Abstract
Purpose
In this study we hypothesized that the mRNA vector Staufen mediates RNA relocalization during meiotic maturation, and by virtue of its interactions with endoplasmic reticulum, provides a possible mechanism by which protein synthesis is regulated.
Methods
We assessed the expression of staufen (STAU) and calreticulin (CALR), the latter adopted as a marker of the endoplasmic reticulum, in human oocytes at different stages of maturation: GV, metaphase MI and MII. Oocytes were subjected to polymerase chain reaction in order to investigate the expression of STAU and CALR. The corresponding protein products were identified by immunofluorescence and confocal laser scanning microscopy.
Results
STAU and CALR were constantly expressed and selectively localized during oocyte maturation. At the GV stage the both proteins displayed a dispersed distribution localization throughout the cytoplasm. Progressing to the MII stage, STAU tended to compartmentalize towards the cortical area of the oocyte clustering in granules of larger sizes. At the MII stage, CALR assumed a pattern reminiscent and possibly coincident with the position of the meiotic spindle.
Conclusions
The changing pattern of STAU distribution during meiotic maturation of human oocytes implicates a novel mechanism for the regulation of protein synthesis based on mRNA localization. Moreover, the unique disposition of CALR at the MII spindle uncovers a physical interaction with endoplasmic reticulum that may mediate cytoskeletal remodelling during oocyte maturation.
Keywords: Oocytes, Oocyte maturation, Staufen, Calreticulin, In vitro fertilization
Introduction
Mammalian oocytes acquire a series of competencies during follicular development that play critical roles at fertilization and subsequent stages of preimplantation embryonic development. Before the mammalian oocyte engages in the fertilization process, it must acquire an array of molecular and cellular assets defining its developmental potential. These properties specify competencies to complete meiosis and initiate mitosis, support monospermic fertilization and egg activation and ensure a timely transition from reliance of gene products of oocyte origin to those derived from the zygotic or embryonic genome [1].
Meiotic maturation requires the acquisition both of nuclear and cytoplasmic competence through a complex mechanism involving most organelles, the cytoskeleton, and molecules that move from nucleus to cytoplasm including newly synthesized mRNAs. STAU, a double strand (ds) RNA-binding protein comprised of five double strand binding domains (dsRBDs), was initially identified in genetic screens for maternal-effect mutants in Drosophila. STAU mutants exhibit multiple embryonic defects including an absence of pole cells, abdomen reduction, and head deformation [2, 3]. These defects are caused, at least in part, by mis-localization of mRNAs, such as oskar and bicoid, in the developing oocyte and embryo [4, 5]. STAU is also required to de-repress mRNAs after localization, implying additional functions in mRNA control. Mammals possess two STAU homologs, STAU1 and STAU2. Together, the results with Drosophila and mammalian proteins thus imply functions in localization, translation and decay [6]. STAU is a protein that recognizes the mRNA 3’UTR terminal and makes possible the movement along the cytoskeleton facilitating the process of anchoring to the specific sites in the cytoplasm where translation takes place, such as in association with endoplasmic reticulum (ER) [7].
The ER undergoes significant changes in organization during maturation reflecting an enhanced ability to release Ca2+. CALR is a multifunctional protein that acts as a major Ca2+-binding (storage) protein in the lumen of the endoplasmic reticulum [8]. The function of binding of Ca2+ of the CALR is crucial to the role played by this ion in important cellular processes, such as cytoskeletal remodelling, and alterations in gene expression and protein synthesis as a sign of cellular stress [9]. In oocytes, Ca2+ transients mediate fertilization through their involvement in maturation allowing the completion of meiosis [10]. In mammalian oocytes, CALR seems to play an important role in calcium homeostasis during oogenesis and development [11]. In this scenario, STAU RNA Binding Protein may play an important role given its function in mRNA translocation. Therefore, we hypothesized that STAU is expressed during oocyte maturation and that the localization of STAU protein changes during the meiotic transition from GV to MII. We also postulated that the pattern of STAU distribution reflects its physical interaction with the ER an organelle directly involved in the process of mRNA translation whose relocalization is central to the process of oocyte maturation.
To test the above hypotheses, we assessed the expression of STAU and CALR, the latter adopted as a marker of the ER in human oocytes at different stages of maturation.
Materials and methods
Oocyte retrieval from patients undergoing assisted reproduction treatments
Oocytes were collected from consenting patients undergoing in-vitro fertilization (IVF) treatments (from January 2008 and December 2011) for different causes of infertility but, in the specific case of this study only oocytes from “normo-ovulatory” patients were considered. Spare eggs, not utilized for the reproductive cycle, were donated for research purposes and fixed or cryopreserved utilizing a slow cooling protocol. Ovarian stimulation was conducted according to a long standard stimulation protocol using gonadotrophin-releasing hormone agonist (GnRH) (Enantone®; Takeda, Osaka, Japan) and recombinant Follicle Stimulating Hormone (r-FSH) (Puregon®; MSD or Gonal F®, Merck Serono, Geneve, Switzerland). Ovulation was triggered with (human Chorionic Gonadotrophin) hCG (Ovitrelle® or Gonasi, ®, MerckSerono, Geneve, Switzerland). Oocyte retrieval was performed transvaginally, under ultrasound guidance, 36 h after hCG injection using a CCD aspiration needle 17 Gauges (CCD, Paris, France).
