Abstract
Purpose
Previous studies identified follicle-stimulating hormone receptor (FSHR) and luteinizing hormone/choriogonadotropin receptor (LHCGR) genes as polycystic ovary syndrome (PCOS) susceptibility loci, which was dependent on the racial/ethnic background of studied population. We investigated the association of genetic variants in FSHR and LHCGR with PCOS in Bahraini Arab women.
Methods
A retrospective case–control study, involving 203 women with PCOS, and 211 age- and ethnically-matched control women. FSHR and LHCGR genotyping was done by allelic exclusion method (real-time PCR).
Results
Significantly lower frequencies of heterozygous LHCGR rs7371084 and FSHR rs11692782 genotype carriers were seen between women with PCOS vs. controls, and increased frequency of heterozygous homozygous LHCGR rs4953616 genotype carriers were detected between women with PCOS compared to control women. Limited linkage disequilibrium was noted among LHCGR and FSHR SNPs, and 2 blocks were constructed: the first (Block 1) spanning 61 kb contained the six tested LHCGR SNPs, and the second (Block 2) spanning 298 kb contained four of the five tested FSHR SNPs. Higher frequency of LHCGR GTCAAG haplotype was seen in women with PCOS compared to controls; the frequencies of the remaining LHCGR haplotypes, and all FSHR haplotypes were similar between cases and controls.
Conclusion
This is the first study to confirm the association of novel LHCGR (rs7371084, rs4953616) and FSHR (rs11692782) SNPs with PCOS. The differential association of LHCGR and FSHR variants with PCOS confirms the racial/ethnic contribution to their association with PCOS.
Keywords: Follicle-stimulating hormone receptor, Luteinizing hormone/ choriogonadotropin receptor haplotypes, Polycystic ovary syndrome
Introduction
Polycystic ovary syndrome (PCOS) is a common endocrine disorder affecting 4–12 % of women in their reproductive age [1, 2], and is characterized by hyperandrogenemia, menstrual irregularities, and polycystic ovarian morphology [1, 3]. PCOS is associated with obesity, impaired glucose tolerance and insulin resistance, and increased risk of developing type 2 diabetes [1, 4], and cardiovascular disease [2, 5]. PCOS is a complex disorder, and both environmental and genetic factors contribute to its pathogenesis. Familial aggregation and genome-wide association studies (GWAS) support the contribution of genetic factors to PCOS etiology [1, 6–8]. PCOS-susceptibility loci include follicle stimulating hormone receptor (FSHR), luteinizing hormone/choriogonadotropin receptor (LHCGR), THADA, and DENND1A [6, 9, 10]. In addition, epigenetic modifications by environmental determinants of PCOS may alter the clinical presentation of PCOS [11].
LH stimulates follicular development, steroidogenesis, and formation of the corpus luteum [10, 12], and ovulation results from a surge in LH levels [13]. LH acts by binding its high-affinity receptor, LHCGR, which also serves as the receptor for human chorionic gonadotropin (hCG). LHCGR gene maps to chromosome 2p16.3, and several polymorphisms throughout the LHCGR gene were identified [10, 14]. On the other hand, binding of FSH to its high affinity receptor (FSHR) stimulates oogenesis, follicle development and gametogenesis, resulting in follicular maturation and proliferation of granulosa cells [12]. Altered FSHR function caused by a number of FSHR genetic variants induces arrest of follicle development, resulting in functional changes, such as primary amenorrhea, hypoplastic ovary, and high FSH serum levels. Altered FSH/LH ratio was linked with insulin resistance [15], and with altered production of and responsiveness to sex hormones, in particular testosterone [16].
Recent GWAS on Han Chinese [6] and European [10] population identified the 2p16.3 region (containing LHCGR and FSHR loci) to be associated with PCOS, with notable differences according to racial background. For example, the LHCGR rs13405728 variant, independently shown to be associated with PCOS in the Chinese [6, 17], is not informative in European-derived population [18–20]. As such, the search of specific PCOS- predisposing loci is influenced by racial background, PCOS phenotype and diagnostic criteria, along with variable fertility of PCOS cases. This study evaluated the association of 5 FSHR and 6 LHCGR SNPs identified as PCOS susceptibility variants in European and non-European populations, with PCOS in Bahraini Arab women.
Subjects and methods
Subjects
A total of 414 unrelated Bahraini Arab women with (n = 203) and without (n = 211) PCOS were recruited from the outpatient obstetrics/gynecology and adult endocrinology clinics in Manama, Bahrain. Control women comprised eumenorrheic university students and employees, or otherwise healthy volunteers. Their total testosterone levels were within the reference range (0.4–3.5 nmol/L), and were studied in the follicular phase of their menstrual cycle. PCOS diagnosis was based on the 2003 Rotterdam Criteria [21], whereby PCOS diagnosis was confirmed when two of the three conditions were met: anovulation, hyperandrogenism, and the presence of polycystic ovary on ultrasound examination.
Exclusion criteria included androgen-producing tumors, 21-hydroxylase-deficiency, nonclassical adrenal hyperplasia, hyperprolactinemia, active thyroid disease, and Cushing’s syndrome. Additional exclusion criteria for cases and controls included extremes of body mass index (BMI) (<18 kg/m2 or >50 kg/m2), recent/current illness, medications likely to affect carbohydrate metabolism or endocrine parameters for at least 3 months before entering the study. The latter included oral contraceptive, anti-hypertensive, lipid-lowering, and anti-inflammatory agents. Demographic data and history of hypertension, diabetes, and hypercholesterolemia were recorded for all subjects. Study participants gave written informed consent prior to entering the study, which was approved by local research and ethics committees.
