Abstract
Mammalian skin is innervated by diverse, unmyelinated C fibers that are associated with senses of pain, itch, temperature, or touch. A key developmental question is how this neuronal cell diversity is generated during development. We reported previously that the runt domain transcription factor Runx1 is required to coordinate the development of these unmyelinated cutaneous sensory neurons, including VGLUT3+ low-threshold c-mechanoreceptors (CLTMs), MrgprD+ polymodal nociceptors, MrgprA3+ pruriceptors, MrgprB4+ c-mechanoreceptors, and others. However, how these Runx1-dependent cutaneous sensory neurons are further segregated is poorly illustrated. Here, we find that the Runx1-dependent transcription factor gene Zfp521 is expressed in, and required for establishing molecular features that define, VGLUT3+ CLTMs. Furthermore, Runx1 and Zfp521 form a classic incoherent feedforward loop (I-FFL) in controlling molecular identities that normally belong to MrgprD+ neurons, with Runx1 and Zfp51 playing activator and repressor roles, respectively (in genetic terms). A knock-out of Zfp521 allows prospective VGLUT3 lineage neurons to acquire MrgprD+ neuron identities. Furthermore, Runx1 might form other I-FFLs to regulate the expression of MrgprA3 and MrgprB4, a mechanism preventing these genes from being expressed in Runx1-persistent VGLUT3+ and MrgprD+ neurons. The evolvement of these I-FFLs provides an explanation for how modality-selective sensory subtypes are formed during development and may also have intriguing implications for sensory neuron evolution and sensory coding.
Keywords: low-threshold c-mechanoreceptors, nociceptors, pruriceptors, Runx1, sensory subtype specification, Zfp521
Introduction
The dorsal root ganglia (DRGs) contain neurons transmitting sensory information related to pain, itch, temperature, and touch and their individual subtypes can be distinguished by differential expression of molecular markers. Expression of MrgprD, a G-protein-coupled receptor, marks neurons required to sense light punctate mechanical pain and chemical itch evoked by β-alanine (Zylka et al., 2005; Cavanaugh et al., 2009; Rau et al., 2009; Liu et al., 2012). MrgprB4+ c-mechanoreceptors detect massage-like stroking (Vrontou et al., 2013). MrgprA3+ nociceptors are dedicated to itch (Liu et al., 2009; Han et al., 2013). The fourth group is the low-threshold c-mechanoreceptors (CLTMs) that are marked by the coexpression of vesicular glutamate transporter 3 (VGLUT3), tyrosine hydroxylase (TH), and the cytokine TAFA4 (Seal et al., 2009; Li et al., 2011; Delfini et al., 2013; Lou et al., 2013). Finally, neurons expressing the transient receptor potential channels, such as TRPM8 and TRPV1, respond to cold and heat stimuli, respectively (Julius, 2013). A challenge in developmental biology is understanding how such cell diversity is generated during development.
The past decades have seen progress in characterizing genetic programs controlling sensory neuron development. C-fiber neurons associated with pain, itch, and thermal sensations are formed during a late wave of neurogenesis (Ma et al., 1999; Bachy et al., 2011; Liu and Ma, 2011; Lallemend and Ernfors, 2012). These neurons are initially marked by the expression of the nerve growth factor receptor TrkA and the runt domain transcription factor Runx1 (Liu and Ma, 2011; Lallemend and Ernfors, 2012). During perinatal and postnatal development, Runx1 expression is switched off in ∼50% of these neurons. Functionally, Runx1 is required for the development of a cohort of sensory neurons that preferentially innervate the skin epidermis and hair follicles (Chen et al., 2006; Yoshikawa et al., 2007; Yang et al., 2013; Moqrich, 2014), as summarized in Figure 1A.
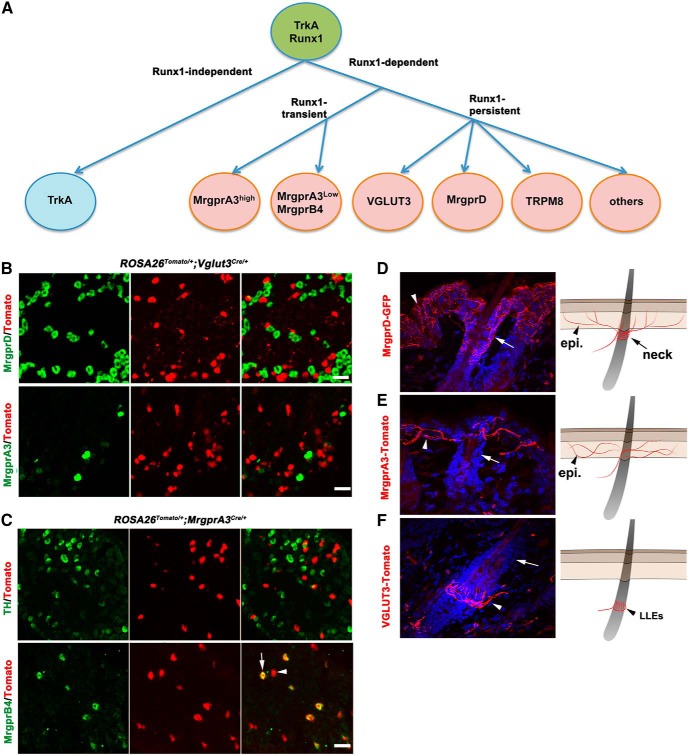
Figure 1.
Segregation of Runx1-dependent cutaneous sensory neurons. A, Schematics showing the segregation of TrkA lineage neurons. B, Transverse sections through P30 lumbar DRG of ROSA26Tomato/+;Vglut3Cre/+ mice. No MrgprD or MrgprA3 mRNA was detected by ISH in VGLUT3-Tomato+ neurons. C, Transverse sections through P30 lumbar DRG of ROSA26Tomato/+;MrgprA3Cre/+ mice. Arrow indicates MrgprB4 mRNA detected in a subset of Tomato+ MrgprA3 lineage neurons. Arrowhead indicates Tomato+ neurons lacking MrgprB4. D–F, Transverse sections through the hairy skin of P60 MrgprD-GFP mice (D), ROSA26Tomato/+;MrgprA3Cre/+ mice (E), or ROSA26Tomato/+;Vglut3Cre/+ mice (F). Arrows indicate the neck region of a hair follicle innervated by MrgprD-GFP+ fibers (D), but not by MrgprA3-Tomato+ fibers (E) or VGLUT3-Tomato+ fibers (F). Arrowheads in D and E indicate epidermal (“epi.”) innervations; arrowhead in F indicates LLEs. The schematics in D–F summarize the innervation patterns. Scale bars, 50 μm.
However, it remains poorly understood how distinct Runx1-dependent cutaneous sensory neuron subtypes are segregated. Several transcription factors, including Brn3a, Islet1, Tlx3, and ZBTB20, also play a role in controlling sensory channels and receptors (Sun et al., 2008; Dykes et al., 2010; Lopes et al., 2012; Zou et al., 2012; Ren et al., 2014); however, all of them are expressed in most, if not all, DRG neurons, so this does not explain how modality-selective sensory subtypes develop.
Here, we report our findings that a Runx1-dependent transcription factor, Zfp521, is expressed in one type of Runx1-dependent neurons: VGLUT3+ CLTMs. The Zfp521 gene encodes a protein composed of 1311 amino acids and 30 Kruppel-like zinc fingers, which has been implicated in controlling bone, adipocyte, and neural development (Hesse et al., 2010; Kamiya et al., 2011; Kang et al., 2012). We demonstrate that Runx1 and Zfp521 form incoherent feedforward loops (I-FFLs) to control the segregation of CLTMs versus MrgprD+ nociceptors. In addition, Runx1 might form other I-FFLs to control the segregation of CLTMs from MrgprA3+ pruriceptors. Our studies therefore provide insight into the mechanisms that govern the emergence of sensory neurons processing distinct modalities.
Materials and Methods
Animals.
The generation of Runx1 floxed mice (Growney et al., 2005), ROSA26-CAG-LSTOPL-tdTomato reporter mice (Madisen et al., 2010), MrgprD-GFP mice (Zylka et al., 2005), MrgprD-Cre mice (Rau et al., 2009), and MrgprA3-Cre mice (Han et al., 2013) has been described previously. The generation of Vglut3-Cre mice was described previously by the Bradford B. Lowell group. The generation of Zfp521 floxed mice will be described in the next section. Genotyping was done by PCR. The following PCR primers were used for the Runx1 mutant and wild-type allele: 5′-GAG TCC CAG CTG TCA ATT CC-3′ and 5′-GGT GAT GGT CAG AGT GAA GC-3′, with the floxed allele producing a larger size of DNA band; for the Vglut3-Cre allele, 5′-TAT CTC ACG TAC TGA CGG TG-3′ and 5′-AGA CTA ATC GCC ATC TTC CAG C-3′; for the ROSA26-CAG-LSTOPL-tdTomato allele, 5′-GGC ATT AAA GCA GCG TAT CC-3′ and 5′-CTG TTC CTG TAC GGC ATG G-3′. Both females and males were used for the studies and all animal procedures were conducted with protocols approved by the animal care and use committees at Dana-Farber Cancer Institute and Hangzhou Normal University.
