Abstract
The rare 6-deoxysugar d-rhamnose is a component of bacterial cell surface glycans, including the d-rhamnose homopolymer produced by Pseudomonas aeruginosa, called A-band O polysaccharide. GDP-d-rhamnose synthesis from GDP-d-mannose is catalyzed by two enzymes. The first is a GDP-d-mannose-4,6-dehydratase (GMD). The second enzyme, RMD, reduces the GMD product (GDP-6-deoxy-d-lyxo-hexos-4-ulose) to GDP-d-rhamnose. Genes encoding GMD and RMD are present in P. aeruginosa, and genetic evidence indicates they act in A-band O-polysaccharide biosynthesis. Details of their enzyme functions have not, however, been previously elucidated. We aimed to characterize these enzymes biochemically, and to determine the structure of RMD to better understand what determines substrate specificity and catalytic activity in these enzymes. We used capillary electrophoresis and NMR analysis of reaction products to precisely define P. aeruginosa GMD and RMD functions. P. aeruginosa GMD is bifunctional, and can catalyze both GDP-d-mannose 4,6-dehydration and the subsequent reduction reaction to produce GDP-d-rhamnose. RMD catalyzes the stereospecific reduction of GDP-6-deoxy-d-lyxo-hexos-4-ulose, as predicted. Reconstitution of GDP-d-rhamnose biosynthesis in vitro revealed that the P. aeruginosa pathway may be regulated by feedback inhibition in the cell. We determined the structure of RMD from Aneurinibacillus thermoaerophilus at 1.8 Å resolution. The structure of A. thermoaerophilus RMD is remarkably similar to that of P. aeruginosa GMD, which explains why P. aeruginosa GMD is also able to catalyze the RMD reaction. Comparison of the active sites and amino acid sequences suggests that a conserved amino acid side chain (Arg185 in P. aeruginosa GMD) may be crucial for orienting substrate and cofactor in GMD enzymes.
Keywords: Aneurinibacillus thermoaerophilus, d-rhamnose, GMD/RMD, Pseudomonas aeruginosa, real-time NMR
The rare sugar d-rhamnose (6-deoxy-d-mannose; d-Rha) has been unambiguously identified only in bacteria, including pathogens of animals [1,2] and plants [3], where it is a component of cell surface polysaccharides. It is also believed to be present in the major viral capsid glycoprotein of Paramecium bursaria chlorella virus-1 (PBCV-1) [4].
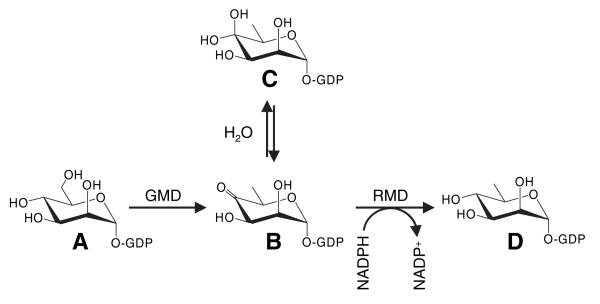
The precursor for d-Rha in glycan biosynthesis is GDP-d-Rha. The biosynthetic pathway for GDP-d-Rha has been elucidated [5] in one bacterial species, the Gram-positive thermophile Aneurinibacillus thermoaerophilus L420-91T, in which d-Rha is a component of the S-layer protein glycan. Two enzymes, GDP-d-mannose-4,6-dehydratase (GMD; EC 4.2.1.47) and GDP-6-deoxy-d-lyxo-hexos-4-ulose-4-reductase (GDP-d-rhamnose forming) (RMD; EC 1.1.1.281), catalyze the conversion of GDP-d-mannose (GDP-d-Man) to GDP-d-Rha (Fig. 1). GMD (note that this enzyme is distinct from GDP-d-Man dehydrogenase, which has also been called GMD in Pseudomonas aeruginosa), catalyzes the dehydration of GDP-d-Man to produce GDP-6-deoxy-d-lyxo-hexos-4-ulose. RMD then reduces the 4-keto moiety to produce GDP-d-Rha [5].
Fig. 1.
The biosynthetic pathway leading to the production of GDP-α-d-Rha in P. aeruginosa. GMD catalyzes the 4,6-dehydration of GDP-d-Man (A), resulting in the production of GDP-6-deoxy-lyxo-hexos-4-ulose (B), which exists in equilibrium with its gem-diol form (C). RMD catalyzes the stereoselective reduction of compound B at C4, resulting in the production of GDP-d-Rha (D). Although GMD can catalyze this final reduction reaction, our data indicate that GMD does so much less rapidly than RMD.
Both proteins are members of the sugar nucleotide-modifying subfamily of the short-chain dehydrogenase/reductase (SDR) family. Members of this large family typically share low sequence identity and can catalyze a wide range of different reactions [6], almost all of which involve oxidoreductase chemistry mediated by a dinucleotide cofactor. GMD is widespread in nature, and catalyzes the first step in the biosynthesis of the 6-deoxy sugars l-fucose [7], 6-deoxy-d-talose [8,9], and d-perosamine [10], as well as d-Rha [5]. For this reason, GMDs from a variety of organisms have been studied [11–15]. Only one RMD, from A. thermoaerophilus, has been purified and characterized in vitro [5]. Bioinformatic analysis indicates that the closest paralog of RMD is GMD. The similarity of these proteins is also suggested by the fact that a number of GMDs are bifunctional, being able to catalyze the same stereospecific reduction as RMD, in addition to their 4,6-dehydratase function [5,16,17].