Biochemical analysis
Peripheral venous blood samples were obtained at 7:00–9:00 am during the early follicular phase of the menstrual cycle (days 2 to 5), after an overnight (>12 h) fasting. FSH, LH, prolactin, total testosterone, progesterone, 17α-hydroxyprogesterone, and thyroid stimulating hormone (TSH) were determined using immunofluorometric assay or radioimmunoassay (coefficients of variation (CV) <5 % for all tests). Free testosterone (FT) and bioactive testosterone (BT) were calculated using online calculator (http://www.issam.ch/freetesto.htm), while free androgen index (FAI) was calculated as per: FAI = 100 × [total testosterone ÷ SHBG] [22]. Glucose was measured by the hexokinase method, and insulin was measured by ELISA according to instructions of the manufacturer (R&D Systems). Indices of insulin resistance included homeostasis model assessment of insulin resistance (HOMA-IR; calculated as per: [fasting glucose (mmol/l)/fasting insulin (μIU/ml)]/22.5) and the revised QUICKI, the latter used since cases and controls spanned several BMI categories.
SNP genotyping
Total genomic DNA was extracted from peripheral blood leukocytes using Illustra blood genomicPrep Mini-Spin kit (GE Healthcare; Buckinghamshire, UK). We selected FSHR (rs46166, rs1007541, rs11692782, rs2055571, and rs1394205) and LHCGR (rs2293275, rs4073366, rs7371084, rs4597581, rs4953616, and rs13405728) SNPs in view of their frequency in Caucasians, and their reported association with PCOS. The FSHR and LHCGR SNPs were genotyped by the allelic discrimination method on StepOne Plus real-time PCR system (Applied Biosystems; Foster City, CA); using commercially available primers obtained from the Assay-on-demand system with well-defined genotype clusters. Genotype frequencies of the five tested FSHR and six LHCGR SNPs were consistent with Hardy-Weinberg equilibrium (Table 2), and the minor allele frequencies (MAF) obtained were comparable to those in the HapMap CEU sample.
Table 2.
Distribution of LHCGR and FSHR alleles in PCOS cases and control women
| Locus | SNP | Positiona | Alleles | Cases | Controls | HWE | χ2 | P c | OR (95 % CI) | Power |
|---|---|---|---|---|---|---|---|---|---|---|
| LHCGR | rs2293275 | 48921375 | G:A | 139 (0.34)b | 141 (0.34)b | 0.43 | 0.00 | 1.00 | 67.5 | |
| rs7371084 | 48939953 | T:C | 16 (0.05) | 41 (0.10) | 1.00 | 8.16 | 0.001 | 0.39 (0.22–0.71) | 72.3 | |
| rs4953616 | 48941428 | T:C | 164 (0.43) | 132 (0.32) | 0.56 | 10.91 | 0.001 | 1.61 (1.21–2.14) | 91.2 | |
| rs4597581 | 48958595 | A:G | 85 (0.21) | 90 (0.21) | 0.21 | 0.02 | 0.89 | 46.8 | ||
| rs13405728 | 48978159 | A:G | 35 (0.09) | 36 (0.09) | 0.39 | 0.44 | 0.51 | 41.1 | ||
| rs4073366 | 48982622 | G:C | 35 (0.09) | 29 (0.07) | 0.87 | 1.14 | 0.29 | 59.1 | ||
| FSHR | rs1007541 | 49209034 | G:A | 35 (0.09) | 36 (0.09) | 1.00 | 0.00 | 1.00 | 42.8 | |
| rs11692782 | 49291893 | A:T | 171 (0.42) | 181 (0.43) | 1.00 | 0.05 | 0.82 | 82.4 | ||
| rs2055571 | 49344814 | G:A | 176 (0.43) | 182 (0.43) | 0.27 | 0.00 | 1.00 | 57.2 | ||
| rs1394205 | 49381585 | G:A | 85 (0.21) | 106 (0.25) | 1.00 | 2.04 | 0.15 | 60.8 | ||
| rs6166 | 49589921 | G:A | 186 (0.46) | 211 (0.50) | 0.89 | 1.45 | 0.23 | 66.2 |
MAF Minor allele frequency, HWE Hardy-Weinberg Equilibrium
aLocation on chromosome based on dbSNP build 125
bMinor allele defined based on frequency in controls
cAdjusted P value, adjusted for BMI, TSH, FT, BT, FAI, SHBG, and fasting insulin
Statistical analysis
Statistical analysis was performed on SPSS v. 20.0 (SPSS Inc., Chicago, IL). Data were expressed as percentages of total (categorical variables), or mean ± SD (continuous variables). Student’s t-test was used to determine differences in means, and Pearson χ2 test was used to assess inter–group significance. For continuous variables that did not follow a normal distribution, we used nonparametric analysis: Mann–Whitney U-test for two group comparisons, or Kruskal–Wallis test for multiple group comparisons; quantitative data described as medians and range values. Genotypes were tested for departures from Hardy–Weinberg equilibrium (HWE) in the control population using Haploview 4.2 (http://www.broadinstitute.org/haploview) [23]. All analyses were conducted assuming additive genetic model. We used CaTS power calculator (www.sph.umich.edu/csg/abecasis/cats) to calculate the power to detect an association between FSHR and LHCGR variants and PCOS in the studied cohort [24]. The parameters used were 203 cases and 211 control women, genotypic relative risk for heterozygote (1/2) and minor allele homozygous (2/2), and MAF for PCOS cases and controls, assuming 6 % PCOS prevalence (www.rightdiagnosis.com/p/pcos/stats-country.htm). Assuming these parameters, we had 73.0 % power to detect an effect at P < 0.01 for LHCGR SNPs, and 71.9 % for FSHR SNPs. Pairwise linkage disequilibrium (LD) values were calculated with Haploview 4.2, which also computed the frequency of common haplotypes (frequency ≥2 %). Logistic regression analysis was performed to determine the adjusted odds ratios (OR) and 95 % confidence intervals (95%CI) associated with the risk of PCOS, after controlling for a number of covariates, taking control women as the reference group. Null hypothesis was rejected at P < 0.05.