Cumulus and corona radiata cells were removed after retrieval by a short exposure to HEPES-buffered medium (Quinn’s Advantage Hepes Medium, Sage IVF Inc, Trumbull, CT, USA) containing 20 IU/ml hyaluronidase (Sage IVF Inc, Trumbull, CT, USA) and gentle aspiration in and out of a hand pipette and mechanically cleaned from the remaining surrounding cumulus cells by aspiration using a denuding pipette (Flexi-PetTM, Cook, Australia) with a 170–140 μl diameter (Denuding needles for Flexi-PetTM; Cook, Australia). The denuded oocytes were then assessed with respect to their meiotic maturation status (GV, MI or MII).
Oocyte cryopreservation
Oocytes were cryopreserved using a slow cooling method with two alternative protocols differing in sucrose concentration [12]. All cryopreservation solutions were prepared as described elsewhere (De Santis et al., 2007b)
Thawing was conducted at room temperature, with a four step procedure and oocytes were then replaced in culture medium at 37 °C for at least 1 h until further evaluation of oocytes degeneration [13, 14].
RNA extraction
Polyadenylated RNA [poly(A)+RNA] was extracted from pools of five oocytes, using Dynabeads mRNA Direct microkit (Invitrogen) according to the manufacturer’s instructions. Briefly, pools were lysed for 10 min at room temperature in 100 μl of lysis buffer (100 mmol Tris–HCl [pH 7.5], 500 mmol LiCl, 10 mmol EDTA [pH 8.0], 1% [w/v] lithium dodecyl sulfate, and 5 mM dithiothreitol [DTT]). After lysis, 20 μl of pre-washed Dynabeads oligo (dT)25 were pipetted into the tube and binding of poly (A)+ RNAs to oligo (dT) was allowed for 5 min at room temperature. The beads were then separated with a magnetic separator, washed twice with 100 μl washing buffer A (10 mM Tris–HCl [pH 7.5], 0.15 mM LiCl, 1 mM EDTA, and 0.1% [w/v] lithium dodecyl sulfate) and three times with 100 μl washing buffer B (10 mM Tris–HCl [pH 7.5], 0.15 M LiCl,1 mM EDTA). Poly (A)+ RNAs were then eluted from the beads by incubation in 10 μl of diethylpyrocarbonatetreated sterile water at 65 °C for 5 min.
RNA reverse transcription
Poly (A)+RNA from each pool of oocytes was reverse transcribed (RT) into cDNA in a total reaction mixture volume of 20 μl containing 8.5 μl sterile water, 1 μl of 10 mM dNTP mix, and 1 μl oligo (dT)12–18 (500 ng/μl). RT was performed with 200 U of SuperScript II reverse transcriptase (Invitrogen, Italy) for 1 h at 42 °C. Enzymes were inactivated at 70 °C for 15 min.
Semi-quantitative polymerase chain reaction (PCR)
Complementary DNA amplifications were carried out in an automated thermal cycler
(iCycler; Bio-Rad), using the appropriate conditions for each set of primers (STAU and 28S; see Table 1). PCR was performed with cDNA equivalents corresponding to 0.25 oocyte. Human DNA was used as positive control. The optimal cycle number at which the transcript was amplified exponentially was established running a linear cycle series, and the number of PCR cycles was kept within this range. RT-PCR products were subjected to electrophoresis on a 2% agarose gel, and the intensity of each band was assessed by densitometric analysis performed with Quantity One software (Bio- Rad). The relative amount of STAU transcript was calculated in arbitrary units by dividing the intensity of the STAU bands by the intensity of the 28S band.
Table 1.
List of primers used for PCR detailing accession number, primer sequence, annealing temperature and expected amplification product size
| GENE | Accession Number | PRIMERS | Tm (°C) | Amplification product |
|---|---|---|---|---|
| Staufen (STAU) | NM_017454.2 | FOR 5’- cacctccgtgtttggtcttt -3’ REV 5’- ggtcacgctgagtaggaagc -3’ |
60 | 183 bp |
| Calreticulin (CALR) | NM_004343.3 | FOR 5’- tctcagttccggcaagttct -3’ REV 5’- tctgagtctccgtgcatgtc -3’ |
60 | 231 bp |
| Homo sapiens heat shock 70 kDa protein 5 (HSPA5) | NM_005347.4 | FOR 5’- tagcgtatggtgctgctgtc -3’ REV 5’- tttgtcaggggtctttcacc -3’ |
60 | 241 bp |
| Nestin (NES) | NM_006617.1 | FOR 5’- aacagcgacggaggtctcta -3’ REV 5’- ttctcttgtcccgcagactt -3’ |
60 | 220 bp |
| Homo sapiens ribosomal protein S28 (RPS28) | NM_001031.4 | FOR 5’- tccatcatccgcaatgtaaa -3’ REV 5’- tgtgacagaccattcccatc -3’ |
60 | 157 bp |
Oocyte gene expression
cDNA amplifications were carried out in an automated thermal cycler (iCycler; Bio-Rad) using the conditions appropriate for each set of primers (Table 1). PCR primers were designed using the oligomer program Primer3 Input. PCR products were run in a 2% agarose gel. 28S was used as an internal control. The expression level for STAU was normalized against 28S. HSPA5 (BIP) was examined to confirm CALR expression, Nestin was amplified as negative control. Human DNA was used as positive control. Amplification products were purified in Spin-X centrifuge tube filters (Corning, Italy), sequenced (SEQLAB, Gottingen, Germany), and aligned using Clustal W version 1.82 software (EMBL-EBI service).
