Abstract
Background
The associations between TNF-α and Interleukin gene polymorphisms and polycystic ovary syndrome (PCOS) risk have been studied in numerous epidemiological studies, but the results remain controversial. To investigate whether these polymorphisms facilitate susceptibility to PCOS, we conducted a comprehensive systematic review and meta-analysis.
Methods
PubMed, Embase, Web of Science, Medline, CNKI, and Google Scholar were searched to obtain the genetic association studies according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. Odds ratios (OR) with corresponding 95 % confidence intervals (CI) were used to assess the strengths of the associations. Funnel plots and Egger’s tests were performed to test for possible publication bias. All statistical analyses were performed using Review Manager 5.2 and STATA11.0.
Results
Eighteen articles were included in the final meta-analysis. The studies involved the following polymorphisms: TNF-α -308G > A, TNF-α -805C > T, TNF-α -1031 T > C, IL-1A -889C > T, IL-1B -511C > T, IL-1B +3953 T > C, IL-6 -174G > C, IL-10 -819C > T, IL-10 -1082A > G, IL-18 -607C > A, and IL-18 -137G > C. Our results show a significant association between PCOS risk and the TNF-α -1031 T > C polymorphism (For TC + CC vs. TT: OR = 2.09, 95 % CI = 1.58–2.76, p < 0.0001. For C allele vs. T allele: OR = 1.67, 95 % CI = 1.33–2.09, p < 0.0001) and between PCOS risk and the IL-6 -174G > C polymorphism (For CC + GC vs. GG: OR = 0.49, 95 % CI = 0.25–0.95, p = 0.03. For CC vs. GG: OR = 0.48, 95 % CI = 0.28–0.80, p = 0.005. For C vs. G: OR = 0.60, 95 % CI = 0.42–0.87, p = 0.007). No associations were found with the other genetic models.
Conclusion
The results of the meta-analysis suggest positive associations between the TNF-α -1031 T > C and IL-6 -174G > C polymorphisms and the risk of PCOS. No associations are found between PCOS risk and the TNF-α -308G > A, TNF-α -805C > T, IL-1A -889C > T, IL-1B -511C > T, IL-1B +3953C > T, IL-10 -819C > T, IL-10 -1082 A > G, IL-18 -607C > A, and IL-18 -137G > C polymorphisms. However, due to the heterogeneity and low quality of the studies related to PCOS polymorphisms in the meta-analysis, the results should be interpreted with caution. Future multi-ethnicity studies of homogeneous populations of PCOS patients with larger sample sizes and well-matched controls are needed.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-015-0449-7) contains supplementary material, which is available to authorized users.
Keywords: Polycystic ovary syndrome, TNF, Interleukin, Polymorphism, Meta-analysis
Introduction
Polycystic ovary syndrome (PCOS) is a common gynecological endocrine disorder that accounts for 70 % of all anovulatory infertility cases. It is associated with obesity, anovulation, hyperandrogenism, hirsutism, and infertility, which affect 6 % to 10 % of reproductive-age women worldwide [1, 2]. PCOS also increases the risk of some diseases such as type 2 diabetes and cardiovascular disease [3, 4]. Genetic, hormonal, and inflammatory factors may participate in the pathogenesis of PCOS. These include altered ovarian synthesis of steroids, hyperinsulinemia, aberrant folliculogenesis, abnormal secretion of gonadotropin, and neuroendocrine abnormalities [1]. Cytokines play an important role in regulating inflammation during these complex processes. Notably, some cytokine polymorphisms may affect the transcription and serum levels, thus changing inflammatory phenomena. Thus, association studies involving probable PCOS-associated cytokine gene polymorphisms and familial or sporadic cases have been performed. The results indicate that PCOS can be heritable, but the etiology and pathogenesis remain elusive.
Cytokines are cell-signaling protein molecules secreted by numerous cells and involved various intercellular communications including inflammatory responses. Low-grade chronic inflammation and imbalances between pro- and anti-inflammatory cytokines are thought to play a role in the pathogenesis of PCOS [5]. Pro-inflammatory cytokines such as tumor necrosis factor α (TNF-α), interleukin 1A (IL-1A), IL-1B, IL-6, and IL-18 are principal mediators of inflammation. Studies on reproductive biology have shown that these pro-inflammatory cytokines influence ovarian function and the processes of ovulation, fertilization, and implantation in women with PCOS. On the other hand, anti-inflammatory cytokines, such as IL-10 and IL-1 receptor antagonist (IL1RA), alter the state of inflammation in PCOS. These cytokine genes can be used as candidate genes to further understand the genetic effects of these cytokines, especially the risks for PCOS susceptibility and functional single nucleotide polymorphisms (SNPs). Therefore, a variety of molecular epidemiological studies have been focused on the association between TNF-α, IL-1A, IL-1B, IL-6, IL-10, or IL-18 polymorphisms and PCOS risk. However, the results from different studies have been inconsistent. To better clarify the association between these reported polymorphisms and the risk of PCOS, we conducted a systematic review and meta-analysis of the results of previously published studies.
Materials and methods
The meta-analysis was conducted according to the latest meta-analysis guidelines (PRISMA), including literature search, data collection, inclusion, etc. The PRISMA checklist was in supplemental Checklist_S1.
Literature search
We searched all the published papers in databases of PubMed, Embase, Web of Science, Medline, CNKI and Google Scholar. The key words were used as follows: ‘polymorphism’, ‘Tumor Necrosis Factor’,‘Tumor Necrosis Factor Alpha’, ‘TNF’, ‘TNF-α’, ‘interleukin’, ‘IL’ and ‘polycystic ovary syndrome’ or ‘PCOS’. Other relevant studies were identified by hand-searching the references of included articles. The search was limited to human studies. Two investigators (Wu and Yang) screened each of the titles, abstracts and full texts to determine inclusion independently. The results were compared and disagreements were resolved by consensus.