Results
Study subjects
The clinical and biochemical characteristics of study subjects are reported in Table 1. While age and waist-hip ratio at examination, menarche, along with fasting glucose, serum lipid profile, total testosterone, FSH and LH serum levels were comparable between PCOS cases and control women, significant differences between were noted in mean BMI (P < 0.001), FT (P = 0.009) and BT (P = 0.004) and FAI (P = 0.015), as well as fasting insulin (P < 0.001) and insulin resistance indices (HOMA-IR, QUICKI). Accordingly, the latter were the covariates that were controlled for in subsequent analysis.
Table 1.
Baseline and endocrine parameters of women with PCOS and control women
| Casesa | Controlsa | P b | |
|---|---|---|---|
| Age (years)c | 28.3 ± 6.1 | 26.9 ± 7.7 | 0.082 |
| BMI (kg/m2)c | 29.9 ± 6.3 | 25.7 ± 5.3 | 2.3 × 10−6 |
| Waist/hip ratio (WHR)c | 0.85 ± 0.12 | 0.83 ± 0.12 | 0.286 |
| LH (IU/L)d | 6.5 (0.8–66.6) | 5.4 (0.4–56.3) | 0.063 |
| FSH (IU/L)d | 5.3 (0.5–16.8) | 5.3 (0.4–15.4) | 0.568 |
| LH/FSHd | 1.3 (0.05–6.12) | 1.0 (0.2–10.9) | 0.060 |
| TSH (μIU/ml)c | 2.7 ± 2.1 | 1.7 ± 0.9 | 0.006 |
| Menarche (years)c | 12.4 ± 1.4 | 12.6 ± 1.4 | 0.247 |
| Total testosterone (nmol/L)c | 1.8 ± 1.0 | 1.7 ± 1.1 | 0.583 |
| Free testosterone (pmol/L) | 25.2 (3.3–92.4) | 18.4 (0.7–49.2) | 0.009 |
| Bioavailable testosterone (pmol/L)d | 684.0 (149.0–2020.0) | 469 (95.9–1480.0) | 0.004 |
| Free androgen index (FAI)d | 5.1 (0.7–320.8) | 3.1 (0.3–151.7) | 0.015 |
| SHBG (nmol/L)d | 20.1 (12.9–185.3) | 57.7 (12.6–189.8) | 1.4 × 10−6 |
| Fasting insulind | 10.2 (1.6–99.4) | 7.2 (1.7–22.1) | 2.6 × 10−4 |
| HOMA-IRc | 3.8 ± 2.7 | 1.9 ± 0.9 | 1.8 × 10−4 |
| QUICKIc | 0.58 ± 0.12 | 0.65 ± 0.11 | 0.001 |
| Total cholesterol (mmol/L)c | 4.8 ± 1.3 | 4.7 ± 1.0 | 0.804 |
| HDL-cholesterol (mmol/L)c | 1.3 ± 0.6 | 1.4 ± 0.5 | 0.498 |
| LDL-cholesterol (mmol/L)c | 2.7 ± 0.9 | 2.6 ± 0.6 | 0.591 |
| Triglycerides (mmol/L)c | 1.5 ± 1.0 | 1.1 ± 0.8 | 0.135 |
aA total of 203 PCOS cases and 211 control women were included
bStudent’s t-test (variables with normal distribution), Mann–Whitney U-test (variables that were not normally distributed)
cMean ± SD
dPercent of total within each group/subgroup
Association studies
Table 2 summarizes the association between LHCGR and FSHR SNPs and PCOS in case–control subjects. The genotypes of the tested LHCGR and FSHR SNPs were in HWE among study participants. Of the tested LHCGR SNPs, minor allele frequency (MAF) of rs7371084 was lower (P = 0.001), while that of rs4953616 was higher (P = 0.001) among women with PCOS compared to control women. MAF of the remaining tested LHCGR were comparable between women with PCOS and control women. On the other hand, MAF of the five tested FSHR variants were comparable between unselected women with PCOS and control women, even before adjusting for covariates.
The distribution of LHCGR and FSHR genotypes between women with PCOS and control women are summarized in Table 3. Genotypes were coded as “1” or “2” according to major or minor allele, respectively. Setting homozygous major allele genotype (1/1) as reference after controlling for BMI, TSH, FT, BT, FAI, SHBG, and fasting insulin (OR = 1.00), significantly lower frequencies of heterozygous (1/2) LHCGR rs7371084 (0.08 vs. 0.18) and FSHR rs11692782 (0.39 vs. 0.57) genotype carriers were seen between women with PCOS vs. control women. On the other hand, increased frequency of heterozygous (1/2) (0.45 vs. 0.38) and homozygous (0.21 vs. 0.31) LHCGR rs4953616 genotype carriers were detected between women with PCOS compared to control women. The distribution of the remaining LHCGR and FSHR genotypes was comparable between women with PCOS and control women.
Table 3.