Gene targeting in embryonic stem cells and generation of Zfp521 floxed mice.
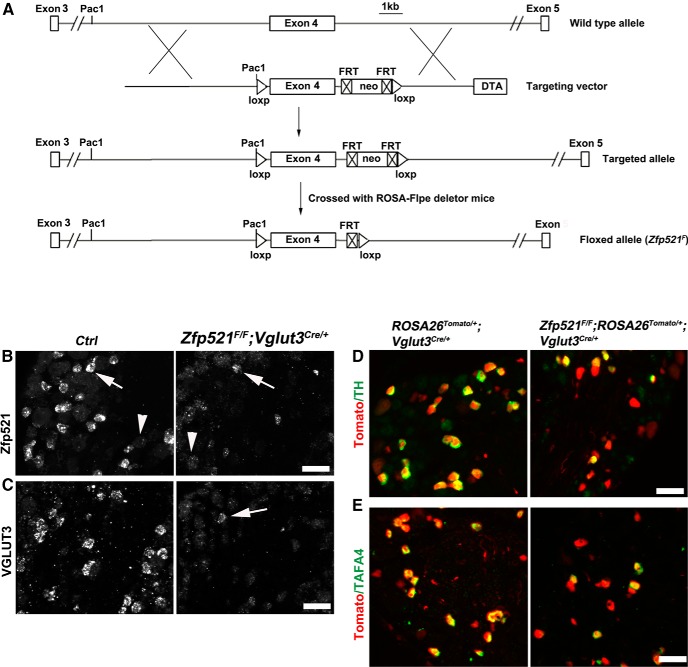
To generate Zfp521 conditional knock-out mice, the 12 kb genomic DNA fragment containing Zfp521 exon 4 was subcloned from a BAC clone using the recombination system developed by Liu et al. (2003). A Neo cassette flanked with LoxP sites was targeted upstream of exon 4 and subsequent excision of the Neo cassette by Cre-mediated recombination leaves behind a single loxP plus a Pac1 restriction enzyme site upstream of exon 4. A new Neo cassette flanked with FRT sites and a downstream LoxP site was then targeted to the region 3′ to the exon 4. A diphtheria toxin (DTA) cassette located at the 3′ end of the recombination arm was used to drive the expression of the DTA for negative selection. Upon linearization, this targeting vector was electroporated into the J1 embryonic stem (ES) cell line (derived from 129Sv/J mouse strain), with G418 included in the culture medium for positive selection. Long-range PCR amplification was performed to screen for targeted ES clones and the targeted ES cells were injected into the blastocysts derived from C57BL/6J females. Chimeric male mice were mated with C57BL/6J females to generate heterozygous mice carrying the targeted allele. These mice were mated with the Flpe deleter mouse line (Rodríguez et al., 2000) to remove the neo cassette, leading to the creation of the mice carrying the Zfp521 floxed allele, in which the exon 4 is flanked with two Loxp sequences (see Fig. 3A). The genotyping for the conditional knock-out mice was performed with the following set of primers: for Zfp521 wild-type and floxed allele, 5′-AGTGAGCATCGCAGATCTGA-3′ and 5′-TGAGGAAAACCTGGTTGTCT 3′, with the floxed allele showing a larger DNA band after gel electrophoresis.
Figure 3.
Generation of Zfp521 CKO mice and impaired development of VGLUT3+ CLTMs. A, Schematics showing the creation of mice carrying the Zfp521 conditional null allele (“floxed allele” or Zfp521F). The “DTA” negative selection cassette drives the expression of the diphtheria toxin gene. The “neo” positive selection cassette was flanked with FRT sites and was removed by crossing with ROSA26-Flpe mice. Exon 4 was flanked with two Loxp sites for conditional knockout after crossing with Cre mouse lines. B, C, ISH with indicated probes on sections through P30 lumbar DRG of wild-type control (“Ctrl”) mice and Zfp521F/F;Vglut3Cre/+ CKO mice. Arrows and arrowheads in B indicate Zfp521high and Zfp521low expression, respectively. Note loss of most Zfp521high neurons in CKO mice. D, E, Double detection of Tomato plus TH or TAFA4 mRNA on sections through P30 lumbar DRG of control littermate mice (ROSA26Tomato/+;Vglut3Cre/+) and Zfp521 CKO mice (Zfp521F/F;ROSA26Tomato/+;Vglut3Cre/+). Note a partial loss of TH and TAFA4 in mutants. Scale bars, 50 μm.
In situ hybridization and immunohistochemistry.
Antisense Zfp521 probe (1.001 kb), Runx1 probe (0.858 kb), VGLUT3 probe (0.771 kb), TH probe (0.788 kb), Piezo2 probe (0.977 kb), MrgprA3 probe (1.15kb), MrgprB4 probe (0.888 kb), MrgprD probe (1.058 kb), P2X3 probe (1.1 kb), and TAFA4 probe (0.75 kb) were amplified from cDNA prepared from adult DRG and labeled with digoxigenin (Roche). The digoxigenin-labeled mGluR5 and SCG10 in situ probes were prepared by in vitro transcription using plasmid DNAs as the templates. For double-color in situ hybridization (ISH), two antisense probes were labeled with digoxigenin and with fluorescein (Roche), respectively. After hybridization, the sections were incubated with peroxidase-conjugated anti-fluorescein antibody (Roche), followed by biotin tyramide (PerkinElmer) treatment and incubation with rhodamine red-conjugated strepavidin (Life Technologies). After taking fluorescent images, alkaline phosphatase (AP)-conjugated anti-DIG antibody (Roche) was applied to the sections overnight and in situ signals were developed with AP reaction. After AP development, bright-field images were taken, converted to pseudofluorescence images, and merged with rhodamine red fluorescent images. ISH/immunohistochemistry and ISH/Tomato double staining was performed as described previously (Liu et al., 2008; Liu et al., 2010; Lou et al., 2013). Rabbit anti-Runx1 antibody (1:100; Abcam) was used for Figure 5.
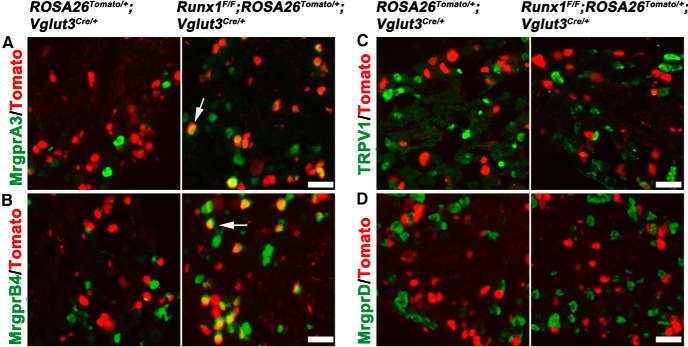
Figure 5.
Prospective VGLUT3 lineage neurons acquire identities of MrgrpA3/B4+ neurons in Runx1 late CKO mice. A–D, Double staining of Tomato with indicated mRNAs (detected by ISH) on sections through P30 lumbar DRG of control littermate mice (ROSA26Tomato/+;Vglut3Cre/+) and Runx1 CKO mice (Runx1F/F;ROSA26Tomato/+;Vglut3Cre/+). Arrows indicate a derepression of gene expression in CKO mice (A, B). C and D indicate no derepression of these two markers. Scale bars, 50 μm.
Hairy skin innervation analyses.
Back skin (1.5 × 1.5 cm2) from 3 ROSA26Tomato/+;Vglut3Cre/+ mice, Zfp521F/F;ROSA26Tomato/+;Vglut3Cre/+mice, Runx1F/F;ROSA26Tomato/+;Vglut3Cre/+mice, ROSA26Tomato/+;MrgprA3Cre/+ mice, and MrgprD-GFP mice were collected. Back skins were fixed in 4% PFA, cryoprotected in 20% sucrose, embedded in OCT, and cut into 30-μm-thick sections. All sections were counterstained with DAPI (Roche, 1 mg/ml) to identify the keratinocyte layers in the epidermis. Tomato+ nerve fibers were directly visualized under a fluorescent microscope, whereas MrgprD-GFP+ fibers were visualized by rabbit anti-GFP immunostaining (1:1000; Invitrogen), as described previously (Yang et al., 2013). To quantify the hair neck region innervation by VGLUT3-Tomato+ fibers and MrgprA3-Tomato+ fibers, skin sections were first screened under the DAPI channel and only “intact” hairs that started with hair follicle and went all the way through the epidermis were selected for subsequent innervation analyses. We used this unbiased method to collect the images of intact hairs from >200 visual fields under a 20× Zeiss fluorescence microscope. The numbers of intact hairs in the visual field and the numbers of hair neck regions that were innervated by Tomato+ or GFP+ fibers were count. At the same time, free nerve endings in the skin epidermis surrounding these intact hairs and the longitudinal lanceolate endings and circumferential endings wrapped around the hair follicles were examined. The experimenters were blinded to genotype on all of these quantitative analyses.
Cell quantification.
Lumbar DRGs (L1–L3) from 2–3 mutant and control mice were dissected bilaterally, fixed, and embedded, and sectioned into sections of 14 μm thickness. For each probe, three sets of control and mutant DRG sections were processed for immunostaining or used for ISH with the marker of interest and positive cells with nuclei were counted.