STAU immunolocalization
Thirty oocytes were fixed in 4% paraformaldehyde in PBS for 30 min at 4 °C and permeabilized with 0.5% Triton X-100 in PBS overnight at 37 °C. They were then incubated in a 0.1 M NH4Cl solution for 10 min at room temperature. Aspecific sites were blocked with a solution of PBS containing 5% bovine albumin serum (BSA). Samples were incubated with anti-human STAU antibody (1:200, Abcam, UK) for 1 h at room temperature and then with the appropriate secondary antibody (Alexafluor; Invitrogen, Italy). Nuclei were stained with 40,6-diamidino-2-phenylindole (DAPI, Sigma, Italy). At the end of the procedure, samples were mounted on glass slides in an anti-fading mounting medium (Pro-Long, Molecular Probes, Italy) and analysed under TCS-NT laser scanning confocal microscope (Leica Microsystems, Heidelberg, Germany). STAU distribution was assessed through equatorial optical sections of 6.7 mm thickness by sequential scanning of at least 40 μm of sample with 2.6 μm step size and controlled oversampling.
Aiming to increase the material available for the project also MII cryopreserved oocytes were thawed (as described above) and the survived cells were fixed and stained with the same protocol illustrated for fresh material. Confocal images were processed using MacBiophotonics ImageJ dedicated software. The area of STAU spots, present in each captured image was measured. STAU spot size was assessed in oocytes at different developmental stages.
CALR immunolocalization
Oocytes were fixed and treated as described above. They were incubated with the anti-human CALR antibody (1:200, Abcam, UK) for 1 h at room temperature and then with the appropriate secondary antibody (Alexafluor; Invitrogen, Italy). Nucleus was stained with Hoechst 33258 (Molecular Probes, Italy). At the end of the procedure, samples were mounted on glass slides in an anti-fading mounting medium (Pro-Long, Molecular Probes, Italy).
Samples were analyzed using the “Alembic” microscopy facility of Vita-Salute San Raffaele University equipped with a Leica TCS SP2 Laser Scanning Confocal Microscope. CALR distribution was assessed through equatorial optical sections of 6.7 mm thickness by sequential scanning of at least 40 μm of sample with 2.6 μm step size and controlled oversampling.
Actin staining
Staining for filamentous actin (F-actin) was carried out simultaneously with that of the other proteins (STAU or CALR). In particular, during the incubation with the secondary antibody, samples were stained with rhodamine-phalloidin (1:200; Molecular Probes) for 1 h at 37 °C. After washing, oocytes were mounted and then images were collected.
Statistical analysis
For molecular analysis all experiments were performed in at least three biologically independents replicates. One-way ANOVA test (SPSS19.1; IBM) was used. Differences were considered statistically significant at P < .05.
Results
STAU expression
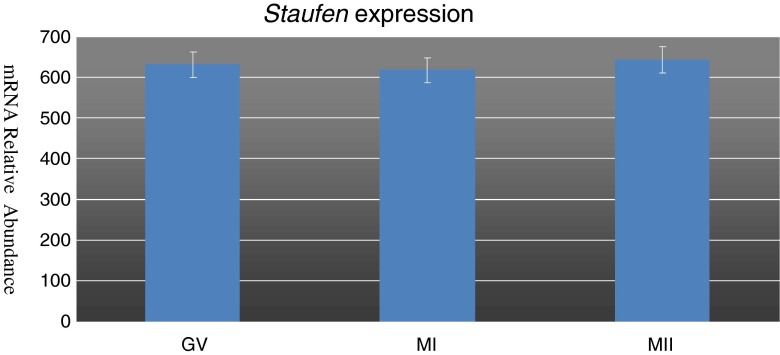
STAU mRNA expression was conducted by molecular analysis performed on human oocytes at the GV, MI and MII stages. In particular, its expression was found at all stages of oocyte maturation and semi-quantivative PCR studies showed a stable expression of the transcript during the maturation process (Fig. 1). This may suggests a possible physiological role for this protein.
Fig. 1.
Graphic representation of Staufen expression levels in human oocytes during the three maturational stages of Germinal Vesicle, Metaphase I and Metaphase II. Values indices are not significantly different (P > 0.05)
Localization of STAU by immunofluorescence confocal microscopy
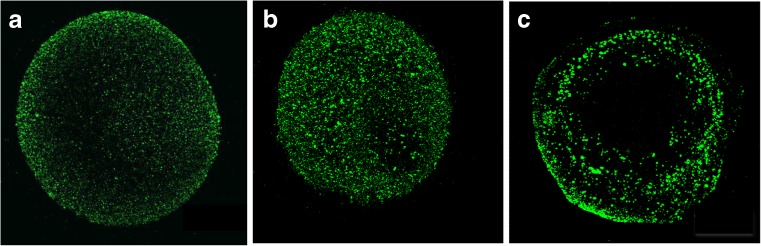
Localization of STAU was performed by immunofluorescence confocal microscopy in human oocytes (N = 30, 10 each) assessed at three stages of maturation: germinal vesicle (GV), metaphase I (MI) and metaphase II (MII). The cytoplasm of all analyzed oocytes was positive for STAU, suggesting that this protein is present throughout the process of maturation.