LHCGR and FSHR genotype frequencies
| 1 / 1 | 1 / 2 | 2 / 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | SNP | Cases | Controls | P a | Cases | Controls | aORb (95 % CI) | Cases | Controls | aOR (95 % CI) |
| LHCGR | rs2293275 | 93 (0.46)c | 102 (0.48) | 0.57 | 81 (0.40) | 74 (0.35) | 1.20 (0.79–1.83) | 29 (0.14) | 35 (0.17) | 0.91 (0.52–1.60) |
| rs7371084 | 184 (0.91) | 172 (0.82) | 0.023 | 17 (0.08) | 37 (0.18) | 0.44 (0.24–0.80) | 2 (0.01) | 2 (0.01) | 0.95 (0.13–6.82) | |
| rs4953616 | 70 (0.34) | 103 (0.49) | 0.007 | 91 (0.45) | 81 (0.38) | 1.66 (1.08–2.56) | 42 (0.21) | 27 (0.13) | 2.30 (1.30–4.08) | |
| rs4597581 | 130 (0.64) | 132 (0.63) | 0.86 | 61 (0.30) | 68 (0.32) | 0.91 (0.60–1.39) | 12 (0.06) | 11 (0.05) | 1.13 (0.48–2.64) | |
| rs13405728 | 168 (0.83) | 170 (0.81) | 0.46 | 35 (0.17) | 40 (0.19) | 0.89 (0.53–1.47) | 0 (0.00) | 1 (0.005) | 0.00 (0.00–NA) | |
| rs4073366 | 169 (0.83) | 183 (0.87) | 0.60 | 32 (0.16) | 27 (0.13) | 1.25 (0.72–2.18) | 2 (0.01) | 1 (0.005) | 2.18 (0.20–24.22) | |
| FSHR | rs1007541 | 170 (0.84) | 177 (0.84) | 1.00 | 31 (0.12) | 32 (0.15) | 0.98 (0.57–1.69) | 2 (0.01) | 2 (0.01) | 1.05 (0.06–16.88) |
| rs11692782 | 78 (0.38) | 60 (0.28) | 8.0 × 10−4 | 79 (0.39) | 121 (0.57) | 0.50 (0.32–0.78) | 46 (0.23) | 30 (0.14) | 1.17 (0.66–2.07) | |
| rs2055571 | 75 (0.37) | 69 (0.33) | 0.20 | 80 (0.39) | 102 (0.48) | 0.72 (0.46–1.13) | 48 (0.24) | 40 (0.19) | 1.09 (0.64–1.88) | |
| rs1394205 | 130 (0.64) | 115 (0.55) | 0.08 | 61 (0.30) | 86 (0.41) | 0.63 (0.41–0.96) | 12 (0.06) | 10 (0.05) | 1.08 (0.45–2.59) | |
| rs6166 | 64 (0.32) | 52 (0.26) | 0.31 | 92 (0.45) | 107 (0.51) | 0.70 (0.44–1.12) | 47 (0.23) | 52 (0.26) | 0.74 (0.43–1.27) | |
Genotypes were coded as per “1” = major allele, “2” = minor allele
a2-way ANOVA
baOR = adjusted OR; covariates that were controlled for were BMI, TSH, FT, BT, FAI, SHBG, and fasting insulin
cNumber of subjects (frequency)
We then investigated the possible association of LHCGR rs7371084 and rs4953616, and FSHR rs11692782 variants with PCOS in obese vs. non-obese women with PCOS and control women. Cases and controls were stratified into obese (BMI ≥30 kg/m2) and non-obese (BMI <30 kg/m2). Results from Table 4 demonstrated association of FSHR rs11692782 with PCOS irrespective of BMI status. On the other hand, the association of LHCGR rs7371084 with PCOS was seen in non-obese (P = 0.002) but not obese (P = 0.695) subjects, while the association of LHCGR rs4953616 with PCOS disappeared when subjects were stratified into non-obese and obese subjects.
Table 4.
Distribution of FSHR and LHCGR variants in women with PCOS and control women according to obesity
| Non-obese (BMI <30 kg/m2) | Obese (BMI ≥30 kg/m2) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Genotype | Cases | Controls | χ2 | P a | Cases | Controls | χ2 | P a |
| FSHR | rs11692782 | A/A | 26 (41.3)b | 26 (24.5) | 9.30 | 0.010 | 4 (36.4) | 4 (20.0) | 6.48 | 0.039 |
| A/T | 24 (38.1) | 66 (62.3) | 25 (37.9) | 14 (70.0) | ||||||
| T/T | 13 (20.6) | 14 (13.2) | 17 (25.8) | 2 (10.0) | ||||||
| LHCGR | rs4953616 | T/T | 21 (33.9) | 50 (47.2) | 5.00 | 0.082 | 20 (31.7) | 11 (55.0) | 3.59 | 0.166 |
| T/C | 26 (41.9) | 43 (40.6) | 31 (49.2) | 6 (30.0) | ||||||
| C/C | 15 (24.2) | 13 (12.3) | 27 (21.6) | 16 (12.7) | ||||||
| LHCGR | rs7371084 | T/T | 62 (96.9) | 85 (80.2) | 9.50 | 0.002 | 57 (89.1) | 17 (85.0) | 0.24 | 0.695 |
| T/C | 2 (3.1) | 21 (19.8) | 7 (10.9) | 3 (15.0) | ||||||
a2-way ANOVA
bNumber of subjects (frequency)
Correlation studies
We evaluated the association of FSHR rs11692782, and LHCGR rs7371084 and rs4953616 with the phenotypic features of women with PCOS. LHCGR rs7371084 was positively associated with BMI, and negatively associated with BT and FAI, while LHCGR rs4953616 was positively associated with menarche, and negatively with total cholesterol and testosterone, and BT (Table 5). On the other hand, FSHR rs11692782 was not associated with any of the examined features (Table 5).
Table 5.