Statistics.
Results are expressed as mean ± SEM. For quantification of histochemistry analysis, data were subjected to the χ2 test. p < 0.05 was accepted as statistically significant.
Results
Segregation of Runx1-dependent cutaneous sensory neuron subtypes
In this study, we studied four subtypes of Runx1-dependent cutaneous sensory neurons: MrgprD+ polymodal nociceptors, MrgprA3+ pruriceptors, MrgprB4+ c-mechanoreceptors, and VGLUT3+ CLTMs (Fig. 1A). To assess the lineage relationship among these neurons, we performed fate-mapping experiments. We first crossed VGLUT3-Cre and MrgprA3-Cre mice each with ROSA26-LSL-tdTomato reporter mice (Madisen et al., 2010; Han et al., 2013), with the resulting double heterozygous mice referred to as ROSA26Tomato/+;VGLUT3Cre/+ and ROSATomato/+;MrgprA3Cre/+, respectively. In these mice, neurons with a developmental history of VGLUT3 or MrgprA3 expression are permanently marked by the expression of the red fluorescent tdTomato protein, so these neurons are referred to as VGLUT3-Tomato+ and MrgprA3-Tomato+ neurons, respectively. In situ hybridization (ISH) on sections from lumbar DRGs showed that VGLUT3-Tomato+ neurons virtually do not express MrgprD, MrgprA3, or MrgprB4 (Fig. 1B and data not shown). MrgprA3-Tomato+ neurons do not express MrgprD (data not shown) or TH (Fig. 1C), a marker for VGLUT3+ CLTMs (Li et al., 2011; Lou et al., 2013), but some of them do express MrgprB4 (Fig. 1C). The association of MrgprB4 with MrgprA3 lineage neurons is consistent with a previous report showing that MrgprA3+ neurons are composed of two subsets distinguished by high or low MrgprA3 expression (MrgprA3high or MrgprA3low), with MrgprB4 expression being confined to MrgprA3low neurons (Liu et al., 2008). Therefore, the expression of MrgprD, VGLUT3, and MrgprA3 marks three nonoverlapped groups of DRG neurons, with MrgprA3+ neurons being additionally subdivided into MrgprA3high neurons that likely represent pruriceptors (Han et al., 2013) and MrgprA3low/MrgprB4+ c-mechanoreceptors that are proposed to be associated with pleasant touch evoked by massage-like mechanical stimuli (Vrontou et al., 2013).
MrgprD+, MrgprA3+, and VGLUT3+ neurons all innervate the hairy skin (Zylka et al., 2005; Liu et al., 2007; Li et al., 2011; Han et al., 2013; Lou et al., 2013), but details of their innervation have not yet been fully characterized. To do this, we first examined MrgprD-GFP reporter mice in which MrgprD+ neurons are marked by the expression of green fluorescent protein (GFP) (Zylka et al., 2005). Immunofluorescent staining against GFP showed that MrgprD-GFP+ fibers densely innervate the skin epidermis and wrap around the “neck” of hair follicles (Fig. 1D, arrow). MrgprA3-Tomato+ neurons from ROSA26Tomato/+;MrgprA3Cre/+ mice, the terminals of which can be directly visualized under a fluorescent microscope, also innervate the epidermis throughout the hairy skin, but, unlike MrgprD-GFP+ fibers, they rarely innervate the neck of hair follicles (Fig. 1E). VGLUT3-Tomato+ fibers form longitudinal lanceolate endings (LLEs) around the middle portion of most hair follicles, an area ventral to the neck region (Fig. 1F). VGLUT3-Tomato+ fibers also innervate the epidermis, but, unlike the broad distribution of MrgprD-GFP+ and MrgprA3-Tomato+ fibers, VGLUT3-Tomato+ epidermal fibers are restricted to the area surrounding a small subset of hairs (see below). In summary, three molecularly distinct Runx1-dependent cutaneous sensory neurons, MrgprD+, MrgprA3+, and VGLUT3+, display distinct innervation patterns in the hairy skin (see schematics in Fig. 1D–F).
Runx1-dependent Zfp521 marks VGLUT3+ CLTMs
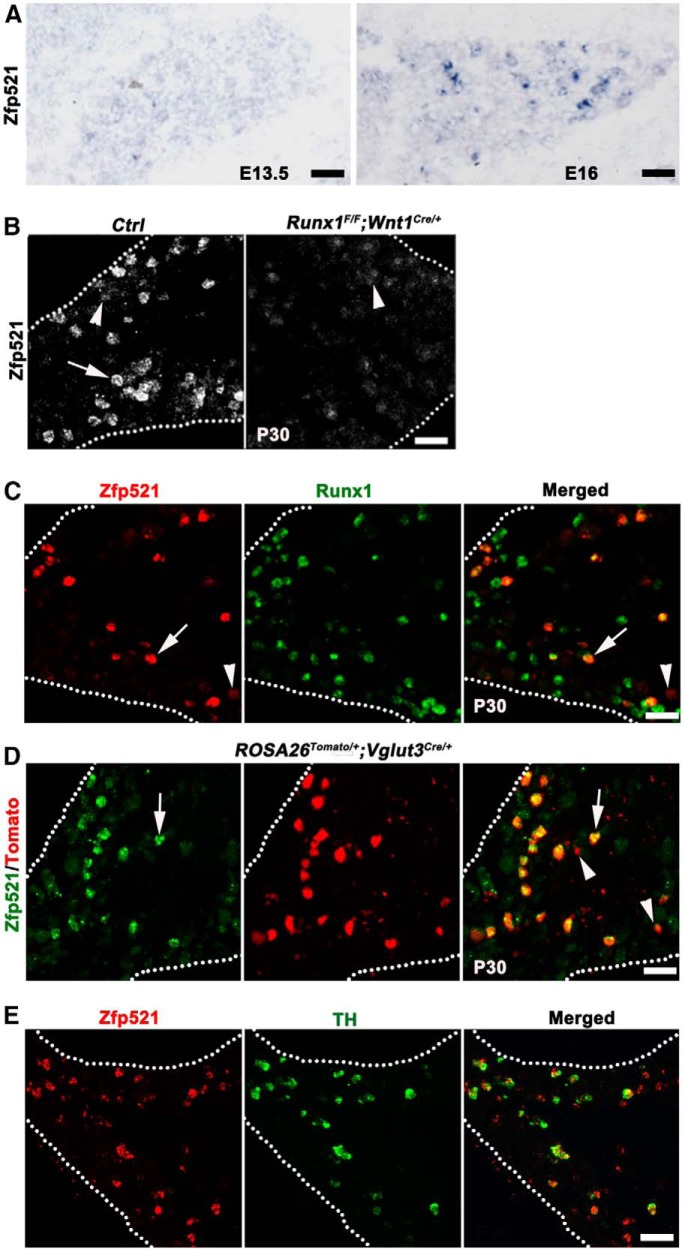
To understand how Runx1-dependent sensory subtypes are specified, we screened the Allen Brain Atlas and found that the zinc finger gene Zfp521 is expressed in a subset of DRG neurons, as well as in the CNS, such as the dorsal spinal cord (http://mousespinal.brain-map.org/imageseries/show.html?id = 100012182). In developing DRGs, ISH showed that high-level Zfp521 expression (Zfp521high) was established at embryonic stages between embryonic day 13.5 (E13.5) and E16 (Fig. 2A). In postnatal day (P30) DRGs, Zfp521high expression is confined in a subset of neurons (Fig. 2B). We next found that the number of Zfp521high neurons in P30 lumbar DRG was greatly reduced in Runx1 conditional knock-out mice (Runx1F/F;Wnt1Cre/+; Fig. 2B), in which Runx1 was removed in the neural crest cells, including sensory precursors (Chen et al., 2006). Double-color ISH of Runx1 and Zfp521 showed that virtually all Zfp521high neurons (283/293 = 96.6%) coexpress Runx1 and 1/3 of Runx1-expressing neurons coexpress Zfp521high (283/821 = 34.5%) in P30 L1–L3 DRGs (Fig. 2C, arrows). In contrast, neurons with low levels of Zfp521 expression do not express Runx1 (Fig. 2C, arrowhead) and, consistently, such low-level expression was still detected in Runx1F/F;Wnt1Cre/+ mice (Fig. 2B, arrowhead). Therefore, Zfp521high expression marks a subset of Runx1-persistent neurons in adult lumbar DRGs.
Figure 2.
Runx1-dependent Zfp521 marks mainly TH+ VGLUT3 lineage neurons. A, B, Zfp521 mRNA detected by ISH on transverse sections through E13.5 or E16 lumbar DRGs of wild-type embryos (A), P30 lumbar DRGs of wild-type control mice (B, “Ctrl”), and Runx1F/F;Wnt1Cre/+ CKO mice (B, right). Arrow and arrowheads in B indicate Zfp521high and Zfp521low expression, respectively. C, Double-color ISH on sections through P30 lumbar DRG of wild-type mice. Arrow indicates neurons coexpressing Zfp521 and Runx1. Arrowhead indicates a Zfp521low neuron not coexpressing Runx1. D, Zfp521 ISH on a section through P30 lumbar DRG of ROSA26Tomato/+;Vglut3Cre/+ mice. Arrows indicate coexpression of Zfp521 mRNA in VGLUT3-Tomato+ neurons. Arrowheads indicate Tomato+ neurons with smallest cell diameters and expressing little or none of Zfp521. E, Double-color ISH on sections through P30 lumbar DRG of wild-type mice indicating coexpression of Zfp521 and TH. Scale bars, 50 μm.