Inclusion and exclusion criteria
Studies included in our analysis met all the following criteria: (a) the studies evaluate the association between polymorphisms of TNF and interleukin and the risk of PCOS, (b) case–control studies with healthy populations as controls and specific diagnostic criteria for PCOS, the National Institute of Health (NIH) criteria or the Rotterdam criteria, and (c) genotype numbers in cases and controls available. Accordingly, the following exclusion criteria were also used: (a) no healthy control population, (b) genotype frequency unavailable, (c) non-conformity with the NIH criteria or the Rotterdam criteria for PCOS, and (d) duplication of previous publications.
Quality assessment for individual studies
The quality of the individual studies was evaluated and scored by two reviewers independently based on the Newcastle-Ottawa Scale (NOS) [6]. Each study was assessed based on three broad perspectives: selection, comparability and exposure, and each satisfactory answer received one point. The NOS ranges between zero (none of the quality criterion was met) up to nine stars (all the quality criteria were met), and the high-quality study was considered as the one with a score equal to or higher than seven. The third reviewer (Yang) examined the results, and a consensus was reached.
Data extraction
For each study, information was extracted including first author, published year, study population, country and genotype numbers in cases and controls. The process of data extraction was independently done by two investigators (Wu and Yu). An agreement was reached after discussion for conflicting data.
Statistical analysis
Associations between polymorphisms and risk of PCOS were calculated by odds ratio (OR) and 95 % confidence interval (95%CI). The data were analyzed under dominant (BB + AB versus AA) and recessive (BB versus AB + AA) genetic models, BB versus AA and B allele versus A allele (A represented major allele and B represented minor allele). Hardy-Weinberg equilibrium (HWE) was evaluated by chi-square test (χ2 test). If p value > 0.05, the genotype distribution of control population conformed to HWE. The heterogeneity was calculated by I2 and p. If p value < 0.05, there was obvious heterogeneity of the data, the random effect model was used. Otherwise, we used the fixed effect model. The pooled ORs were performed on the dominant and recessive models, respectively. The significance of pooled ORs was tested by Z test (p < 0.05 was considered significant). Publication bias was analyzed by Begg’s funnel plot and Egger’s test. All above statistical analysis were performed using Review Manager 5.2 and Stata 11.0.
Results
Search results
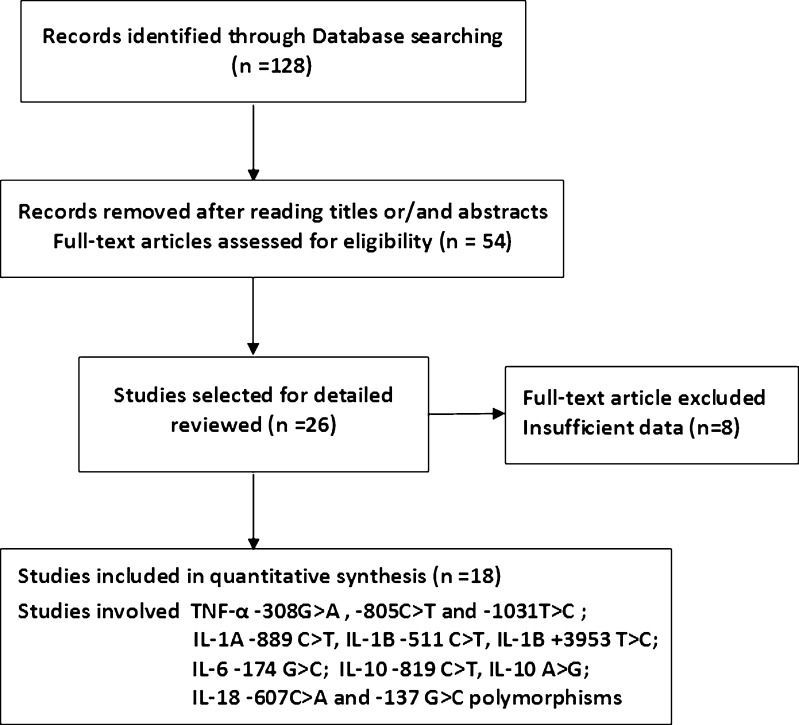
Searches of PubMed, Embase, Web of Science, Medline, CNKI, and Google Scholar revealed 128 potentially relevant articles. After careful reading the abstracts and titles, 26 potential articles were included [7–32]. Eight publications were excluded for insufficient or uncorrelated data according to our inclusion criteria [9, 11, 10, 14, 13, 23, 24, 30]. Finally, 18 articles met the inclusion criteria and were selected [8, 7, 27, 12, 28, 18, 15, 17, 20, 19, 22, 21, 25, 26, 29, 31, 16]. The study selection process is shown in Fig. 1.
Fig. 1.
Flow diagram of the study selection process
Study characteristics
The analysis of TNF-α involved 3 polymorphisms: −308G > A (rs1800629), −805C > T (rs1799724), and -1031 T > C (rs1799964). For -308G > A, 582 PCOS cases and 563 controls from 4 articles [32, 31, 17, 27] were included. For -805C > T, 87 cases and 115 controls were analyzed from the study by Korhonen [29]. Finally, for the -1031 T > C polymorphism, 500 PCOS cases and 450 controls from 2 articles were included [28, 27].