Correlation between FSHR and LHCGR SNPs and PCOS parameters
| FSHR rs11692782 | LHCGR rs7371084 | LHCGR rs4597581 | ||||
|---|---|---|---|---|---|---|
| r | P | r | P | r | P | |
| BMI | 0.050 | 0.560 | 0.184 | 0.031 | −0.002 | 0.978 |
| WHR | 0.024 | 0.779 | 0.102 | 0.233 | −0.030 | 0.733 |
| Menarche | −0.104 | 0.230 | 0.002 | 0.980 | 0.201 | 0.021 |
| Insulin | −0.128 | 0.204 | −0.050 | 0.628 | 0.065 | 0.526 |
| HOMA-IR | −0.155 | 0.155 | 0.068 | 0.544 | 0.172 | 0.117 |
| HDL | −0.064 | 0.619 | −0.052 | 0.691 | 0.102 | 0.425 |
| LDL | −0.054 | 0.671 | −0.140 | 0.283 | −0.229 | 0.072 |
| Cholesterol | −0.043 | 0.740 | −0.060 | 0.649 | −0.289 | 0.024 |
| Triglycerides | −0.028 | 0.824 | −0.051 | 0.689 | −0.223 | 0.074 |
| LH | 0.086 | 0.411 | −0.010 | 0.923 | −0.053 | 0.620 |
| FSH | −0.155 | 0.143 | 0.183 | 0.088 | −0.024 | 0.827 |
| SHBG | −0.120 | 0.248 | −0.074 | 0.480 | 0.070 | 0.502 |
| Testosterone | 0.097 | 0.287 | −0.012 | 0.894 | −0.217 | 0.018 |
| FT | −0.115 | 0.492 | −0.261 | 0.119 | −0.033 | 0.847 |
| BT | −0.084 | 0.614 | −0.293 | 0.018 | −0.258 | 0.011 |
| FAI | 0.005 | 0.977 | −0.333 | 0.013 | −0.040 | 0.803 |
r Spearman correlation index
Haplotype analysis
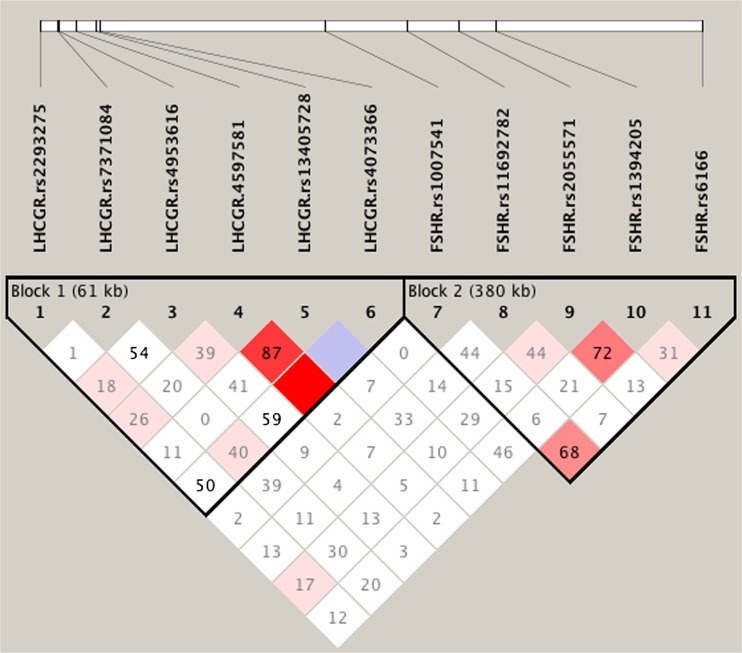
Haploview analysis demonstrated limited linkage disequilibrium (LD) among the LHCGR and FSHR SNPs (Fig. 1). Two blocks were constructed: the first spanning 61 kb (Block 1) contained the six tested LHCGR SNPs, and the second spanning 298 kb (Block 2) contained the five tested FSHR SNPs. Within Block 1, the majority of 6-locus haplotype diversity (frequencies ≥0.04) was captured by 8 of the possible 64 haplotypes. Higher frequency of haplotype GTCAAG was seen in women with PCOS compared to controls (P = 0.026), thus assigning a susceptibility nature to it (Table 6). The frequencies of the remaining LHCGR haplotypes in Block 1, and all FSHR haplotypes contained in Block 2 were similar between cases and controls (Table 6).
Fig. 1.
Linkage disequilibrium (LD) map of the FSHR and LHCGR SNPs genotyped by Haploview. The positions of the SNPs (Build 37.3) are indicated along with the basic gene structure, and displayed above the Haploview output. The relative LD between specific pair of SNPs is indicated by the color scheme, which represents the LD relationships. This is based on D’ values (normalized linkage disequilibrium measure or D) multiplied by 100; D’ is calculated as D divided by the theoretical maximum for the observed allele frequencies. Values approaching zero indicate absence of LD, and those approaching 100 indicate complete LD. The square colored red represent varying degrees of LD < 1 and LOD (logarithm of odds) > 2 scores; darker shades indicating stronger LD
Table 6.