As described in Figure 1A, Runx1-dependent neurons include a cohort of cutaneous sensory neuron subtypes. To determine which subtypes of sensory neurons express Zfp521, we performed a series of double staining. We first found that Zfp521 is not expressed in classic nonpeptidergic neurons labeled by the isolectin B4 (IB4) or in classic peptidergic neurons marked by expression of the calcitonin gene-related peptide (data not shown). This expression pattern is reminiscent of that of VGLUT3+ CLTMs (Seal et al., 2009; Li et al., 2011; Delfini et al., 2013; Lou et al., 2013). To test this directly, we performed Zfp521 ISH on lumbar DRG sections from P30 ROSA26Tomato/+;VGLUT3Cre/+ reporter mice, in which VGLUT3 lineage neurons are marked by Tomato expression. We found that 79.5% (265/333) of Zfp521high neurons are Tomato+ and 73.0% (1105/1512) of VGLUT3-Tomato+ neurons express Zfp521 (Fig. 2D, arrows). In thoracic DRGs, Zfp521high was detected in ∼61% of VGLUT3-Tomato+ neurons (data not shown). Note that those Tomato+ neurons with the smallest cell diameters either do not express or express low levels of Zfp521 (Fig. 2D, arrowheads). We reported previously that VGLUT3 lineage DRG neurons are heterogeneous, with 85% and 15% of them expressing VGLUT3 persistently and transiently, respectively (Lou et al., 2013). The VGLUT3-persistent portion represents CLTMs that express Runx1 persistently, 2/3 of which can be marked by the expression of TH (Lou et al., 2013). We found here that >80% (154/192) of Zfp521high neurons coexpress TH (Fig. 2E). It should be noted that there is an early wave of TH expression in embryonic DRG, the expression of which is downregulated during postnatal development (Lagerström et al., 2010). We found that these early TH+ neurons do not express Zfp521 (data not shown). Therefore, Zfp521high expression selectively marks the TH+ subset of VGLUT3+ CLTMs that emerge mainly during postnatal development and their lineage relationship with embryonic TH+ neurons remains unclear. The association of Zfp521 with VGLUT3+ CLTMs is supported by recent gene expression profiling analyses (Usoskin et al., 2014).
Requirement of Zfp521 for VGLUT3+ CLTM development
To determine the role of Zfp521 in controlling the specification of these neurons, we generated a mouse line containing a conditional null allele of Zfp521, which is referred to as Zfp521F/F (Fig. 3A). In this allele, the 3.3 kb exon 4 is flanked by two loxP sequences and can be removed by Cre-mediated DNA recombination. By crossing Zfp521F/+ mice with VGLUT3-Cre mice and after further crossing, we created Zfp521 conditional knock-out (CKO) mice in which Zfp521 is selectively removed in VGLUT3 lineage neurons. These CKO mice are referred to as Zfp521F/F;Vglut3Cre/+. To label the VGLUT3 lineage neurons with the Tomato reporter, we further generated CKO mice that carry the ROSA26-LSL-tdTomato reporter allele, referred to as Zfp521F/F;ROSA26Tomato/+;Vglut3Cre/+ mice. ISH using the deleted exon 4 as the probe revealed a marked loss of Zfp521high neurons (Fig. 3B). The detection of a few remaining Zfp521high neurons is consistent with the fact that not all Zfp521high neurons belong to the VGLUT3 lineage.
We next examined how the development of CLTMs was affected in the Zfp521 knock-out. ISH revealed marked decrease of VGLUT3 expression in lumbar DRGs of Zfp521F/F;Vglut3Cre/+ CKO mice compared with the wild-type control (Fig. 3C). Only few VGLUT3+ neurons remained in the mutant DRGs (Fig. 3C, arrow), consistent with the fact that Zfp521high is not expressed in all of VGLUT3 lineage neurons (Fig. 2). The loss of VGLUT3 expression was not due to loss of VGLUT3 lineage cells that were marked by Tomato expression because the percentage of Tomato+ neurons over the pan-neuronal marker SCG10 (Stein et al., 1988) (detected by ISH) in L1–L3 DRGs was comparable between Zfp521F/F;ROSA26Tomato/+;Vglut3Cre/+ CKO mice and ROSA26Tomato/+;Vglut3Cre/+ control mice (19.6 ± 1.2% in control, and 19.1 ± 2.4% in CKO mice, p > 0.05). In these CKO mice, Zfp521 knock-out occurs after the onset of VGLUT3-Cre expression. The loss of VGLUT3 in Zfp521F/F;Vglut3Cre/+ CKO mice therefore suggests that Zfp521 is required to maintain VGLUT3 expression. Future knock-out of Zfp521 at early embryonic stages is needed to determine whether the initiation of VGLUT3 expression is dependent on Zfp521.
We reported previously that Runx1 controls other molecular features in VGLUT3+ CLTMs in addition to VGLUT3 itself (Lou et al., 2013). We first examined TH expression and found that the percentage of TH+ neurons in VGLUT3-Tomato+ neurons was significantly reduced in Zfp521 mutant mice (Fig. 3D), from 59.9% (904/1560) in ROSA26Tomato/+;Vglut3Cre/+ control mice to 32.8% (800/2437) in Zfp521F/F;ROSA26Tomato/+;Vglut3Cre/+ CKO mice, or a 47% loss of TH+ neurons (χ2 test, p < 0.001). TH expression was also reduced by ∼38% in thoracic DRG (data not shown). TAFA4, a cytokine involved with pain modulation, is expressed mainly in TH+ CLTMs (Delfini et al., 2013). We found that TAFA4 expression in VGLUT3-Tomato+ neurons is also reduced in Zfp521 CKO mice (Fig. 3E), from 55.1% (826/1499) in ROSA26Tomato/+;Vglut3Cre/+ control mice to 25.1% (231/920) in Zfp521F/F;ROSATomato/+;Vglut3Cre/+ CKO mice, or a 54.4% loss (χ2 test, p < 0.001). The loss of TH and TAFA4 expression in Zfp521 CKO mice is, however, less than that seen in Runx1 CKO mice (Runx1F/F;ROSATomato/+;Vglut3Cre/+), in which only 15.8% (235/1484) and 8.0% (81/1014) of Tomato+ neurons retain TH and TAFA4 expression, respectively (data not shown), compared with the 32.8% and 25.1% retained in Zfp521 CKO mice, respectively (χ2 test, p < 0.001). The difference between Zfp521 versus Runx1 CKO mice is also indicated by the observation that expression of Piezo2 in VGLUT3-Tomato+ neurons, which is dependent on Runx1 (Lou et al., 2013), is largely unaffected in Zfp521F/F;ROSA26Tomato/+;Vglut3Cre/+ CKO mice (data not shown). Furthermore, skin innervation by VGLUT3 lineage neurons, which is impaired in Runx1 CKO mice (see below), is largely unchanged in Zfp521 CKO mice (see Fig. 6C). Therefore, Zfp521 plays an essential role in establishing a portion of Runx1-dependent molecular features in VGLUT3+ CLTMs, including the expression of VGLUT3 and a portion of TH and TAFA4, but Runx1 uses Zfp521-independent pathways to control Piezo2, a separate portion of TH and TAFA4, and skin innervations.
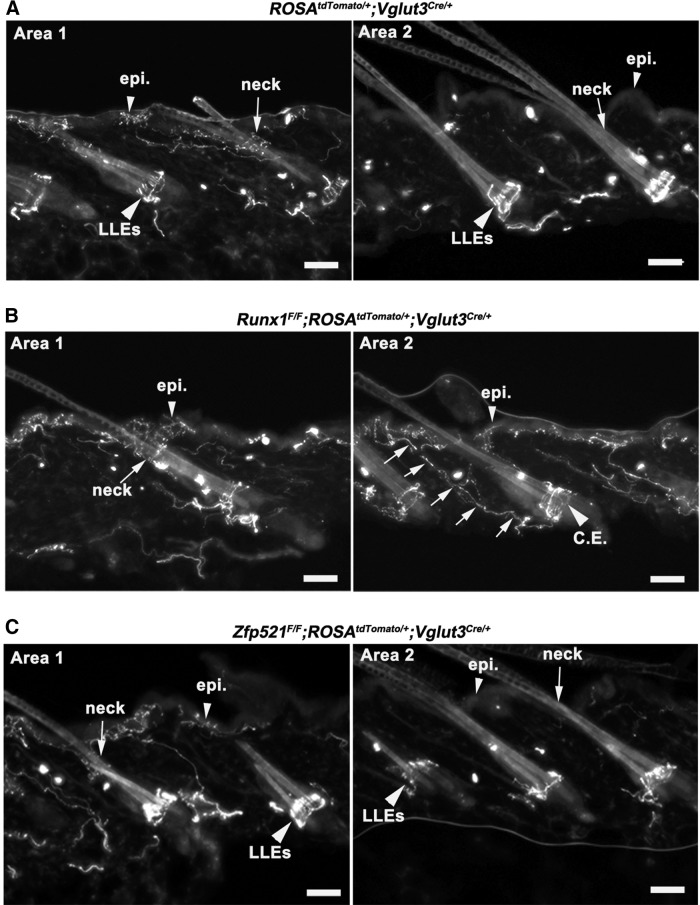
Figure 6.