The analysis of the interleukin system involved the genes encoding IL-1A, IL-1B, IL-6, IL-10, and IL-18, which together contain 8 SNP sites. For IL-1A -889C > T (rs1800587), 310 cases and 279 controls from 2 studies [25, 20] were analyzed. For IL-1B -511C > T (rs16944), we looked at 482 cases and 421 controls from 4 studies [25, 8, 18, 16]. For the IL-1B +3953C > T polymorphism (rs1143634) in exon 5, 223 cases and 188 controls from 2 studies were included [18, 25]. For IL-6 -174G > C (rs1800795), 416 cases and 569 controls from 5 studies were analyzed [7, 17, 19, 21, 26]. For IL-10 -819C > T polymorphism (rs1800871) we looked at 91 cases and 75 controls from 1 article [22], and we included 188 cases and 170 cases from 2 studies for the IL-10 -1082A > G polymorphism (rs1800896) [17, 22]. For the IL-18 -607C > A polymorphism (rs1946518), we analyzed 118 cases and 79 controls from 1 study [15], and for IL-18 -137G > C (rs187238), we included 244 cases and 192 controls from 2 studies [12, 15].
The populations in the selected studies come mainly from Australia, China, Turkey, India, Finland, Austria, and Korea. The methods for detecting polymorphisms included PCR-SSCP, PCR-RFLP, AOS-PCR, and sequencing. The characteristics of the included studies are listed in Table 1.
Table 1.
Characteristics of included studies in this meta-analysis for polymorphisms of TNF-α and Interleukin system genes
| SNPs | First author | Year | Country | Method | Ethnicity | Case/Control | HWE | Association | Score | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | Total | ||||||||||
| TNF-α -308 G > A polymorphism rs1800629 | |||||||||||||
| Milner, C. R. | 1999 | Australia | PCR-SSCP | unknown | 59/63 | 23/42 | 2/3 | 84/108 | Y(0.19) | N | 7 | [32] | |
| Mao, W. | 2000 | China | PCR-RFLP | Asians | 88/37 | 29/13 | 1/4 | 118/54 | Y(0.09) | N | 6 | [31] | |
| Vural, P. | 2010 | Turkey | PCR-RFLP | Caucasians | 78/77 | 16/15 | 3/3 | 97/95 | Y(0.07) | N | 8 | [17] | |
| Deepika, M. L. | 2013 | India | PCR-RFLP | Asians | 10/10 | 270/293 | 3/3 | 283/306 | N(<0.001) | N | 6 | [27] | |
| TNF-α -805 C > T polymorphism rs1799724 | |||||||||||||
| Korhonen, S. | 2002 | Finland | PCR-RFLP | Caucasians | 74/96 | 11/16 | 2/3 | 87/115 | N(0.04) | N | 6 | [29] | |
| TNF-α -1031 T > C polymorphism rs1799964 | |||||||||||||
| Deepika, M. L. | 2013 | India | PCR-RFLP | Asians | 107/162 | 170/139 | 6/5 | 283/306 | N(<0.001) | Y | 6 | [27] | |
| Yun, J. H. | 2011 | China | Sequencing | Asians | 144/122 | 71/22 | 2/0 | 217/144 | Y(0.32) | Y | 7 | [28] | |
| IL-6 -174 G > C polymorphism rs1800795 | |||||||||||||
| Walch, K. | 2004 | Austria | Sequencing | Caucasians | 24/43 | 30/25 | 8/16 | 62/84 | N(0.002) | N | 6 | [26] | |
| Erdogan, M. | 2008 | Turkey | PCR-RFLP | Caucasians | 36/31 | 25/73 | 4/11 | 65/115 | N(<0.001) | Y | 6 | [21] | |
| Erdogan, M. | 2009 | Turkey | PCR-RFLP | Caucasians | 57/32 | 26/75 | 5/12 | 88/119 | N(<0.001) | Y | 6 | [19] | |
| Vural, P. | 2010 | Turkey | PCR-RFLP | Caucasians | 59/46 | 34/42 | 4/7 | 97/95 | Y(0.54) | N | 8 | [17] | |
| Tumu, V. R. | 2013 | India | Sequencing | Asians | 69/77 | 31/73 | 4/6 | 104/156 | N(0.02) | Y | 6 | [7] | |
| IL-1A −889 C > T polymorphism rs1800587 | |||||||||||||
| Kolbus, A. | 2007 | Austria | Sequencing | Caucasians | 42/55 | 56/41 | 7/6 | 105/102 | Y(0.65) | Y | 8 | [25] | |
| Wang, B. | 2009 | China | PCR-RFLP | Asians | 171/137 | 32/40 | 2/0 | 205/177 | Y(0.09) | N | 6 | [20] | |
| IL-1B −511 C > T polymorphism rs16944 | |||||||||||||
| Kolbus, A. | 2007 | Austria | Sequencing | Caucasians | 43/40 | 47/48 | 15/14 | 105/102 | Y(0.95) | N | 8 | [25] | |
| Mu, Y. | 2010 | China | N/A | Asians | 64/26 | 76/87 | 60/64 | 200/177 | Y(0.68) | Y | 8 | [16] | |
| Yang, Y. | 2010 | China | PCR-RFLP | Asians | 30/34 | 56/26 | 32/26 | 118/86 | N(<0.001) | Y | 6 | [18] | |
| Xia, Y. H. | 2013 | China | PCR-RFLP | Asians | 13/18 | 21/31 | 25/7 | 59/56 | Y(0.26) | N | 7 | [8] | |