Haplotype frequencies across FSHR and LHCGR SNPs
| Blocka | Haplotypeb | Frequency | Case, control frequencies | χ2 | P c |
|---|---|---|---|---|---|
| 1 (LHCGR) | G T T A A G | 0.297 | 0.285, 0.308 | 0.530 | 0.466 |
| G T C A A G | 0.137 | 0.165, 0.111 | 4.933 | 0.026 | |
| A T T A A G | 0.097 | 0.089, 0.104 | 0.563 | 0.453 | |
| A T C G A G | 0.074 | 0.082, 0.067 | 0.694 | 0.405 | |
| A T C A A G | 0.067 | 0.070, 0.064 | 0.101 | 0.750 | |
| G T C G A G | 0.052 | 0.059, 0.045 | 0.803 | 0.370 | |
| G T T G A G | 0.044 | 0.031, 0.056 | 2.927 | 0.087 | |
| G T T A G G | 0.040 | 0.032, 0.048 | 1.468 | 0.226 | |
| 2 (FSHR) | A G G G | 0.220 | 0.223, 0.217 | 0.044 | 0.835 |
| A G G A | 0.187 | 0.188, 0.185 | 0.013 | 0.910 | |
| T A G G | 0.092 | 0.105, 0.080 | 1.525 | 0.217 | |
| T G G G | 0.084 | 0.086, 0.083 | 0.022 | 0.882 | |
| T A G A | 0.077 | 0.072, 0.081 | 0.202 | 0.653 | |
| T A A A | 0.071 | 0.062, 0.080 | 0.908 | 0.341 | |
| A A A A | 0.060 | 0.058, 0.062 | 0.060 | 0.806 | |
| T A A G | 0.047 | 0.043, 0.052 | 0.346 | 0.556 |
a LHCGR SNP within Block 1 haplotypes were: rs2293275, rs7371084, rs4953616, rs4597581, rs13405728, and rs4073366. FSHR SNP within Block 2 haplotypes were: rs11692782, rs2055571, rs1394205, and rs6166
bUnderlined indicate minor allele
cAdjusted P value, adjusted for BMI, TSH, FT, BT, FAI, SHBG, and fasting insulin
Discussion
We analyzed the association of PCOS with 11 SNPs from LHCGR and FSHR previously tested for their association with PCOS in European [18–20, 25], and non-European [17, 26–29] populations. This case–control sample included 203 women with PCOS and 211 age- and ethnically-matched (Bahraini Arab) control women. PCOS was diagnosed according to the 2003 Rotterdam criteria [21], and confirmed by clinical, biochemical, and radiological assessment.
Recent GWAS studies on Han Chinese [6, 28] and women of European ancestry [10] identified FSHR as PCOS susceptibility locus. FSHR gene variants were linked with and PCOS according to some [27, 28], but not all [19, 25, 30], studies. This was largely due to the ethnic/racial background of studied participant, supported by the association of rs2268361 with PCOS in Chinese [28], but not Dutch [19] PCOS cases, and by the association of rs6166 with PCOS in Chinese [27], but not Dutch [19], UK [31], or Turkish [30] women. Of the five tested FSHR SNPs, rs11692782 was negatively associated with PCOS, but at the genotype not at the allele level. While not addressed here, this may be attributed to the limited number of cases and/or control subjects.
Among the LHCGR tested variants, rs7371084 was negatively associated, while rs4953616 was positively associated with PCOS. These remained significant after applying Bonferroni’s correction, indicating that it was not a spurious finding. Previous studies that evaluated the association of LHCGR variants with PCOS focused on rs13405728, but with conflicting outcomes, and a racial influence was evident. In our study, rs13405728 was not associated with PCOS among Bahraini Arab subjects, which was in agreement with recent studies on Dutch [19], and USA women of European ancestry [18, 20], but in sharp contrast to two independent Chinese studies, which confirmed its strong association with PCOS in Chinese [6] and in Han Chinese [17] subjects. In addition to rs13405728, rs2293275 was not associated with PCOS among Bahraini Arab (this study) and Dutch [25] subjects, with comparable case:control MAF recorded for Bahraini Arab (0.34:0.34) and Dutch (0.40:0.42) cases and controls.
While BMI was higher in women with PCOS compared to control women, WHR and hence visceral adiposity was similar between both groups. An influence of obesity on the association of FSHR and LHCGR variants with PCOS was noted, highlighted by the association of LHCGR rs7371084, and to a lesser extent rs4953616, with PCOS in non-obese subjects. This was in contrast to the association of FSHR rs11692782 with PCOS, which was not affected by obesity. The latter was in agreement with Chinese [29] and Greek [32] studies, which documented lack of contribution of obesity to the association of FSHR Ala307Thr (rs6165) and Ser680Asn (rs6166) variants with PCOS. This may suggest no direct contribution of LHCGR variants to PCOS susceptibility, as the presence of obesity-related metabolic genetic and non-genetic factors becomes required for the influence of LHCGR polymorphisms to be more pronounced.
We evaluated the correlation between FSHR rs11692782, and LHCGR rs7371084 and rs4953616 variants and the severity of the phenotypic features of PCOS. The significant associations were seen for both LHCGR polymorphisms, more so than FSHR rs11692782, which was associated with a marginal and not significant reduction in FSH levels (P = 0.276). Since FSH levels in women with PCOS are usually within reference values [33], this questions the contribution of altered FSH sensitivity to PCOS. This was supported by the finding that rs6166 (Ser680Asn), linked with higher basal FSH levels here (P = 0.040) and elsewhere [33, 34] was not associated with PCOS or associated features in Bahraini Arabs. This was in contrast to LHCGR rs7371084 and rs4953616 variants, which apparently modulate the phenotype of women with PCOS predominantly at the level of hyperandrogenism (markedly higher testosterone, BT, and FAI). However, the contribution of these and related variants to the phenotype of PCOS may be marginal, and hence may require the association of other genetic variants for the phenotypic changes to be more pronounced [35].