Also, comparative analysis of GV, MI and MII oocytes suggested a reorganization of STAU localization during maturation. Representative images of STAU distribution at the different maturation stages are shown in Fig. 2a, b and c. At the GV stage the protein displayed a dispersed localization throughout the cytoplasm. At the MI stage STAU tended to compartmentalize towards the cortical area of the oocyte clustering in granules of larger sizes. The preferential localization at the cortex was even more apparent in MII oocytes where a discrete number of granules predominantly occupied the peripheral part of the ooplasm. Quantitative assessment showed that the mean area of STAU positive granules increased progressively during maturation, being 1.61x103 μm2 (1.6 ± 0.1), 2.01×103 μm2 (2.0 ± 0.2) and 3.10×103 μm2 (3.1 ± 0.2) in GV, MI and MII oocytes, respectively.
Fig. 2.
Staufen distribution (GREEN Immunostaining) at the different maturation stages GV (a), MI (b) and MII (c)
CALR expression
Molecular analysis was extended to ascertain the expression of CALR. For comparison, CALR expression was analysed in association with that of STAU and HSPA5. The protein encoded by the latter gene is a member of the heat shock protein 70 (HSP70) family. It is localized in the lumen of the endoplasmic reticulum (ER), and is involved in protein folding and assembly occurring in the ER. As