| IL-1B +3953 C > T polymorphism rs1143634 | |||||||||||||
| Kolbus, A. | 2007 | Austria | PCR-RFLP | Caucasians | 48/60 | 46/37 | 11/5 | 105/102 | Y(0.82) | N | 7 | [25] | |
| Yang, Y. | 2010 | China | PCR-RFLP | Asians | 114/74 | 4/12 | 0/0 | 118/86 | Y(0.49) | Y | 7 | [18] | |
| IL-10 -1082 A > G polymorphism rs1800896 | |||||||||||||
| Karadeniz, M. | 2008 | Turkey | PCR-RFLP | Caucasians | 44/31 | 37/34 | 10/10 | 91/75 | Y(0.89) | N | 7 | [22] | |
| Vural, P. | 2010 | Turkey | ASO-PCR | Caucasians | 25/25 | 57/54 | 15/16 | 97/95 | Y(0.15) | N | 7 | [17] | |
| IL-10 -819 C > T polymorphism rs1800871 | |||||||||||||
| Karadeniz, M. | 2008 | Turkey | PCR-RFLP | Caucasians | 36/36 | 45/31 | 10/8 | 91/75 | Y(0.73) | N | 7 | [22] | |
| IL-18 -607 C > A polymorphism rs1946518 | |||||||||||||
| Yang, Y. | 2010 | China | PCR-SSCP | Asians | 39/22 | 61/48 | 18/9 | 118/79 | N(0.03) | N | 6 | [15] | |
| IL-18 -137 G > C polymorphism rs187238 | |||||||||||||
| Yang, Y. | 2010 | China | PCR-SSCP | Asians | 92/63 | 25/16 | 1/0 | 118/79 | Y(0.32) | N | 7 | [15] | |
| Kim, J. W. | 2012 | Korea | PCR-RFLP | Asians | 88/83 | 37/29 | 1/1 | 126/113 | Y(0.37) | N | 7 | [12] | |
HWE Hardy-Weinberg equilibrium, Y yes, N no
Quantitative synthesis
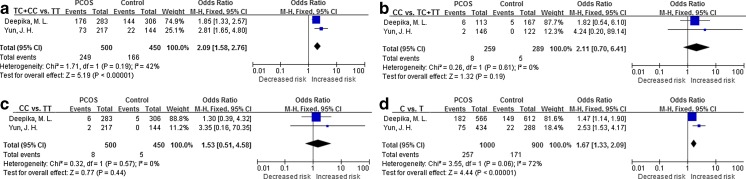
Three SNP sites in TNF-α were analyzed. To study the first (−308G > A), the data from 4 populations were combined. The results showed no evidence of association between this SNP and susceptibility to PCOS, suggesting that it is not a risk factor for PCOS. For the second polymorphism,-805C > T, 1 population was reanalyzed and no significant association with PCOS was found. For the last polymorphism, −1031 T > C, the data from 2 populations were combined and significant associations were found (Fig. 2).
Fig. 2.
Forest plot of the TNF-α -1031 T > C polymorphism and PCOS risk. The C allele increased the risk of PCOS. a, TC + CC vs. TT model; b, CC vs. TC + TT model; c, CC vs. TT model; d, C allele vs. T allele model
Our analysis of the IL-1 system involved the IL-1A -889C > T, IL-1B -511C > T, and IL-1B +3953 T > C polymorphisms. For IL-1A -889C > T, 2 populations were analyzed, and no significant associations were detected. The analysis of the IL-1B -511C > T polymorphism included 4 populations, and no significant associations were found. For IL-1B +3953 T > C, 2 populations were included, and no significant associations were found.
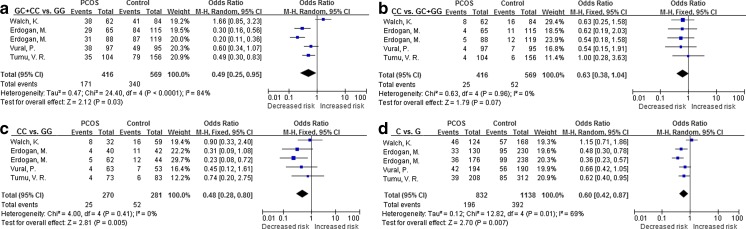
For the IL-6 -174G > C polymorphism, the data from 5 populations were combined and significant associations were found in the dominant model CC + GC vs. GG (OR = 0.49, 95 % CI = 0.25–0.95, p = 0.03), model CC vs. GG (OR = 0.48, 95 % CI = 0.28–0.80, p = 0.005), and model C vs. G (OR = 0.60, 95 % CI = 0.42–0.87, p = 0.007). However, in the recessive model, CC vs. GC + GG, no association was detected (OR = 0.63, 95 % CI = 0.38–1.04, p = 0.07). A forest plot of the IL-16 -174G > C polymorphism is shown in Fig. 3.
Fig. 3.
Forest plot of IL-6 -174G > C polymorphisms and PCOS risk, the allele C was a protective factor against PCOS risk. a, GC + CC vs. GG model; b, CC vs. GC + GG model; c, CC vs. GG model; d, C allele vs. G allele model
We analyzed 2 polymorphism sites in IL-10. In the first, IL-10 -819C > T, 1 population was reanalyzed, and no associations were found. For IL-10 -1082, the data for 2 populations were combined, and no associations were found with any of the polymorphisms.
Two polymorphisms in IL-18 were reanalyzed in the present study. For IL-18 -607C > A, 1 population was reanalyzed and no significant associations were found. For the IL-18 -137G > C polymorphism, the data from 2 populations were combined, and no significant associations were found for all genetic models.
In total, we identified significant associations between PCOS and the TNF-α -1031 T > C and IL-6 -174G > C polymorphisms. The main results of meta-analysis are shown in Figs. 2, 3, and Table 2.
Table 2.