We analyzed the linkage disequilibrium pattern between FSHR and LHCGR SNPs, since the interaction of variants within a haplotype is more informative than single variants in determining disease susceptibility, including PCOS. Hapolview analysis demonstrated moderate-weak LD among the studied LHCGR and FSHR SNPs, with notable heterogeneity in the haplotypes obtained, evidenced by the concentration of most of the 6-locus LHCGR haplotypes in 8 out of the possible 64 haplotypes, the frequencies of which exceeding the >4 % threshold. Apart from LHCGR GTCAAG haplotype, which was enriched in women with PCOS, thus assigning a susceptibility nature to it, no other LHCGR or FSHR haplotypes (Block 1 and Block 2) were identified. Given the multi-factorial nature of PCOS, it is possible that additional genetic factors, including other LHCGR/FSHR SNPs, copy number variants, altered upstream promoter methylation, and regulatory factors in LHCGR/FSHR and nearby and distant genes, may all influence PCOS risk.
Our study demonstrated the association of FSHR rs11692782, and LHCGR rs7371084 and rs4953616 with PCOS. Our study has several strengths. It was adequately powered, that cases and controls were ethnically matched (only Bahraini Arabs were included), and potential covariates were controlled for in single SNP and haplotype analysis. The present study is limited by the study design (retrospective case–control study), and in the selection criteria for control women. Considering that affecting 4–12 % of otherwise healthy women will develop PCOS [1, 2], it is likely that results obtained may in fact underestimate the real difference between cases and controls. Follow up studies on additional FSHR and LHCGR variants, and populations of related and distant ethnic origin are needed to confirm the association of FSHR and LHCGR variants with increased risk of PCOS.
Acknowledgments
Funding statement
This work was supported by grant from Arabian Gulf University REC funds.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Capsule We investigated the association of genetic variants in FSHR and LHCGR as polycystic ovary syndrome (PCOS) susceptibility loci with PCOS in 203 Bahraini women with PCOS, and 211 age- and ethnically-matched control women. We demonstrate the association of novel LHCGR (rs7371084, rs4953616) and FSHR (rs11692782) variants with PCOS, thereby confirming the racial/ethnic contribution to their association with PCOS.
References
- 1.Azziz R, Woods KS, Reyna R, Key TJ, Knochenhauer ES, Yildiz BO. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab. 2004;89:2745–9. doi: 10.1210/jc.2003-032046. [DOI] [PubMed] [Google Scholar]
- 2.Goodarzi MO, Azziz R. Diagnosis, epidemiology, and genetics of the polycystic ovary syndrome. Best Pract Res Clin Endocrinol Metab. 2006;20:193–205. doi: 10.1016/j.beem.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Sam S, Dunaif A. Polycystic ovary syndrome: syndrome XX? Trends Endocrinol Metab. 2003;14:365–70. doi: 10.1016/j.tem.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Dunaif A. Insulin resistance in women with polycystic ovary syndrome. Fertil Steril. 2006;86(Suppl 1):S13–4. doi: 10.1016/j.fertnstert.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Scicchitano P, Dentamaro I, Carbonara R, Bulzis G, Dachille A, Caputo P, et al. Cardiovascular risk in women with PCOS. Int J Endocrinol Metab. 2012;10:611–8. doi: 10.5812/ijem.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–9. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 7.Dumesic DA, Abbott DH, Padmanabhan V. Polycystic ovary syndrome and its developmental origins. Rev Endocr Metab Disord. 2007;8:127–41. doi: 10.1007/s11154-007-9046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joe-kechebelu NN, Mbamara SU, Ikechebelu JI. Familial trend in polycystic ovarian syndrome: report of two cases. Ann Afr Med. 2013;12:182–4. doi: 10.4103/1596-3519.117630. [DOI] [PubMed] [Google Scholar]
- 9.Du J, Zhang W, Guo L, Zhang Z, Shi H, Wang J, et al. Two FSHR variants, haplotypes and meta-analysis in Chinese women with premature ovarian failure and polycystic ovary syndrome. Mol Genet Metab. 2010;100:292–5. doi: 10.1016/j.ymgme.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Mutharasan P, Galdones E, Peñalver Bernabé B, Garcia OA, Jafari N, Shea LD, et al. Evidence for chromosome 2p16.3 polycystic ovary syndrome susceptibility locus in affected women of European ancestry. J Clin Endocrinol Metab. 2013;98:E185–90. doi: 10.1210/jc.2012-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hickey TE, Legro RS, Norman RJ. Epigenetic modification of the X chromosome influences susceptibility to polycystic ovary syndrome. J Clin Endocrinol Metab. 2006;91:2789–91. doi: 10.1210/jc.2006-0069. [DOI] [PubMed] [Google Scholar]
- 12.Shoham Z, Jacobs HS, Insler V. Luteinizing hormone: its role, mechanism of action, and detrimental effects when hypersecreted during the follicular phase. Fertil Steril. 1993;59:1153–61. doi: 10.1016/s0015-0282(16)55968-8. [DOI] [PubMed] [Google Scholar]
- 13.Dufau ML. The luteinizing hormone receptor. Annu Rev Physiol. 1998;60:461–96. doi: 10.1146/annurev.physiol.60.1.461. [DOI] [PubMed] [Google Scholar]