Skin innervation in control, Runx1 CKO, and Zfp521 CKO mice. A, Hairy skin innervation of VGLUT3 lineage neurons in P60 control mice (ROSA26Tomato/+;Vglut3Cre/+). Left, Innervation of Tomato+ fibers surrounding a cluster of hairs in the skin “Area 1,” with the arrowhead indicating epidermal (“epi.”) innervation and the arrow indicating the neck region of a hair follicle. Right, Skin “Area 2” in which VGLUT3-Tomato+ fibers only form the “LLEs” (large arrowhead) without innervation to the skin epidermis (small arrowhead). B, Hairy skin innervation of VGLUT3 lineage neurons in P60 Runx1 CKO mice (Runx1F/F;ROSA26Tomato/+;Vglut3Cre/+). Arrow in left panel indicates normal innervation to the hair follicle neck region of the skin “Area 1”; arrowhead indicates normal epidermal innervation. For the right panel, large arrowhead indicates Tomato+ fibers being converted from LLEs to circumferential endings (“C.E.”) in skin “Area 2”; arrows indicate fibers growing toward skin surface and terminating the epidermis (small arrowhead). There was no typical innervation to the hair neck (B, right). C, The innervation pattern was not changed in Zfp521F/F;ROSA26Tomato/+;Vglut3Cre/+ CKO mice compared with wild-type controls (A), including: (1) the retaining of “LLEs” (large arrowheads) and (2) restricted innervation to the skin epidermis (small arrowhead, left) and the neck region (arrow, left) only around a small subset of hairs (“area 1”), but not around most hairs (“area 2”). Scale bars, 50 μm.
CLTMs acquired molecular identities of MrgprD+ neurons in Zfp521 CKO mice
Because Zfp521 is a downstream target of Runx1, it is not surprising that Runx1 expression itself is unaffected in Zfp521 CKO mice (Fig. 4A). As illustrated in Figure 1A, Runx1 is required for the development of a large cohort of cutaneous sensory neurons. We next investigated whether, in the absence of Zfp521, Runx1 could drive prospective CLTMs to differentiate into other types of Runx1-dependent neurons. To test this, we looked at the expression of markers of other cutaneous sensory neuron subtypes in VGLUT3-Tomato+ neurons in control and Zfp521 CKO mice.
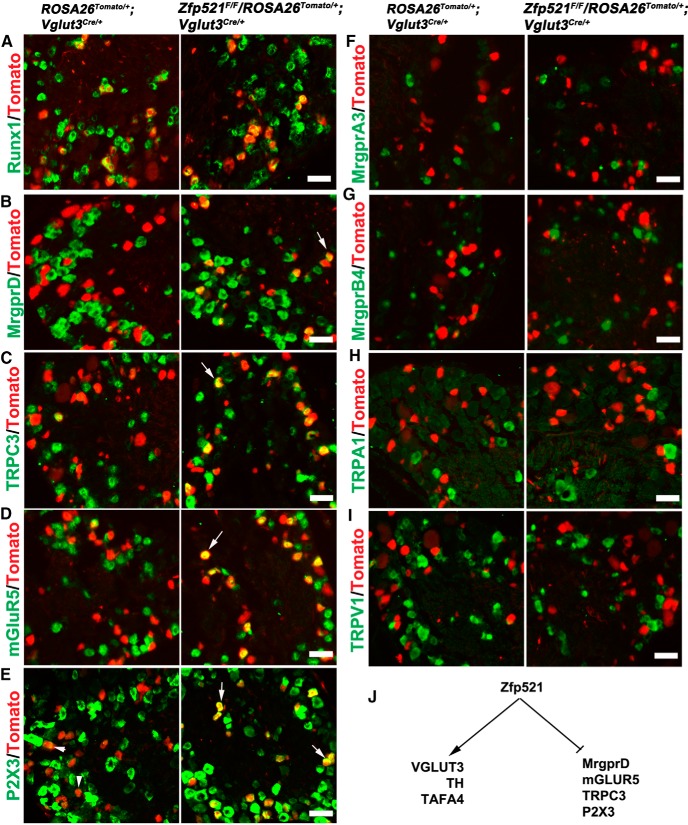
Figure 4.
Prospective VGLUT3 lineage neurons acquire molecular features of MrgprD+ nociceptors in Zfp521 CKO mice. A–I, Double staining of Tomato with indicated mRNAs detected by ISH on sections through P30 lumbar DRG of control littermate mice (ROSA26Tomato/+;Vglut3Cre/+) and Zfp521 CKO mice (Zfp521F/F;ROSA26Tomato/+;Vglut3Cre/+). Arrows indicate derepression of gene expression in CKO mice (B–E). Arrowheads in E indicate low P2X3 expression in VGLUT3-Tomato+ neurons in control mice. F–I indicate no derepression of these markers. J, Summary of dual functions of Zfp521. Scale bars, 50 μm.
Like VGLUT3+ CLTMs, a majority of MrgprD+ nociceptors express Runx1 persistently (Liu et al., 2008). These neurons are marked by the expression of a series of channels and receptors, including MrgprD itself, the ATP-gated ion channel P2X3, the glutamate receptor mGluR5, and the transient receptor potential channel TRPC3, with the expression of all of these genes being dependent on Runx1 (Dong et al., 2001; Chen et al., 2006; Abdel Samad et al., 2010). In Rosa26Tomato/+;Vglut3Cre/+ control mice, expression of these four genes in VGLUT3-Tomato+ neurons (Fig. 4B–E, left) is either not detectable, such as MrgprD (Fig. 4B), or is detected at low levels, such as P2X3 (Fig. 4E, arrowheads; in comparison with surrounding neurons with high-level P2X3 expression). In Zfp521F/F;Rosa26Tomato/+;Vglut3Cre/+ CKO mice, the expression of MrgprD, TRPC3, and mGLUR5 is expanded into, and the P2X3 expression levels are elevated in, a portion of Tomato+ neurons (Fig. 4B–E, right). Quantitative analyses showed that MrgprD mRNA was detected in <2% (25/1370) of VGLUT3-Tomato+ in control mice, but the percentage was increased to 34.8% (493/1418) in Zfp521 CKO mice (χ2 test, p < 0.001). A similar expansion of MrgprD expression was observed in thoracic DRG (data not shown). Notably, no expansion was observed for those sensory channels and receptors that mark other types of Runx1-dependent neurons listed in Figure 1A, such as MrgprA3, MrgprB4, TRPA1, and TRPV1 (Fig. 4F–I). For example, in Zfp52F/F;Rosa26Tomato/+;Vglut3Cre/+ mice, only 0.9% (8/877) of Tomato+ neurons expressed MrgprA3, comparable to the 2.0% (22/1087) observed in Rosa26Tomato/+;Vglut3Cre/+ control mice (χ2 test, p > 0.05). It should be noted that, whereas deep RNA sequencing was able to reveal TRPA1 expression in the TH+ subset of VGLUT3+ CLTMs (Usoskin et al., 2014), this expression does not reach levels detectable by our in situ hybridization in either wild-type or CKO mice (Fig. 4H). It should also be pointed out that TRPM8 is expressed in the small-diameter TH− subset of VGLUT3+ CLTMs (Draxler et al., 2014) and no obvious loss or expansion of TRPM8 expression was observed in Zfp52F/F;Rosa26Tomato/+;Vglut3Cre/+ mice compared with control littermates (data not shown), consistent with the enrichment of Zfp521high in TH+ CLTMs.
Zfp521, therefore, has dual functions (Fig. 4J). First, it is required to establish and/or maintain features that are uniquely associated with VGLUT3+ CLTMs, such as the full expression of VGLUT3, TH, and TAFA4. Second, it acts as a genetic repressor of Runx1 that prevents VGLUT3+ CLTMs from acquiring molecular identities that normally belong to a different type of Runx1-dependent neurons: MrgprD+ polymodal nociceptors. As shown in Figure 7A and explained in the Discussion (see below), the dual roles of Runx1 in both activating and repressing (through Zfp521) MrgprD indicate that Runx1 and Zfp521 form a classic incoherent feedforward loop (Alon, 2007) to control the segregation of VGLUT3+ CLTMs that express Zfp521 versus MrgprD+ nociceptors that lack Zfp521.
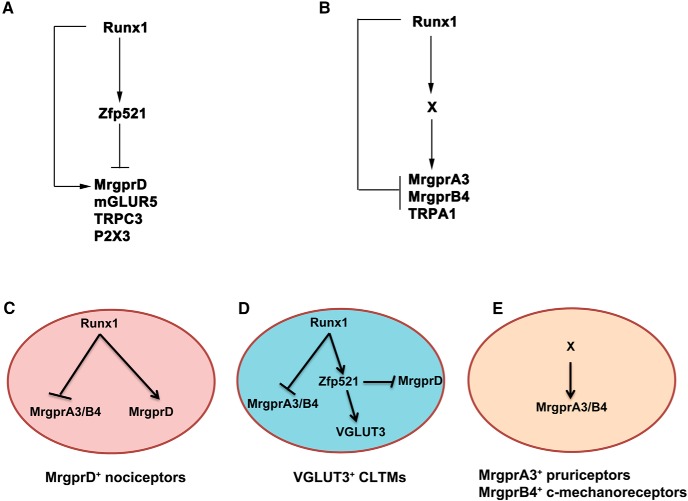
Figure 7.