Meta-analyses of TNF-α and Interleukin system genes polymorphisms and PCOS risk in overall analysis
| SNPs Position | Case/Control | BB + AB vs. AA | BB vs. AB + AA | BB vs. AA | B allele vs. A allele | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95 % CI) | model | P value | OR (95 % CI) | model | P value | OR (95 % CI) | model | P value | OR (95 % CI) | model | P value | ||
| TNF-α -308 G > A | 582/563 | 0.77 [0.54, 1.10] | Fixed | 0.15 | 0.63 [0.27, 1.47] | Fixed | 0.29 | 0.58 [0.24, 1.38] | Fixed | 0.22 | 0.91 [0.75, 1.10] | Fixed | 0.31 |
| TNF-α -805 C > T | 87/115 | 0.89 [0.41, 1.91] | Fixed | 0.76 | 0.88 [0.14, 5.37] | Fixed | 0.89 | 0.86 [0.14, 5.31] | Fixed | 0.88 | 0.89 [0.45, 1.77] | Fixed | 0.74 |
| TNF-α -1031 T > C | 500/450 | 2.09 [1.58, 2.76] | Fixed | <0.001* | 1.53 [0.51, 4.58] | Fixed | 0.44 | 2.11 [0.70, 6.41] | Fixed | 0.19 | 1.67 [1.33, 2.09] | Fixed | <0.001* |
| IL-6 -174 G > C | 416/569 | 0.49 [0.25, 0.95] | Random | 0.03* | 0.63 [0.38, 1.04] | Fixed | 0.07 | 0.48 [0.28, 0.80] | Fixed | 0.005* | 0.60 [0.42, 0.87] | Random | 0.007* |
| IL-1A −889 C > T | 310/279 | 1.09 [0.43, 2.75] | Random | 0.86 | 1.42 [0.51, 3.96] | Fixed | 0.51 | 1.79 [0.62, 5.21] | Fixed | 0.28 | 1.08 [0.79, 1.47] | Fixed | 0.65 |
| IL-1B −511 C > T | 482/421 | 1.00 [0.45, 2.19] | Random | 0.99 | 1.25 [0.62, 2.53] | Random | 0.52 | 1.19 [0.44, 3.19] | Random | 0.73 | 1.11 [0.67, 1.84] | Random | 0.69 |
| IL-1B +3953 C > T | 223/188 | 0.65 [0.09, 4.89] | Random | 0.67 | 2.27 [0.76, 6.78] | Fixed | 0.14 | 2.75 [0.89, 8.46] | Fixed | 0.08 | 0.65 [0.10, 4.38] | Random | 0.66 |
| IL-10 -1082 A > G | 188/170 | 0.87 [0.56, 1.36] | Fixed | 0.55 | 0.86 [0.48, 1.56] | Fixed | 0.62 | 0.82 [0.42, 1.60] | Fixed | 0.57 | 0.90 [0.67, 1.22] | Fixed | 0.51 |
| IL-10 -819 C > T | 91/75 | 1.41 [0.76, 2.62] | Fixed | 0.28 | 1.03 [0.39, 2.77] | Fixed | 0.95 | 1.25 [0.44, 3.53] | Fixed | 0.67 | 1.22 [0.77, 1.93] | Fixed | 0.40 |
| IL-18 -607 C > A | 118/79 | 0.78 [0.42, 1.46] | Fixed | 0.44 | 1.40 [0.59, 3.30] | Fixed | 0.44 | 1.13 [0.43, 2.93] | Fixed | 0.80 | 0.97 [0.65, 1.46] | Fixed | 0.89 |
| IL-18 -137 G > C | 244/192 | 1.16 [0.75, 1.80] | Fixed | 0.50 | 1.31 [0.17, 10.15] | Fixed | 0.80 | 1.35 [0.17, 10.57] | Fixed | 0.77 | 1.15 [0.77, 1.72] | Fixed | 0.50 |
*represents significant association
Population-based subgroup analysis
For IL-1A -889C > T, we combined the data for the Chinese and Indian populations and the Australian and Turkish populations. The corresponding pooled ORs of the genetic models were not materially altered. For the IL-6 -174G > C polymorphism, we combined the data from 4 Caucasian populations from Turkey and Austria, and the OR results were similar. For IL-1B -511C > T, the data for the populations of Asians from China and India were combined, and similar results were obtained. Unfortunately, subgroup analysis of other polymorphisms was not possible due to lack of published data.
Sensitivity analysis
Sensitivity analyses were conducted to determine whether modification of the inclusion criteria of the meta-analysis affected the final results. The included studies were limited to those conforming to HWE, detected method. In addition, we also performed sensitivity analysis by randomly removing 1 to 2 studies. Overall, the corresponding pooled ORs were not materially altered. In addition, random effects model were retested in all primal fixed models and corresponding pooled ORs were not materially altered. This suggested that our overall results of this meta-analysis were statistically robust.
Sensitivity analysis
Sensitivity analyses were conducted to determine whether modification of the inclusion criteria of the meta-analysis affected the final results. The included studies were limited to those conforming to the Hardy-Weinberg Equilibrium (HWE), detected method. In addition, we also performed sensitivity analysis by randomly excluding 1 to 2 studies. Overall, the corresponding pooled ORs were not materially altered. In addition, the random effects model was retested in all primal fixed models, and the corresponding pooled ORs were not materially altered. This suggested that the results of this meta-analysis are statistically robust.
Publication bias
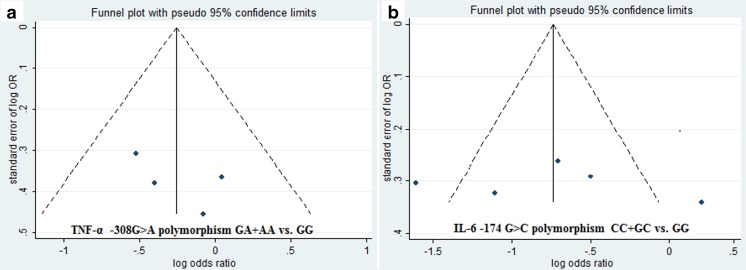
Begg’s funnel plot and Egger’s regression test were performed to assess the potential publication bias of the overall analyses. These processes need at least 4 included articles. For TNF-α -308G > A, IL-1B -511C > T, and IL-6 -174G > C, no obvious asymmetry was observed according to a visual assessment of the funnel plot (Fig. 4). Moreover, the results of Egger’s regression test did not provide any statistical evidence for publication bias (p > 0.05). Both the Begg's funnel plot and Egger's test showed that there was no obvious publication bias in overall analysis.
Fig. 4.