- 14.Rousseau-Merck MF, Atger M, Loosfelt H, Milgrom E, Berger R. The chromosomal localization of the human follicle-stimulating hormone receptor gene (FSHR) on 2p21-p16 is similar to that of the luteinizing hormone receptor gene. Genomics. 1993;15:222–4. doi: 10.1006/geno.1993.1041. [DOI] [PubMed] [Google Scholar]
- 15.Nawrocka J, Starczewski A. Effects of metformin treatment in women with polycystic ovary syndrome depends on insulin resistance. Gynecol Endocrinol. 2007;23:231–7. doi: 10.1080/09513590701260193. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, Sallinen K, Zhou S, Su Y, Pöllänen P, Erkkola R. Androgen excess contributes to altered growth hormone/insulin-like growth factor-1 axis in nonobese women with polycystic ovary syndrome. Fertil Steril. 2000;73:730–4. doi: 10.1016/S0015-0282(99)00634-2. [DOI] [PubMed] [Google Scholar]
- 17.Cui L, Zhao H, Zhang B, Qu Z, Liu J, Liang X, et al. Genotype-phenotype correlations of PCOS susceptibility SNPs identified by GWAS in a large cohort of Han Chinese women. Hum Reprod. 2013;28:538–44. doi: 10.1093/humrep/des424. [DOI] [PubMed] [Google Scholar]
- 18.Jones MR, Chua AK, Mengesha EA, Taylor KD, Chen YD, Li X, et al. Metabolic and cardiovascular genes in polycystic ovary syndrome: a candidate-wide association study (CWAS) Steroids. 2012;77(4):317–22. doi: 10.1016/j.steroids.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louwers YV, Stolk L, Uitterlinden AG, Laven JS. Cross-ethnic meta-analysis of genetic variants for polycystic ovary syndrome. J Clin Endocrinol Metab. 2013;98:E2006–12. doi: 10.1210/jc.2013-2495. [DOI] [PubMed] [Google Scholar]
- 20.Welt CK, Styrkarsdottir U, Ehrmann DA, Thorleifsson G, Arason G, Gudmundsson JA, et al. Variants in DENND1A are associated with polycystic ovary syndrome in women of European ancestry. J Clin Endocrinol Metab. 2012;97:E1342–7. doi: 10.1210/jc.2011-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rotterdam ESHRE/ASRM-Sponsored PCOS consensus workshop group Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS) Hum Reprod. 2004;19:41–7. doi: 10.1093/humrep/deh098. [DOI] [PubMed] [Google Scholar]
- 22.Vermeulen A, Verdonck L, Kaufman JM. A critical evaluation of simple methods for the estimation of free testosterone in serum. J Clin Endocrinol Metab. 1999;84:3666–72. doi: 10.1210/jcem.84.10.6079. [DOI] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 25.Valkenburg O, Uitterlinden AG, Piersma D, Hofman A, Themmen AP, de Jong FH, et al. Genetic polymorphisms of GnRH and gonadotrophic hormone receptors affect the phenotype of polycystic ovary syndrome. Hum Reprod. 2009;24:2014–22. doi: 10.1093/humrep/dep113. [DOI] [PubMed] [Google Scholar]
- 26.Fu L, Zhang Z, Zhang A, Xu J, Huang X, Zheng Q, et al. Association study between FSHR Ala307Thr and Ser680Asn variants and polycystic ovary syndrome (PCOS) in Northern Chinese Han women. J Assist Reprod Genet. 2013;30:717–21. doi: 10.1007/s10815-013-9979-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu BH, Park JM, Baek KH. Genetic variations of follicle stimulating hormone receptor are associated with polycystic ovary syndrome. Int J Mol Med. 2010;26:107–12. doi: 10.3892/ijmm_00000441. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Zhao H, Shi Y, Cao Y, Yang D, Li Z, et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. 2012;44:1020–5. doi: 10.1038/ng.2384. [DOI] [PubMed] [Google Scholar]
- 29.Wu XQ, Xu SM, Liu JF, Bi XY, Wu YX, Liu J. Association between FSHR polymorphisms and polycystic ovary syndrome among Chinese women in north China. J Assist Reprod Genet. 2014;31:371–7. doi: 10.1007/s10815-013-0166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unsal T, Konac E, Yesilkaya E, Yilmaz A, Bideci A, Ilke Onen H, et al. Genetic polymorphisms of FSHR, CYP17, CYP1A1, CAPN10, INSR, SERPINE1 genes in adolescent girls with polycystic ovary syndrome. J Assist Reprod Genet. 2009;26:205–16. doi: 10.1007/s10815-009-9308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohiyiddeen L, Salim S, Mulugeta B, McBurney H, Newman WG, Pemberton P, et al. PCOS and peripheral AMH levels in relation to FSH receptor gene single nucleotide polymorphisms. Gynecol Endocrinol. 2012;28:375–7. doi: 10.3109/09513590.2011.633649. [DOI] [PubMed] [Google Scholar]
- 32.Anagnostou E, Drakakis P, Marinopoulos S, Mavrogianni D, Loutradis D. The impact of genetics profile (gene polymorphisms) in obese non-PCOS women entering an IVF/ICSI program. Curr Drug Targets. 2013;14:850–5. doi: 10.2174/1389450111314080004. [DOI] [PubMed] [Google Scholar]
- 33.Laven JS, Imani B, Eijkemans MJ, Fauser BC. New approach to polycystic ovary syndrome and other forms of anovulatory infertility. Obstet Gynecol Surv. 2002;57:755–67. doi: 10.1097/00006254-200211000-00022. [DOI] [PubMed] [Google Scholar]
- 34.Simoni M, Nieschlag E, Gromoll J. Isoforms and single nucleotide polymorphisms of the FSH receptor gene: implications for human reproduction. Hum Reprod Update. 2002;8:413–21. doi: 10.1093/humupd/8.5.413. [DOI] [PubMed] [Google Scholar]
- 35.Simoni M, Tempfer CB, Destenaves B, Fauser BC. Functional genetic polymorphisms and female reproductive disorders: part I: polycystic ovary syndrome and ovarian response. Hum Reprod Update. 2008;14:459–84. doi: 10.1093/humupd/dmn024. [DOI] [PMC free article] [PubMed] [Google Scholar]