CALR interacts with other ER proteins, it may play a key role in monitoring protein transport through the cell. 28S RNA was detected as housekeeping gene and therefore as a positive control, while Nestin (whose expression is exclusive of nerve cells) was adopted as a negative control. Expression of both CALR and HSPA5 mRNAs was observed in MII oocytes, suggesting, although not proving, the presence of mechanism of sorting, folding and assembly of proteins at the mature stage (Table 2).
Table 2.
CALR expression in human MII oocytes evaluated together with STAU, HSPA5, 28S and Nestin
| GENE | MII OOCYTES |
|---|---|
| Staufen | + |
| Calreticulin | + |
| HSPA5 (BIP) | + |
| Nestin | - |
| 28S | + |
CALR localization
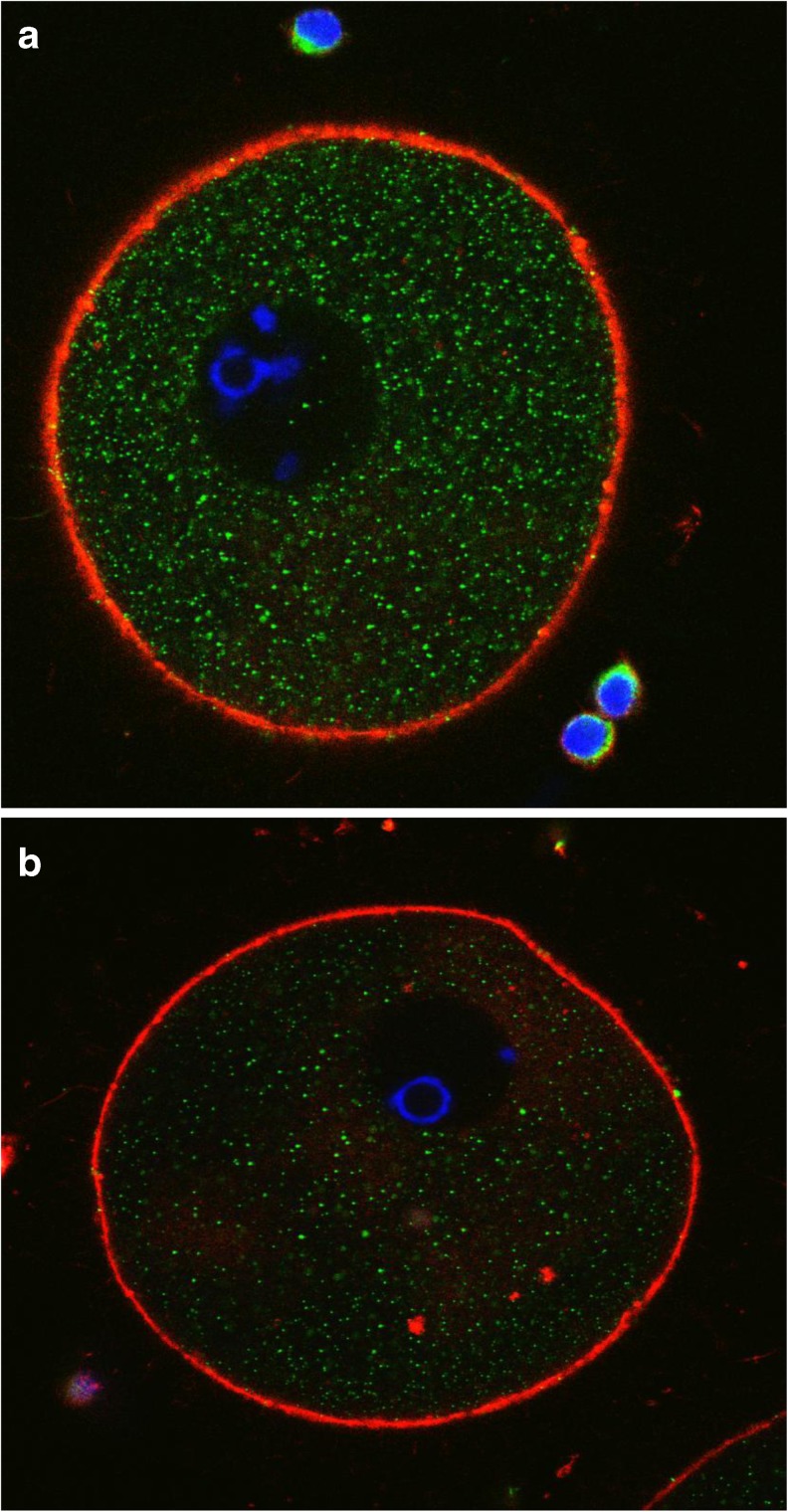
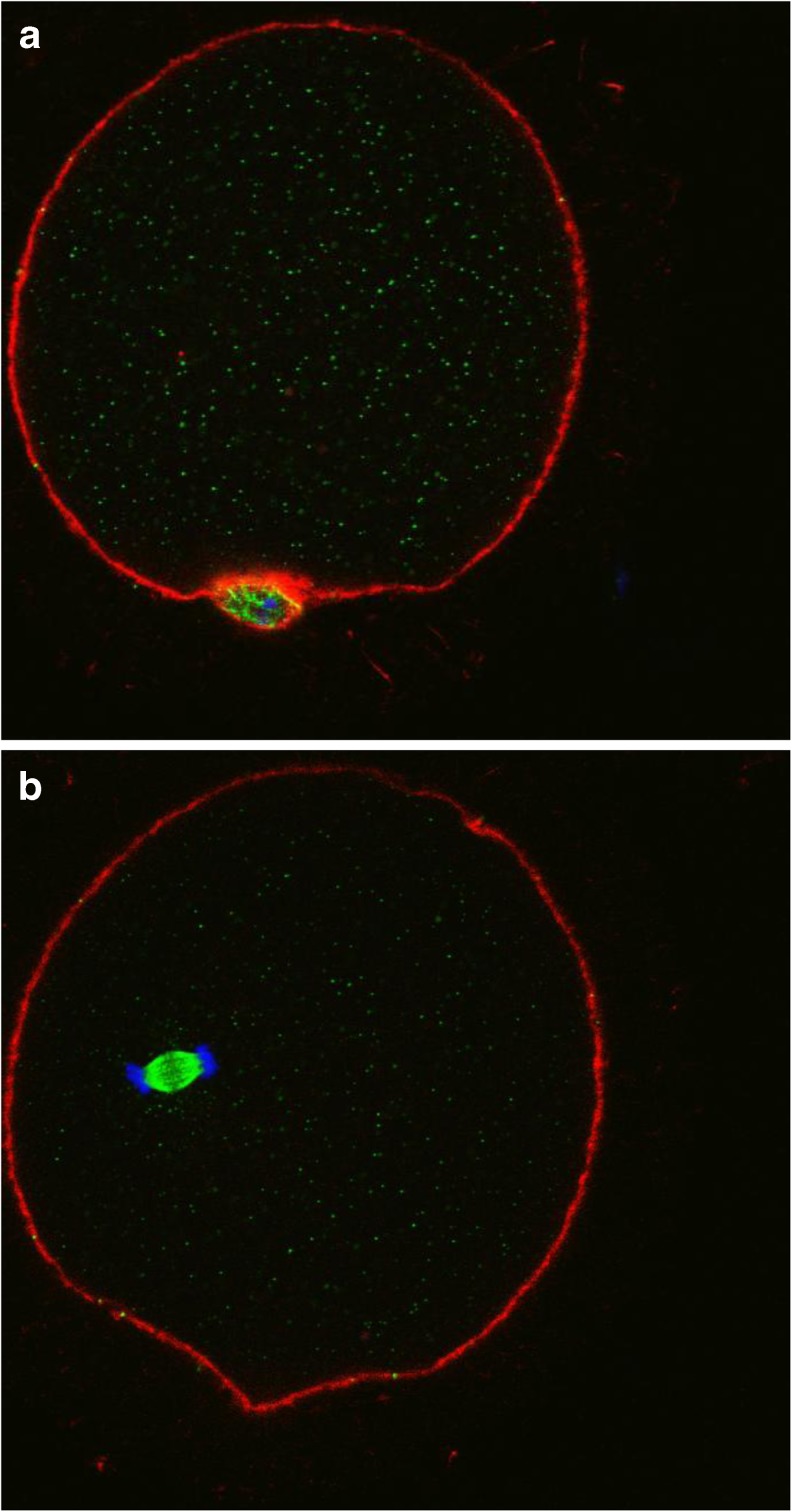
CALR presence was assessed by immunofluorescence confocal microscopy in oocytes at different maturation stages. Figure 3a and b are representative of results obtained in GV oocytes. Optical sections were acquired from the equatorial plane of each sample. The cytoplasm displayed a green staining characterized by a finely granular distribution corresponding to the localization of CALR. In addition to being evenly distributed throughout the cytoplasm, CALR was also found closely surrounding, but not included in, the nucleus (Fig. 3b), suggesting a co-localization with the ER.