Funnel plot for dominant genetic models. a, TNF-α -308G > A polymorphism; b IL-6 -174G > C polymorphism
Discussion
PCOS is a heterogeneous endocrine disorder that is characterized by oligo- or anovulation, clinical and/or biochemical signs of hyperandrogenism, and polycystic ovaries. The etiology involves multiple genetic and epigenetic alterations that are still indistinct, but the imbalance between pro- and anti-inflammatory cytokines and polymorphisms in cytokine genes may play roles in the etiology of PCOS [17]. Identification of crucial genetic markers such as inflammation-related genes could help clarify the pathogenesis of PCOS. Many genetic studies have been performed to investigate the associations between PCOS risk and TNF-α, IL-1, IL-6, IL-10, and IL-18 polymorphisms. However, inconclusive results have made it difficult to find functional SNPs and build a bigger picture of how these genes affect PCOS risk. The present study was performed to make the most of the available data by reanalyzing the associations between these gene polymorphisms and PCOS.
At present, association studies involving inflammation-related genes and PCOS have mainly focused on TNF-α -308G > A (rs1800629), TNF-α -805C > T (rs1799724), TNF-α -1031 T > C (rs1799964), IL-1A -889C > T (rs1800587), IL-1B -511C > T (rs16944), IL-1B +3953 T > C (rs1143634), IL-6 -174G > C (rs1800795), IL-10 -819C > T (rs1800871), IL-10-1082A > G (rs1800896), IL-18 -607C > A (rs1946518), and IL-18 -137G > C (rs187238). Our meta-analysis provides the most comprehensive analysis on the relationships between theses polymorphisms and PCOS. For the pro-inflammatory cytokines IL-1A, IL-1B, and IL-18 and the anti-inflammatory cytokine IL-10, we did not find any positive associations. This is consistent with previous reports. However, some clinical observations suggest that the levels of these cytokines are increased in women with PCOS. There are some reasons that could explain the inconsistencies between genetic association analyses and the clinical phenomena. First, the sample size of the present association study is likely insufficient for detecting all the associations between these SNPs and PCOS. Second, genetic and phenotypic heterogeneity makes complex diseases, such as PCOS, hard to analyze. In our meta-analysis, we were not able to measure the plasma cytokine concentrations of individuals with PCOS and other known or unknown polymorphisms linked with these reported loci. Finally, inflammatory cytokines are intermediate molecules involving physiological or pathology reaction indicated the elusive pathogenesis for PCOS.
In the present meta-analysis, we reviewed multiple inflammatory cytokine genes systematically for the first time. The results synthesize our current knowledge of these polymorphisms and PCOS. Notably, significant associations were detected in the IL-6 -174G > C polymorphism and the TNF-α -1031 T > C polymorphism for the first time. For the IL-6 -174G > C polymorphism, the C allele was a protective factor for PCOS risk. The protective effect on PCOS susceptibility was found in the dominant model. The models allele C versus G and CC versus CG further confirmed the associations. However, no association was found in the recessive models and we believe that this may be due to insufficient sample sizes. Most women with PCOS have insulin resistance, hyperinsulinemia, and elevated serum IL-6 levels. A recent meta-analysis suggested that the serum IL-6 levels of PCOS patients may be influenced by metformin, and early application of metformin therapy may relieve chronic low-grade inflammation in women with PCOS [33]. Moreover, a similar positive association with the IL-6 -174 site was found for Alzheimer’s disease and cancer, which are also recognized as inflammation-related diseases [34]. Consistent results between genetic studies and clinical observations suggest that strong associations exist between this IL-6 polymorphism and PCOS. The major G allele was a risk factor for PCOS susceptibility. In addition, a strong association was also identified with the TNF-α -1031 T > C polymorphism, while no associations were found with the -308G > A or -805C > T polymorphisms. These results suggest that the −1310 site may be a true functional site for PCOS susceptibility and may affect the plasma level of protein. For the TNF-α -1031 T > C polymorphism, the minor C increased the risk of PCOS in dominant models and allele comparison, while no associations were found in recessive and CC versus TT models. The results should be interpreted with caution because only two studies were used in the meta-analysis.
We conducted a systematic review and found several significant associations between inflammatory cytokines and PCOS; however, some limitations should be acknowledged. First, the sample size in most of the included studies was small, and repeated studies for a single SNP were insufficient. This tends to increase the probability of false positives or false negatives. Second, the studies included in our meta-analysis were limited to published articles. We did not track any unpublished articles to obtain data for the analysis, and thus the data were incomplete. Third, PCOS is a multifactorial disease and potential gene–gene and gene-environment interactions should be considered. We did not carry out subgroup analysis based on other factors such as race, diet, smoking status, age, or sex due to the lack of sufficient data. Fourth, insufficient data regarding gene polymorphisms limited haplotype meta-analysis of PCOS risk. Lastly, the influence of bias in the present analysis could not be completely avoided since studies with positive results are generally easier to publish than those with negative results.
Notably, the controls of quite a few studies were not in HWE, and most of the included studies had a quality score of less than seven. These data need to be carefully evaluated, although the results of limited HWE or methods showed that the corresponding pooled ORs were not materially altered in the sensitivity analysis. In light of this, we suggest that several conditions be met in future studies: 1) homogeneous groups of PCOS patients and well-matched controls should be recruited; 2) patients should be diagnosed using the newest diagnostics guide; 3) the controls should be in HWE; 4) the results of fragment analysis should be verified by sequencing; 5) sample sizes should be large; 6) gene–gene and gene-environment interactions should be considered, and the related indices should be collected and analyzed, especially the associations between SNPs and levels of cytokines.