I-FFLs control sensory subtype segregation. A, B, Runx1 and the downstream factors form I-FFLs in controlling a distinct set of sensory channels and receptors. For details, see Discussion. The nature of the Runx1-dependent X event is unknown. C–E, Molecular mechanisms leading to the segregation of three types of sensory neurons. Transient Runx1 expression in MrgprA3/B4+ neurons is not illustrated (E). The arrows (“→”) or “—I” indicate the necessity of upstream factors for the activation or repression of the downstream events, not necessarily indicating a sufficiency to do so. For details, see the Discussion.
CLTMs acquired identities of MrgprA3+ and/or MrgprB4+ neurons in Runx1 late CKO mice
Runx1-dependent cutaneous sensory neurons can be divided into Runx1-persistent and Runx1-transient subpopulations (Fig. 1A). Runx1-persistent neurons include VGLUT3+ CLTMs, MrgprD+ nociceptors, and others; Runx1-transient neurons include MrgprA3+ pruriceptors and MrgprB4+ c-mechanoreceptors. The above studies show that, upon the conditional knock-out of Zfp521 by crossing with VGLUT3-Cre mice, in which Cre expression in CLTMs is established at neonatal stages (Lou et al., 2013), prospective VGLUT3+ CLTMs are competent to acquire molecular features of MrgprD+ neurons. We next investigated whether, upon the neonatal loss of the more upstream transcription factor Runx1 in Runx1F/F;ROSA26Tomato/+;Vglut3Cre/+ CKO mice, which led to a loss of Zfp521 expression as well (data not shown), prospective VGLUT3+ CLTMs are competent to adopt a cell fate that normally undergoes transient Runx1 expression, such as MrgprA3+ and/or MrgprB4+ neurons. To test this, we performed ISH to analyze MrgprA3 and MrgprB4 expression in the lumbar DRG of Runx1F/F;ROSA26Tomato/+;Vglut3Cre/+ mice versus ROSA26Tomato/+;Vglut3Cre/+ control mice. We found that the percentage of Tomato+ neurons expressing MrgprA3 increased from virtually none in ROSA26Tomato/+;Vglut3Cre/+ control mice (8/877 = 0.9%) to 34.1% (767/2249) in Runx1F/F;ROSA26Tomato/+;Vglut3Cre/+ mice (χ2 test, p < 0.001; Fig. 5A). Tomato+ neurons expressing MrgprB4 also dramatically increased, from nearly none (1/1523 = 0.1%) in control mice to 51.3% (835/1627) in CKO mice (χ2 test, p < 0.001; Fig. 5B). In contrast, expression of other Runx1-dependent genes enriched in Runx1-persistent neurons, such as TRPV1 (Fig. 5C), MrgprD (Fig. 5D), and TRPM8 (data not shown), was not derepressed in Tomato+ neurons in Runx1F/F;ROSATomato/+;Vglut3Cre/+ CKO mice. Therefore, upon the loss of Runx1 at perinatal/postnatal stages, a significant proportion of VGLUT3 lineage neurons are competent to acquire molecular identities that normally belong to MrgprA3+ or MrgprB4+ neurons.
We next investigated whether the changes that we observed in the molecular phenotype of VGLUT3 lineage neurons in our conditional knock-out lines are reflected in the patterns of skin innervation and terminal morphologies. There are three types of VGLUT3-Tomato+ fibers in the hairy skin. The first one represents fibers from VGLUT3-transient myelinated mechanoreceptors that form the Merkel-cell neurite complexes. As reported previously, this type of innervation is unaffected in Runx1F/F;ROSA26Tomato/+;Vglut3Cre/+ CKO mice (Lou et al., 2013). The second type forms free nerve endings in the epidermis surrounding 17% (64/361) of hairs (often in clusters) (Fig. 6A, left). Little innervation was detected in the epidermis surrounding the remaining majority of hairs in control mice (Fig. 6A, right, small arrowhead). Note that, within the innervated hair clusters, VGLUT3-Tomato+ fibers also innervated the neck region of the hair follicle (Fig. 6A, left, arrow), as Mrgprd-GFP+ fibers do (Fig. 1). In Runx1F/F;ROSA26Tomato/+;Vglut3Cre/+ CKO mice, innervation in the neck region of hair follicles and the epidermis surrounding a small subset of hairs is still observed (Fig. 6B, left), indicating that this type of innervation is independent of Runx1. The third type of VGLUT3-Tomato+ fibers forms longitudinal lanceolate endings (LLEs) in most hair follicles (Fig. 6A, right, large arrowhead). In Runx1F/F;Rosa26Tomato/+;Vglut3Cre/+ CKO mice, LLEs are lost and became circumferential endings (Fig. 6B, right, large arrowhead), as reported previously (Lou et al., 2013). Strikingly, we noticed that Tomato+ fibers originating from the base of the circumferential endings started to grow superficially (Fig. 6B, right, arrows) and eventually entered the skin epidermis (Fig. 6B, right, small arrowhead). As a result, epidermal innervation is now observed throughout the hairy skin. Notably, these fibers do not innervate the hair neck region (Fig. 6B, right), a pattern mimicking that of MrgprA3-Tomato+ fibers described in Figure 1. Therefore both molecularly and anatomically, a portion of VGLUT3 lineage neurons acquire features belonging to MrgprA3+ neurons in Runx1 CKO mice. In contrast, no innervation defects were observed upon knock-out of Zfp521 in Zfp521F/F;Rosa26Tomato/+;Vglut3Cre/+ CKO mice (Fig. 6C), indicating that Zfp521 only controls molecular identities of VGLUT3+ CLTMs, but not afferent innervations in the skin.
Discussion
Zfp521 is necessary for proper development of VGLUT3+ c-mechanoreceptors
The runt domain transcription factor Runx1 coordinates the development of a large cohort of sensory neurons innervating the skin (Fig. 1A). Our studies support that Runx1 may operate in a cell-context-dependent manner to control the expression of downstream transcription factors that in turn specify sensory neuron subtypes. Specifically, we found that Zfp521, a Runx1-dependent transcription factor, is expressed predominantly in the TH+ subset of VGLUT3+ CLTMs, but not in other Runx1-dependent sensory neurons. Zfp521 controls certain Runx1-dependent features in these neurons (Lou et al., 2013), including the expression of VGLUT3 and a portion of TH and TAFA4. However, the expression of the mechanically gated ion channel Piezo2, which is lost in Runx1 CKO mice (Lou et al., 2013), is unaffected in Zfp521 CKO mice (data not shown). Furthermore, in Runx1 CKO mice, the lanceolate endings formed by TH+ CLTMs are transformed to become circumferential endings (Lou et al., 2013) and also form free nerve endings that grow into the skin epidermis, a pattern reminiscent of MrgprA3+ neurons (Fig. 6). In contrast, such innervation changes do not occur in Zfp521 CKO mice (Fig. 6). Therefore, Runx1 uses both Zfp521-dependent and Zfp521-independent pathways to control distinct features of VGLUT3+ CLTMs. The loss of molecular identities, without changing skin innervations, also supports distinct genetic programs controlling these developmental processes, as suggested previously (Gorokhova et al., 2014).
I-FFLs control sensory submodality segregation
I-FFLs are recurring regulatory motifs observed from bacteria to mammals (Alon, 2007). A salient feature of these regulatory loops is that a transcription factor (or a signal) operates through two opposing legs (activation vs repression) in controlling a target gene (Alon, 2007). To our knowledge, our studies reveal for the first time that I-FFLs, in genetic terms, are used to control the segregation of a cohort of mammalian sensory neuron subtypes that innervate distinct parts of the skin, including VGLUT3+ CLTMs, MrgprD+ polymodal nociceptors, MrgprA3+ pruriceptors, and MrgprB4+ c-mechanoreceptors (summarized in Fig. 7).
First, Runx1 and Zfp521 form an I-FFL in controlling the molecular identities belonging to MrgprD+ polymodal nociceptors. We reported previously that Runx1 is required to establish a large molecular program that defines MrgprD+ neurons, such as the expression of MrgprD, P2X3, mGLUR5, and TRPC3 (Dong et al., 2001; Chen et al., 2006; Abdel Samad et al., 2010), thereby forming a positive leg for this I-FFL (Fig. 7A). Here, we found that Zfp521, a Runx1-dependent transcription factor, acts as a genetic repressor that prevents Runx1 from activating this molecular program in VGLUT3+ CLMRs (Fig. 4), thereby forming an opposing repressor leg for this I-FFL (Fig. 7A). It should be noted that, temporally, expression of Zfp521 is established at E13.5–E16, preceding that of MrgprD, which occurs at E16.5 (Chen et al., 2006; Liu et al., 2008). This precocious onset of the Zfp521-mediated repression leg may provide a mechanism that preserves a subset of DRG neurons to develop into prospective VGLUT3+ CLTMs rather than MrgprD+ neurons. Consistently, MrgrpD lineage neurons, which were marked by crossing MrgprD-Cre mice (Rau et al., 2009) with ROSA26-LSL-tdTomato reporter mice, do not include the TH+ subset of VGLUT3+ CLTMs (data not shown), indicating no transient MrgprD expression in VGLUT3+ CLMRs.