Fig. 3.
a and b: human oocytes at the GV stage stained by anti-Calreticulin antibody. CALR signal is excluded from the nucleus and relatively weak. Immunostaining: actin (RED), chromosomes (BLUE) and CALR (GREEN)
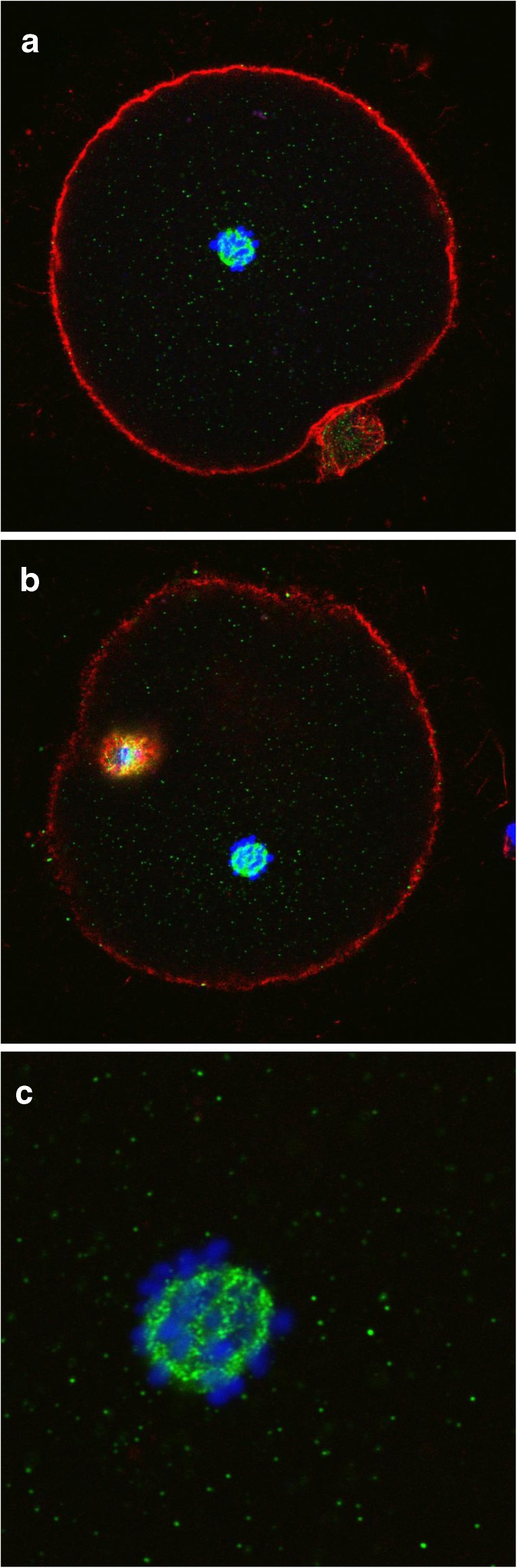
Figure 4a and b are indicative of the patterns observed in MII oocytes. In Fig. 4a, the chromosomes are in a condensed status and closely clustered, probably held together by the meiotic spindle (not visible by the staining used for these experiments). The chromosomes appeared atypically far from the cortex. The green staining, corresponding to CALR, was detectable in a weakly diffused pattern and characterized by small dots dispersed throughout the cytoplasm. Also, CALR was concentrated in the immediate vicinity of the chromosomes where it assumed a distribution that was highly reminiscent of the meiotic spindle. Figure 4b is representative of an MII stage oocyte where the first polar body is not perfectly distinguishable because occupying a position in a different geometric plane. However, again, the chromosomes are confined in a small area away from the cortex. The staining for CALR appears homogenous and weak throughout the cytoplasm, while it is intensely and specifically localized around the chromosomes with a morphology reminiscent of the meiotic spindle. Figure 4c shows even more clearly the chromosomes and CALR distribution.
Fig. 4.
a, b and c: Human oocytes at the MII stage. In A the first polar body (PB1) is evident at the 5 o’clock position and in B the PB1 remains on another observational plane. c represents a particular of the CALR staining with the detail of chromosomes with respect to CALR localization. Immunostaining: actin (RED), chromosomes (BLUE) and CALR (GREEN)
Figure 5a and b is representative of two distinct focal planes of a fresh oocyte. Image A shows the first polar body at the 6 o’clock position and a weak and diffuse green staining specific for CALR. The b image displays the chromosomes arranged in two groups according to a pattern that is consistent with the anaphase II stage. This suggests that prior to fixation the oocyte underwent a partial event of spontaneous activation in the absence of extrusion of the second polar body. The green staining specific for CALR, in addition to a diffuse distribution, assumes a pattern extraordinarily similar to the morphology of the meiotic spindle in correspondence of the area that separates the two sets of chromosomes [15]. The spindle appears detached from the cortex but this is unlikely to have been caused by cryopreservation.