In conclusion, our present meta-analysis suggests positive associations between the TNF-α -1031 T > C rs1799964 and IL-6 -174G > C polymorphisms and the risk of PCOS. No associations were found with TNF-α -308G > A, TNF-α -805C > T, IL-1A -889C > T, IL-1B -511C > T, IL-1B +3953, IL-10 -819 C > T, IL-10 A > G, IL-18 -607C > A, or IL-18-137G > C. However, due to the heterogeneity and poor quality of the studies included in the meta-analysis, the results should be interpreted with caution. Future studies should involve large sample sizes, multiple ethnicities, homogeneous populations of PCOS patients, and well-matched controls.
Electronic supplementary material
(DOC 71 kb)
Acknowledgments
No funding was provided for the analysis.
Conflict of interest
None Declared.
Footnotes
Capsule The twelve SNPs for TNF-α and Interleukin genes: TNF-α -1031T > C and IL-6 -174G > C are associated with PCOS risk, and no associations are found in TNF-α -308G > A, TNF-α -805C > T, IL-1A -889C > T, IL-1B -511C > T, IL-1B +3953C > T, IL-10 -819C > T, IL-10 -1082 A > G, IL-18 -607C > A, and IL-18 -137G > C polymorphisms.
References
- 1.Goodarzi MO, Dumesic DA, Chazenbalk G, Azziz R. Polycystic ovary syndrome: etiology, pathogenesis and diagnosis. Nat Rev Endocrinol. 2011;7(4):219–31. doi: 10.1038/nrendo.2010.217. [DOI] [PubMed] [Google Scholar]
- 2.Brassard M, AinMelk Y, Baillargeon JP. Basic infertility including polycystic ovary syndrome. Med Clin N Am. 2008;92(5):1163–92. doi: 10.1016/j.mcna.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Mani H, Levy MJ, Davies MJ, Morris DH, Gray LJ, Bankart J, et al. Diabetes and cardiovascular events in women with polycystic ovary syndrome: a 20-year retrospective cohort study. Clin Endocrinol (Oxf) 2013;78(6):926–34. doi: 10.1111/cen.12068. [DOI] [PubMed] [Google Scholar]
- 4.Dokras A. Cardiovascular disease risk factors in polycystic ovary syndrome. Semin Reprod Med. 2008;26(1):39–44. doi: 10.1055/s-2007-992923. [DOI] [PubMed] [Google Scholar]
- 5.Ojeda-Ojeda M, Murri M, Insenser M, Escobar-Morreale HF. Mediators of low-grade chronic inflammation in polycystic ovary syndrome (PCOS) Curr Pharm Des. 2013;19(32):5775–91. doi: 10.2174/1381612811319320012. [DOI] [PubMed] [Google Scholar]
- 6.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 7.Tumu VR, Govatati S, Guruvaiah P, Deenadayal M, Shivaji S, Bhanoori M. An interleukin-6 gene promoter polymorphism is associated with polycystic ovary syndrome in South Indian women. J Assist Reprod Genet. 2013;30(12):1541–6. doi: 10.1007/s10815-013-0111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia YH, Yao L, Zhang ZX. Correlation between IL-1beta, IL-1Ra gene polymorphism and occurrence of polycystic ovary syndrome infertility. Asian Pac J Trop Med. 2013;6(3):232–6. doi: 10.1016/S1995-7645(13)60030-9. [DOI] [PubMed] [Google Scholar]
- 9.Kosova G, Urbanek M. Genetics of the polycystic ovary syndrome. Mol Cell Endocrinol. 2013;373(1–2):29–38. doi: 10.1016/j.mce.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deligeoroglou E, Vrachnis N, Athanasopoulos N, Iliodromiti Z, Sifakis S, Iliodromiti S, et al. Mediators of chronic inflammation in polycystic ovarian syndrome. Gynecol Endocrinol. 2012;28(12):974–8. doi: 10.3109/09513590.2012.683082. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Qiao J, Li MZ. Correlation between interleukin-1 and the obesity of polycystic ovary syndrome. Zhonghua Fu Chan Ke Za Zhi. 2012;47(1):9–13. [PubMed] [Google Scholar]
- 12.Kim JW, Lee MH, Park JE, Yoon TK, Lee WS, Shim SH. Association of IL-18 genotype with impaired glucose regulation in Korean women with polycystic ovary syndrome. Eur J Obstet Gynecol Reprod Biol. 2012;161(1):51–5. doi: 10.1016/j.ejogrb.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Kawamura S, Maesawa C, Nakamura K, Nakayama K, Morita M, Hiruma Y, et al. Predisposition for borderline personality disorder with comorbid major depression is associated with that for polycystic ovary syndrome in female Japanese population. Neuropsychiatr Dis Treat. 2011;7:655–62. doi: 10.2147/NDT.S25504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YS, Tsai SJ, Lin MW, Yang CT, Huang MF, Wu MH. Interleukin-6 as an early chronic inflammatory marker in polycystic ovary syndrome with insulin receptor substrate-2 polymorphism. Am J Reprod Immunol. 2011;66(6):527–33. doi: 10.1111/j.1600-0897.2011.01059.x. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Qiao J, Li MZ. Association of polymorphisms of interleukin-18 gene promoter region with polycystic ovary syndrome in chinese population. Reprod Biol Endocrinol. 2010;8:125. doi: 10.1186/1477-7827-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mu Y, Liu J, Wang B, Wen Q, Wang J, Yan J, et al. Interleukin 1 beta (IL-1beta) promoter C [−511] T polymorphism but not C [+3953] T polymorphism is associated with polycystic ovary syndrome. Endocrine. 2010;37(1):71–5. doi: 10.1007/s12020-009-9268-x. [DOI] [PubMed] [Google Scholar]
