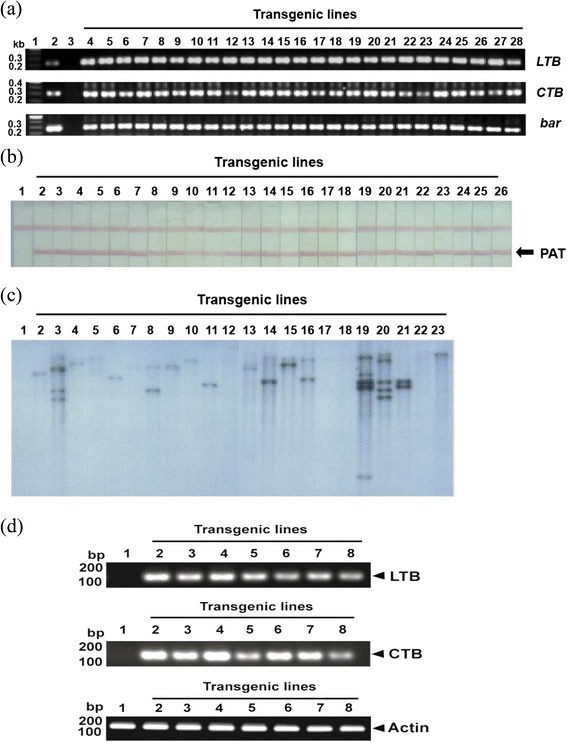
Detection of LTB, CTB and bar genes and phosphoacetyl transferase (PAT) in putative transgenic plant leaf tissues. (a) PCR amplification of LTB (217 bp), CTB (292 bp) and bar (302 bp) from transgenic rice genomic DNA. Lane 1 is the size marker. Lane 2 is the binary vector (pMJ103-LTB-CTB). Lane 3 is the wild-type. Lanes 4–28 are transgenic rice plants. (b) Lateral flow assay using the Trait LL lateral flow test kit (Strategic diagnostics). Lane 1 is the wild-type. Lanes 2–26 are transgenic rice plants. (c) Southern hybridization analysis of chromosomal DNA in transgenic rice. Genomic DNA from wild-type untransformed or independent transgenic rice T0 lines carrying the pMJ103-LTB-CTB construct was hybridized with a probe specific for the LTB coding region. A total of 10 μg of total leaf genomic DNA from transgenic rice plants was digested with XhoI, which cuts at a single site within the binary vector pMJ103-LTB-CTB. (d) LTB and CTB gene transcripts in transgenic plants were detected by RT-PCR. Plant RNA was isolated from selected transgenic plant rice seeds and RT-PCR was performed using a primer pair that specifically amplified 107- and 111-bp DNA fragments of the LTB and CTB genes (lanes 2 to 8). Lane 1 shows the RNA of non-transgenic plants used as a negative control.