Fig. 5.
a and b: Human oocyte at the MII stage observed on two different focal planes. A with PB1 at “6” h position and B with PB barely distinguishable on a different focal plane but with chromosomes neatly arranged at the periphery of a structure evocative of the meiotic spindle. Immunostaining: actin (RED), chromosomes (BLUE) and CALR (GREEN)
Discussion
The early stages of development are governed by proteins and RNA synthesized and accumulated during oogenesis. A wide number of proteins and factors is involved in the process of transport of mRNAs from the oocyte nucleus to the cytoplasm.[16] One of these is represented by STAU, whose function is to bind the mature mRNA and transport it to specific sites of the cytoskeleton, where it promotes anchorage for the translation of the corresponding protein [7].
There are several studies on the characterization of STAU in oocytes from various species such as murine [17] bovine [18] and porcine [19] but there is little information in humans.
Cellular localization of STAU is mainly cytoplasmic and to a lesser extent nuclear. STAU is typically present in highly polarized cells such as epithelial cells, neurons and oocytes. During maturation oocyte transcription is repressed and therefore translation relies on mRNAs already present in the cytoplasm or derived from the nucleus. The role of STAU in the localization and transport of mRNA has been studied in Drosophila oocytes [20, 21], Xenopus leavis [22] and in zebrafish [23]. Other important studies have been conducted in higher species such as murine [17], bovine [18] and porcine [19]. In particular, immunolocalization studies carried out on pig oocytes showed a scattered distribution of STAU during the germinal vesicle (GV) stage, while starting from the MI stage STAU localization becomes restricted to the inner region and the oocyte cortex. At the MII stage, STAU granules appears much larger in diameter and spread throughout the cytoplasm, except the area surrounding the spindle. This compartmentalization is reminiscent of the reorganization of the endoplasmic reticulum described in oocytes of different species [24, 25]. In in vivo matured oocytes, ER elements are associated in clusters of 2–3 μm preferentially localized in the cortex but also discernible in more internal regions. Therefore, also in the human, the ER undergoes significant changes during maturation in its distribution that reflect an enhanced ability to release Ca 2+ [26].
Based on this evidence, we tested the hypothesis that STAU is expressed in human oocytes during maturation and physically interacts with the ER. CALR was adopted as a marker for the localization of the ER. STAU RNA is present at the GV, MI and MII stages. Also, the present results show that at the GV stage, CALR distribution appears finely granular and ubiquitous throughout the cytoplasm, with a moderate accumulation around the GV. This pattern is similar to that of STAU observed at the same stage of maturation. In MII stage oocytes, STAU localization is organized in larger granules. On the contrary, CALR staining appears coincident with the presumptive position and morphology of the meiotic spindle, as suggested by the relative position of the chromosomes. Although this finding was not expected, it does not exclude the possibility that STAU dynamics reflects the one of ER in order to assist the process of translation. It is possible that CALR, while being a very important component of the ER, is segregated in a specific ER subdomain that interacts with the meiotic spindle. In fact, in the mouse it has been shown that at the MII stage the ER acquires a highly specific conformation enveloping the meiotic spindle [26]. The significance of this association is not fully understood, but it is plausible that the dense microtubular array forming the meiotic spindle is locally and specifically regulated by Ca2+ ions released from a subdomain of the ER selectively enriched in CALR. Therefore to confirm the association of STAU with the ER more specific markers of this organelle should be tested.
In conclusion, the present data support the notion that STAU distribution undergoes a specific rearrangement during meiotic maturation in the human oocyte. The specific patterns observed at the different maturation stage are compatible with an involvement in the general process of mRNA translation. Finally, CALR specific localization in association with the meiotic spindle suggests the hypothesis of different ER subdomains and an important involvement of Ca2+ in the regulation of the meiotic spindle. This opens entirely new scenarios for future studies on the ER-cytoskeleton physical and regulative interaction in the human oocyte.
Acknowledgments
F.G., G.P., S.M., and T.A.L.B. are members of the COST Action FA1201 Epiconcept: Epigenetics and Periconception environment. Part of the experiments are supported by Carraresi Foundation. L.D.S. thanks the “ Alembic” microscopy facility for supporting confocal analysis.
Footnotes
Capsule Staufen RNA-binding protein undergoes a specific rearrangement during meiotic maturation in human oocytes implying an involvement in the process of mRNA translation. Calreticulin, adopted as a marker for the endoplasmic reticulum, shows a specific localization in association with the meiotic spindle suggesting a regulative function of Ca2+.