Second, Runx1 might form a different I-FFL to control the expression of MrgprA3 and MrgprB4 (Fig. 7B). For this I-FFL, Runx1 itself acts as a genetic repressor but might also form a positive leg by establishing an unknown downstream event “X.” The negative leg is clearly indicated by the derepression of MrgprA3/B4 in VGLUT3+ CLTMs upon Runx1 knock-out at neonatal stages (Fig. 5). A previous study further showed that Runx1 uses its c-terminal repression domain to suppress MrgprA3/B4 (Liu et al., 2008). The existence of the positive leg and the intermediate X event is suggested from the following observations. MrgprA3/B4 expression in DRGs is completely lost in Runx1F/F;Wnt1Cre/+ mice (Chen et al., 2006), in which Runx1 is knocked out in early sensory precursors, but such loss does not occur if Runx1 is knocked out at E16–E17 (Chen et al., 2006; Abdel Samad et al., 2010). The simplest interpretation is that the X event is established at embryonic stages, which is required for subsequent activation of MrgprA3/B4 that normally occurs at neonatal stages (Dong et al., 2001; Chen et al., 2006; Abdel Samad et al., 2010). Once the X event is established, Runx1 itself is no longer required for subsequent MrgprA3/B4 expression.
By forming these genetic I-FFLs, the same Runx1 transcription factor is now able to control the segregation of multiple cutaneous sensory neuron subtypes (Fig. 7C–E). First, the possible existence of the second I-FFL (Fig. 7B) for controlling MrgprA3/B4 expression, with Runx1 itself serving as the dominant repression leg, may explain why these two sensory receptors are normally expressed in neurons with transient Runx1 expression and are excluded from neurons with persistent Runx1 expression, such as VGLUT3+ CLTMs and MrgprD+ polymodal nociceptors. Second, the I-FFL formed by Runx1 and Zfp521 (Fig. 7A), with Zfp521 serving as the dominant repression leg, allows Runx1-persistent neurons to be further segregated into two subtypes: VGLUT3+ CLTMs that express Zfp521 and MrgrpD+ polymodal nociceptors that do not express Zfp521. Such control mechanisms can explain phenotypic changes observed in various knock-out mice. In Zfp521 CKO mice, the removal of the Zfp521-mediated repression leg unmasks the Runx1-mediated activation leg, allowing prospective VGLUT3 lineage neurons to acquire molecular identities of MrgrpD+ neurons. In Runx1F/F;ROSA26Tomato/+;Vglut3Cre/+ mice, neonatal knock-out of Runx1 leads to a loss of CLTM neuron identities (due to the loss of both Runx1 and Runx1-dependent Zfp521), an inability to acquire MrgprD+ neuron identities (due to the loss of Runx1-mediated activation), and a derepression of MrgprA3 and MrgprB4 (due to a loss of Runx1-mediated repression plus the establishment of the putative X event by Runx1 at earlier embryonic stages). As a result, prospective VGLUT3 lineage neurons acquire features that normally belong to MrgprA3+ pruriceptors or MprgprB4+ c-mechanoreceptors in Runx1 CKO mice.
Implication on sensory coding and sensory neuron evolution
In principle, the relative strength of the positive and negative legs of I-FFLs could be tuned in different cellular contexts or species. This modulation could have intriguing implication on sensory coding and sensory neuron evolutions. If the repression legs are completely dominant (i.e., Runx1-mediated repression of MrgprA3 and Zfp521-mediated repression of MrgprD), it could lead to nonoverlapped molecular expression, such as VGLUT3, MrgprD, and MrgprA3, at least at levels detected by in situ hybridization. This could form a genetic basis for the existence of modality-selective sensory neurons, such as the dedication of MrgprA3+ neurons for itch (Liu et al., 2009; Han et al., 2013). However, if the repression leg is not completely dominant, it could create variegated expression levels for the same receptors in different sensory populations, as does P2X3 (Fig. 4). Such variegated expression might form a basis for the “intensity” and “population” encoding strategies for sensory information processing (Ma, 2010, 2012; Prescott et al., 2014). For example, low dosage of a chemical stimulus might selectively activate neurons with highest receptor expression, whereas high dosage stimuli might additionally activate neurons with low receptor expression. According to the population coding theory, combinatorial activation of two or multiple types of sensory afferents could then generate new sensations in the brain (Ma, 2010, 2012; Prescott et al., 2014).
From an evolutionary point of view, a tuning of the relative strength of the activation versus the repression legs could create cross-species variability on the expression of sensory channels and receptors. For example, MrgprD+ noiceptors do not express the itch-related MrgprA genes in mice (Dong et al., 2001), but they do coexpress MrgprA in rats (Zylka et al., 2003), which could simply be caused by a reduction in the repression leg and/or an increase in the activation leg of the I-FFLs in rats. In addition, whereas TH+ CLTMs represent 6–19% of DRG neurons at different axial levels in mice (Li et al., 2011; Lou et al., 2013), none or <1% of rat DRG neurons express TH (Price and Mudge, 1983). Therefore, a given stimulus may activate different constituents of sensory neurons in different species and, according to the population coding hypothesis, it could evoke different behaviors and perceptions (Ma, 2010, 2012; Prescott et al., 2014). Indeed, cheek injection of histamine evokes pure itch-indicating scratching responses in mice (Shimada and LaMotte, 2008), but evokes mainly pain-indicating wiping in rats (Klein et al., 2011). In addition, nape injection of chloroquine evokes robust scratching in mice (Han et al., 2013), but nearly none in rats (Klein et al., 2011). Such cross-species variability may contribute to the debate regarding to what degree a given animal species can faithfully model human pain and itch (Mogil, 2009).
Footnotes
Work done in the laboratory of Y.L. was supported by National Natural Science Foundation of China (Grant 31171071). Work done in the laboratory of Q.M. was supported by the National Institutes of Health (National Institute of Dental and Craniofacial Research Grant R01 DE018025 and National Institute of Neurological Disorders and Stroke Grant P01 NS047572). We thank Nancy Speck and Gary Gilliland for the floxed Runx1 mice, Mark Zylka and the Mutant Mouse Regional Resource Centers for the MrgprD-GFP and MrgprD-Cre mice, Xinzhong Dong for the MrgprA3-Cre mice, Susan Dymecki for the ROSA26-Flpe deletor mice, the Allen Brain Institute and the Jackson Laboratory for the Rosa26LSL-tdTomato mice, and the Intellectual and Developmental Disabilities Research Center Mouse Gene Manipulation Core at Boston Children's Hospital for the generation of Zfp521 floxed mice.
The authors declare no competing financial interests.