- 17.Vural P, Degirmencioglu S, Saral NY, Akgul C. Tumor necrosis factor alpha (−308), interleukin-6 (−174) and interleukin-10 (−1082) gene polymorphisms in polycystic ovary syndrome. Eur J Obstet Gynecol Reprod Biol. 2010;150(1):61–5. doi: 10.1016/j.ejogrb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y, Qiao J, Tang RX, Li MZ. Genotype and haplotype determination of interleukin (IL) 1 beta (g. -511C > T and g. +3954C > T) and IL-1RN in polycystic ovary syndrome. Fertil Steril. 2010;94(1):384–6. doi: 10.1016/j.fertnstert.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 19.Erdogan M, Karadeniz M, Berdeli A, Tamsel S, Yilmaz C. The relationship of the interleukin-6–174 G > C gene polymorphism with cardiovascular risk factors in Turkish polycystic ovary syndrome patients. Int J Immunogenet. 2009;36(5):283–8. doi: 10.1111/j.1744-313X.2009.00867.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Zhou S, Wang J, Liu J, Ni F, Liu C, et al. Lack of association between interleukin-1a gene (IL-1a) C (−889) T variant and polycystic ovary syndrome in Chinese women. Endocrine. 2009;35(2):198–203. doi: 10.1007/s12020-009-9152-8. [DOI] [PubMed] [Google Scholar]
- 21.Erdogan M, Karadeniz M, Berdeli A, Alper G, Caglayan O, Yilmaz C. The relationship of the interleukin-6–174 G > C gene polymorphism with oxidative stress markers in Turkish polycystic ovary syndrome patients. J Endocrinol Investig. 2008;31(7):624–9. doi: 10.1007/BF03345614. [DOI] [PubMed] [Google Scholar]
- 22.Karadeniz M, Erdogan M, Zengi A, Tamsel S, Berdeli A, Saygili F, et al. Polymorphism of the interleukin-10 gene in polycystic ovary syndrome. Int J Immunogenet. 2008;35(2):119–23. doi: 10.1111/j.1744-313X.2007.00746.x. [DOI] [PubMed] [Google Scholar]
- 23.Kolbus A, Walch K, Szabo L, Huber JC, Nagele F, Unfried G. A polymorphism of the interleukin 1 receptor antagonist is not associated with polycystic ovary syndrome in Caucasian women. Fertil Steril. 2006;85(2):523–5. doi: 10.1016/j.fertnstert.2005.07.1317. [DOI] [PubMed] [Google Scholar]
- 24.Mohlig M, Spranger J, Osterhoff M, Ristow M, Pfeiffer AF, Schill T, et al. The polycystic ovary syndrome per se is not associated with increased chronic inflammation. Eur J Endocrinol. 2004;150(4):525–32. doi: 10.1530/eje.0.1500525. [DOI] [PubMed] [Google Scholar]
- 25.Kolbus A, Walch K, Nagele F, Wenzl R, Unfried G, Huber JC. Interleukin-1 alpha but not interleukin-1 beta gene polymorphism is associated with polycystic ovary syndrome. J Reprod Immunol. 2007;73(2):188–93. doi: 10.1016/j.jri.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Walch K, Grimm C, Zeillinger R, Huber JC, Nagele F, Hefler LA. A common interleukin-6 gene promoter polymorphism influences the clinical characteristics of women with polycystic ovary syndrome. Fertil Steril. 2004;81(6):1638–41. doi: 10.1016/j.fertnstert.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 27.Deepika ML, Reddy KR, Yashwanth A, Rani VU, Latha KP, Jahan P. TNF-alpha haplotype association with polycystic ovary syndrome - a South Indian study. J Assist Reprod Genet. 2013;30(11):1493–503. doi: 10.1007/s10815-013-0080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yun JH, Choi JW, Lee KJ, Shin JS, Baek KH. The promoter −1031(T/C) polymorphism in tumor necrosis factor-alpha associated with polycystic ovary syndrome. Reprod Biol Endocrinol. 2011;9:131. doi: 10.1186/1477-7827-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korhonen S, Romppanen EL, Hiltunen M, Mannermaa A, Punnonen K, Hippelainen M, et al. Lack of association between C-850 T polymorphism of the gene encoding tumor necrosis factor-alpha and polycystic ovary syndrome. Gynecol Endocrinol. 2002;16(4):271–4. doi: 10.1080/713603095. [DOI] [PubMed] [Google Scholar]
- 30.Peral B, San Millan JL, Castello R, Moghetti P, Escobar-Morreale HF. Comment: the methionine 196 arginine polymorphism in exon 6 of the TNF receptor 2 gene (TNFRSF1B) is associated with the polycystic ovary syndrome and hyperandrogenism. J Clin Endocrinol Metab. 2002;87(8):3977–83. doi: 10.1210/jcem.87.8.8715. [DOI] [PubMed] [Google Scholar]
- 31.Mao W, Yu L, Chen Y. Study on the relationship between a polymorphism of tumor necrosis factor-alpha gene and the pathogenesis of polycystic ovary syndrome. Zhonghua Fu Chan Ke Za Zhi. 2000;35(9):536–9. [PubMed] [Google Scholar]
- 32.Milner CR, Craig JE, Hussey ND, Norman RJ. No association between the −308 polymorphism in the tumour necrosis factor alpha (TNFalpha) promoter region and polycystic ovaries. Mol Hum Reprod. 1999;5(1):5–9. doi: 10.1093/molehr/5.1.5. [DOI] [PubMed] [Google Scholar]
- 33.Xu X, Du C, Zheng Q, Peng L, Sun Y. Effect of metformin on serum interleukin-6 levels in polycystic ovary syndrome: a systematic review. BMC Womens Health. 2014;14:93. doi: 10.1186/1472-6874-14-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai L, Liu D, Guo H, Wang Y, Bai Y. Association between polymorphism in the promoter region of Interleukin 6 (−174 G/C) and risk of Alzheimer’s disease: a meta-analysis. J Neurol. 2012;259(3):414–9. doi: 10.1007/s00415-011-6164-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 71 kb)