References
- 1.Albertini DF, Sanfins A, Combelles CM. Origins and manifestations of oocyte maturation competencies. Reprod BioMed Online. 2003;6:410–5. doi: 10.1016/S1472-6483(10)62159-1. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–91. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 3.Schupbach T, Wieschaus E. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol. 1986;113:443–8. doi: 10.1016/0012-1606(86)90179-X. [DOI] [PubMed] [Google Scholar]
- 4.St Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-O. [DOI] [PubMed] [Google Scholar]
- 5.Ferrandon D, Elphick L, Nusslein-Volhard C, St JD. Staufen protein associates with the 3’UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–32. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 6.Legendre JB, Campbell ZT, Kroll-Conner P, Anderson P, Kimble J, Wickens M. RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in C. elegans. J Biol Chem. 2013;288:2532–45. doi: 10.1074/jbc.M112.397349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2220–30. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelebart P, Opas M, Michalak M. Calreticulin, a Ca2 + −binding chaperone of the endoplasmic reticulum. Int J Biochem Cell Biol. 2005;37:260–6. doi: 10.1016/j.biocel.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 9.Wang WA, Groenendyk J, Michalak M. Calreticulin signaling in health and disease. Int J Biochem Cell Biol. 2012;44:842–6. doi: 10.1016/j.biocel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Zhang DX, Li XP, Sun SC, Shen XH, Cui XS, Kim NH. Involvement of ER-calreticulin-Ca2+ signaling in the regulation of porcine oocyte meiotic maturation and maternal gene expression. Mol Reprod Dev. 2010;77:462–71. doi: 10.1002/mrd.21166. [DOI] [PubMed] [Google Scholar]
- 11.Mao L, Lou H, Lo Y, Wang N, Jin F. Behaviour of cytoplasmic organelles and cytoskeleton during oocyte maturation. Reprod BioMed Online. 2014;28:284–99. doi: 10.1016/j.rbmo.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 12.De Santis L, Cino I, Rabellotti E, Papaleo E, Calzi F, Fusi FM, et al. Oocyte cryopreservation: clinical outcome of slow-cooling protocols differing in sucrose concentration. Reprod BioMed Online. 2007;14:57–63. doi: 10.1016/S1472-6483(10)60764-X. [DOI] [PubMed] [Google Scholar]
- 13.De Santis L, Cino I, Coticchio G, Fusi FM, Papaleo E, Rabellotti E, et al. Objective evaluation of the viability of cryopreserved oocytes. Reprod BioMed Online. 2007;15:338–45. doi: 10.1016/S1472-6483(10)60348-3. [DOI] [PubMed] [Google Scholar]
- 14.De Santis L, Coticchio G, Paynter S, Albertini D, Hutt K, Cino I, et al. Permeability of human oocytes to ethylene glycol and their survival and spindle configurations after slow cooling cryopreservation. Hum Reprod. 2007;22:2776–83. doi: 10.1093/humrep/dem240. [DOI] [PubMed] [Google Scholar]
- 15.Bromfield J, Coticchio G, Hutt K, Sciajno R, Borini A, Albertini DF. Meiotic spindle dynamics in human oocytes following slow-cooling cryopreservation. Reprod BioMed Online. 2009;24:2114–23. doi: 10.1093/humrep/dep182. [DOI] [PubMed] [Google Scholar]
- 16.Labreque R, Sirard MA. The study of mammalian oocyte competence by transcriptome analysis: progress and challenges. Mol Hum Reprod.20:103–16 [DOI] [PubMed]
- 17.Saunders PT, Pathirana S, Maguire SM, Doyle M, Wood T, Bownes M. Mouse staufen genes are expressed in germ cells during oogenesis and spermatogenesis. Mol Hum Reprod. 2000;6:983–91. doi: 10.1093/molehr/6.11.983. [DOI] [PubMed] [Google Scholar]
- 18.Calder MD, Madan P, Watson AJ. Bovine oocytes and early embryos express Staufen and ELAVL RNA-binding proteins. Zygote. 2008;16:161–8. doi: 10.1017/S096719940700456X. [DOI] [PubMed] [Google Scholar]
- 19.Brevini TA, Cillo F, Antonini S, Gandolfi F. Cytoplasmic remodelling and the acquisition of developmental competence in pig oocytes. Anim Reprod Sci. 2007;98:23–38. doi: 10.1016/j.anireprosci.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 20.Gavis ER, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell. 1992;71:301–13. doi: 10.1016/0092-8674(92)90358-J. [DOI] [PubMed] [Google Scholar]
- 21.Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–63. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- 22.Yoon YJ, Mowry KL. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development. 2004;131:3035–45. doi: 10.1242/dev.01170. [DOI] [PubMed] [Google Scholar]
- 23.Bateman MJ, Cornell R, D’Alencon C, Sandra A. Expression of the zebrafish Staufen gene in the embryo and adult. Gene Expr Patterns. 2004;5:273–8. doi: 10.1016/j.modgep.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Ebner T, Moser M, Shebl O, Sommerguber M, Tews G. Prognosis of oocytes showing aggregation of smooth endoplasmic reticulum. Reprod BioMed Online. 2008;16:113–8. doi: 10.1016/S1472-6483(10)60563-9. [DOI] [PubMed] [Google Scholar]
- 25.Otsuki J, Okada A, Morimoto K, Nagai Y, Kubo H. The relationship between pregnancy outcome and smooth endoplasmic reticulum clusters in MII human oocytes. Hum Reprod. 2004;19:1591–7. doi: 10.1093/humrep/deh258. [DOI] [PubMed] [Google Scholar]
- 26.De Santis L, Brevini TA. Organelle rearrangement in the maturing oocyte. In: Coticchio G, Albertini DF, De Santis L, editors. Oogenesis. London: Springer; 2013. [Google Scholar]