References
- Abdel Samad O, Liu Y, Yang FC, Kramer I, Arber S, Ma Q. Characterization of two Runx1-dependent nociceptor differentiation programs necessary for inflammatory versus neuropathic pain. Mol Pain. 2010;6:45. doi: 10.1186/1744-8069-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Bachy I, Franck MC, Li L, Abdo H, Pattyn A, Ernfors P. The transcription factor Cux2 marks development of an A-delta sublineage of TrkA sensory neurons. Dev Biol. 2011;360:77–86. doi: 10.1016/j.ydbio.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Cavanaugh DJ, Lee H, Lo L, Shields SD, Zylka MJ, Basbaum AI, Anderson DJ. Distinct subsets of unmyelinated primary sensory fibers mediate behavioral responses to noxious thermal and mechanical stimuli. Proc Natl Acad Sci U S A. 2009;106:9075–9080. doi: 10.1073/pnas.0901507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Broom DC, Liu Y, de Nooij JC, Li Z, Cen C, Samad OA, Jessell TM, Woolf CJ, Ma Q. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron. 2006;49:365–377. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Delfini MC, Mantilleri A, Gaillard S, Hao J, Reynders A, Malapert P, Alonso S, François A, Barrere C, Seal R, Landry M, Eschallier A, Alloui A, Bourinet E, Delmas P, Le Feuvre Y, Moqrich A. TAFA4, a chemokine-like protein, modulates injury-induced mechanical and chemical pain hypersensitivity in mice. Cell Rep. 2013;5:378–388. doi: 10.1016/j.celrep.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Dong X, Han S, Zylka MJ, Simon MI, Anderson DJ. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106:619–632. doi: 10.1016/S0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- Draxler P, Honsek SD, Forsthuber L, Hadschieff V, Sandkühler J. VGluT3+ primary afferents play distinct roles in mechanical and cold hypersensitivity depending on pain etiology. J Neurosci. 2014;34:12015–12028. doi: 10.1523/JNEUROSCI.2157-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes IM, Lanier J, Eng SR, Turner EE. Brn3a regulates neuronal subtype specification in the trigeminal ganglion by promoting Runx expression during sensory differentiation. Neural Dev. 2010;5:3. doi: 10.1186/1749-8104-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorokhova S, Gaillard S, Urien L, Malapert P, Legha W, Baronian G, Desvignes JP, Alonso S, Moqrich A. Uncoupling of molecular maturation from peripheral target innervation in nociceptors expressing a chimeric TrkA/TrkC receptor. PLoS Genet. 2014;10:e1004081. doi: 10.1371/journal.pgen.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, Speck NA, Gilliland DG. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Ma C, Liu Q, Weng HJ, Cui Y, Tang Z, Kim Y, Nie H, Qu L, Patel KN, Li Z, McNeil B, He S, Guan Y, Xiao B, Lamotte RH, Dong X. A subpopulation of nociceptors specifically linked to itch. Nat Neurosci. 2013;16:174–182. doi: 10.1038/nn.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse E, Saito H, Kiviranta R, Correa D, Yamana K, Neff L, Toben D, Duda G, Atfi A, Geoffroy V, Horne WC, Baron R. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J Cell Biol. 2010;191:1271–1283. doi: 10.1083/jcb.201009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius D. TRP channels and pain. Annu Rev Cell Dev Biol. 2013;29:355–384. doi: 10.1146/annurev-cellbio-101011-155833. [DOI] [PubMed] [Google Scholar]
- Kamiya D, Banno S, Sasai N, Ohgushi M, Inomata H, Watanabe K, Kawada M, Yakura R, Kiyonari H, Nakao K, Jakt LM, Nishikawa S, Sasai Y. Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature. 2011;470:503–509. doi: 10.1038/nature09726. [DOI] [PubMed] [Google Scholar]
- Kang S, Akerblad P, Kiviranta R, Gupta RK, Kajimura S, Griffin MJ, Min J, Baron R, Rosen ED. Regulation of early adipose commitment by Zfp521. PLoS Biol. 2012;10:e1001433. doi: 10.1371/journal.pbio.1001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Carstens MI, Carstens E. Facial injections of pruritogens or algogens elicit distinct behavior responses in rats and excite overlapping populations of primary sensory and trigeminal subnucleus caudalis neurons. J Neurophysiol. 2011;106:1078–1088. doi: 10.1152/jn.00302.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerström MC, Rogoz K, Abrahamsen B, Persson E, Reinius B, Nordenankar K, Olund C, Smith C, Mendez JA, Chen ZF, Wood JN, Wallén-Mackenzie A, Kullander K. VGLUT2-dependent sensory neurons in the TRPV1 population regulate pain and itch. Neuron. 2010;68:529–542. doi: 10.1016/j.neuron.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemend F, Ernfors P. Molecular interactions underlying the specification of sensory neurons. Trends Neurosci. 2012;35:373–381. doi: 10.1016/j.tins.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Li L, Rutlin M, Abraira VE, Cassidy C, Kus L, Gong S, Jankowski MP, Luo W, Heintz N, Koerber HR, Woodbury CJ, Ginty DD. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell. 2011;147:1615–1627. doi: 10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Vrontou S, Rice FL, Zylka MJ, Dong X, Anderson DJ. Molecular genetic visualization of a rare subset of unmyelinated sensory neurons that may detect gentle touch. Nat Neurosci. 2007;10:946–948. doi: 10.1038/nn1937. [DOI] [PubMed] [Google Scholar]
- Liu Q, Tang Z, Surdenikova L, Kim S, Patel KN, Kim A, Ru F, Guan Y, Weng HJ, Geng Y, Undem BJ, Kollarik M, Chen ZF, Anderson DJ, Dong X. Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell. 2009;139:1353–1365. doi: 10.1016/j.cell.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Sikand P, Ma C, Tang Z, Han L, Li Z, Sun S, LaMotte RH, Dong X. Mechanisms of itch evoked by β-alanine. J Neurosci. 2012;32:14532–14537. doi: 10.1523/JNEUROSCI.3509-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ma Q. Generation of somatic sensory neuron diversity and implications on sensory coding. Curr Opin Neurobiol. 2011;21:52–60. doi: 10.1016/j.conb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yang FC, Okuda T, Dong X, Zylka MJ, Chen CL, Anderson DJ, Kuner R, Ma Q. Mechanisms of compartmentalized expression of Mrg class G-protein-coupled sensory receptors. J Neurosci. 2008;28:125–132. doi: 10.1523/JNEUROSCI.4472-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Abdel Samad O, Duan B, Zhang L, Tong Q, Lopes C, Ji RR, Lowell BB, Ma Q. VGLUT2-dependent glutamate release from peripheral nociceptors is required to sense pain and suppress itch. Neuron. 2010;68:543–556. doi: 10.1016/j.neuron.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes C, Liu Z, Xu Y, Ma Q. Tlx3 and Runx1 act in combination to coordinate the development of a cohort of nociceptors, thermoceptors, and pruriceptors. J Neurosci. 2012;32:9706–9715. doi: 10.1523/JNEUROSCI.1109-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou S, Duan B, Vong L, Lowell BB, Ma Q. Runx1 controls terminal morphology and mechanosensitivity of VGLUT3-expressing C-mechanoreceptors. J Neurosci. 2013;33:870–882. doi: 10.1523/JNEUROSCI.3942-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. Labeled lines meet and talk: population coding of somatic sensations. J Clin Invest. 2010;120:3773–3778. doi: 10.1172/JCI43426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. Population coding of somatic sensations. Neurosci Bull. 2012;28:91–99. doi: 10.1007/s12264-012-1201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Fode C, Guillemot F, Anderson DJ. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes and Development. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogil JS. Animal models of pain: progress and challenges. Nat Rev Neurosci. 2009;10:283–294. doi: 10.1038/nrn2606. [DOI] [PubMed] [Google Scholar]
- Moqrich A. Peripheral pain-sensing neurons: from molecular diversity to functional specialization. Cell Rep. 2014;6:245–246. doi: 10.1016/j.celrep.2014.01.018. [DOI] [PubMed] [Google Scholar]
- Prescott SA, Ma Q, De Koninck Y. Normal and abnormal coding of somatosensory stimuli causing pain. Nat Neurosci. 2014;17:183–191. doi: 10.1038/nn.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Mudge AW. A subpopulation of rat dorsal root ganglion neurones is catecholaminergic. Nature. 1983;301:241–243. doi: 10.1038/301241a0. [DOI] [PubMed] [Google Scholar]
- Rau KK, McIlwrath SL, Wang H, Lawson JJ, Jankowski MP, Zylka MJ, Anderson DJ, Koerber HR. Mrgprd enhances excitability in specific populations of cutaneous murine polymodal nociceptors. J Neurosci. 2009;29:8612–8619. doi: 10.1523/JNEUROSCI.1057-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren AJ, Wang K, Zhang H, Liu A, Ma X, Liang Q, Cao D, Wood JN, He DZ, Ding YQ, Yuan WJ, Xie Z, Zhang WJ. ZBTB20 regulates nociception and pain sensation by modulating TRP channel expression in nociceptive sensory neurons. Nat Commun. 2014;5:4984. doi: 10.1038/ncomms5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Seal RP, Wang X, Guan Y, Raja SN, Woodbury CJ, Basbaum AI, Edwards RH. Injury-induced mechanical hypersensitivity requires C-low threshold mechanoreceptors. Nature. 2009;462:651–655. doi: 10.1038/nature08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada SG, LaMotte RH. Behavioral differentiation between itch and pain in mouse. Pain. 2008;139:681–687. doi: 10.1016/j.pain.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R, Orit S, Anderson DJ. The induction of a neural-specific gene, SCG10, by nerve growth factor in PC12 cells is transcriptional, protein synthesis dependent, and glucocorticoid inhibitable. Dev Biol. 1988;127:316–325. doi: 10.1016/0012-1606(88)90318-1. [DOI] [PubMed] [Google Scholar]
- Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;11:1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2014;8:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- Vrontou S, Wong AM, Rau KK, Koerber HR, Anderson DJ. Genetic identification of C fibres that detect massage-like stroking of hairy skin in vivo. Nature. 2013;493:669–673. doi: 10.1038/nature11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang FC, Tan T, Huang T, Christianson J, Samad OA, Liu Y, Roberson D, Davis BM, Ma Q. Genetic control of the segregation of pain-related sensory neurons innervating the cutaneous versus deep tissues. Cell Rep. 2013;5:1353–1364. doi: 10.1016/j.celrep.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Senzaki K, Yokomizo T, Takahashi S, Ozaki S, Shiga T. Runx1 selectively regulates cell fate specification and axonal projections of dorsal root ganglion neurons. Dev Biol. 2007;303:663–674. doi: 10.1016/j.ydbio.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Zou M, Li S, Klein WH, Xiang M. Brn3a/Pou4f1 regulates dorsal root ganglion sensory neuron specification and axonal projection into the spinal cord. Dev Biol. 2012;364:114–127. doi: 10.1016/j.ydbio.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka MJ, Dong X, Southwell AL, Anderson DJ. Atypical expansion in mice of the sensory neuron-specific Mrg G protein-coupled receptor family. Proc Natl Acad Sci U S A. 2003;100:10043–10048. doi: 10.1073/pnas.1732949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka MJ, Rice FL, Anderson DJ. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron. 2005;45:17–25. doi: 10.1016/j.neuron.2004.12.015. [DOI] [PubMed] [Google Scholar]