P. aeruginosa is a Gram-negative, opportunistic pathogen that accounts for approximately one in 10 of hospital-acquired infections [18]. It also establishes chronic lung infections in cystic fibrosis patients, in whom it is a major cause of morbidity and mortality. This bacterium produces a cell surface polymer known as A-band O polysaccharide, which consists of a linear d-Rha homopolymer attached to lipopolysaccharide [19]. The function of A-band lipopolysaccharide (LPS) in infection has not been defined, but this molecule is produced by the majority of P. aeruginosa isolates, and is maintained on the cell surface in chronic infections. A-band O polysaccharide is apparently immunologically invisible to the host in the initial stages of infection, but becomes a major antigen over time as other LPS forms are selectively lost. The appearance of antibodies against A band in host serum correlates with extended duration of disease and reduced lung function [20].
An eight-gene cluster encodes functions for synthesis and export of A-band O polysaccharide [21,22], and contains genes for the expression of GMD and RMD homologs, gmd (originally called gca) and rmd, respectively. Genetic evidence supports the annotation of these genes, but their functions have not been confirmed biochemically. The gmd gene was identified in a 1 kb region on plasmid pFV36 that could restore A-band synthesis in the A-band-deficient P. aeruginosa strain, rd7513. This region encodes a protein of approximately 37 kDa, and conferred the ability to Escherichia coli lysates to synthesize 14C-labeled GDP-Rha from labeled GDP-Man [23]. Mutation of rmd in P. aeruginosa abrogated A-band O polysaccharide production [24], and coexpression in Saccharomyces cerevisiae of rmd from P. aeruginosa and gmd from Helicobacter pylori enabled the yeast cell lysates to convert GDP-Man to GDP-Rha [25].
A specific question about the activity of RMD arises from early work on 6-deoxyhexose biosynthesis in Pseudomonas. A soil isolate known as ‘strain GS’ produces a capsular polysaccharide containing d-Rha and 6-deoxy-d-talose, two residues that differ only in the stereochemistry at C4. A cellular fraction was able to nonstereospecifically reduce the ketone in GDP-6-deoxy-d-lyxo-hexos-4-ulose, thereby producing both epimers: GDP-d-Rha and GDP-6-deoxy-d-talose [26]. That study did not establish whether this activity was due to an RMD homolog or to more than one enzyme, but biochemical characterization of P. aeruginosa RMD will show whether or not this enzyme is a stereospecific reductase.
Crystal structures have been determined for the GMDs from P. aeruginosa [27], E. coli [28], Arabidopsis thaliana [29], and PBCV-1 [30]. Up to now, no RMD structure has been reported.
Here, we report the biochemical characterization of purified His6-tag fusions of GMD and RMD from P. aeruginosa, and the structural characterization of RMD from A. thermoaerophilus.
Results
Purification and stability of His6-GMD and His6-RMDPa
We purified N-terminally His6-tagged fusions of P. aeruginosa GMD and RMD (His6-GMD and His6-RMDPa, respectively) in two chromatography steps to greater than 95% purity (as judged by Coomassie-stained SDS/PAGE; not shown). In 25% glycerol, both enzymes retained activity after storage for more than 1 year at −80 °C. We made qualitative observations that His6-GMD lost activity slowly in the course of enzyme–substrate incubations, particularly at 37 °C, but 4,6-dehydratase activity was still detectable after incubation for 16 h at 25 °C (not shown). Addition of BSA (10 mg·mL−1) and glycerol [10% (v/v)] improved the stability of His6-GMD during incubations at 37 °C, but these additives were not routinely included in assays, as the proteins were stable during the time-scales of the experiments, and both additives prevented accurate measurement of reaction products by capillary electrophoresis (CE) or NMR.
CE analysis of His6-GMD functions
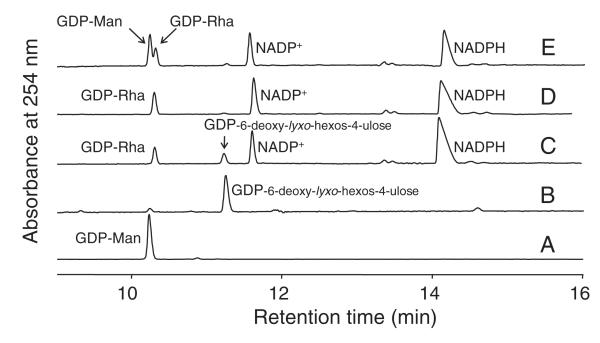
CE is a technique that is able to resolve closely related sugar nucleotides, and was our method of choice for initial in vitro characterization of these enzymes. Analysis of the products that were formed after incubation of His6-GMD with GDP-d-Man indicated that this enzyme catalyzes quantitative 4,6-dehydration of this substrate to GDP-6-deoxy-d-lyxo-hexos-4-ulose (Fig. 2A,B). The chemical structures of sugar nucleotide compounds in these reactions were unambiguously identified by NMR spectroscopy (see below). The proposed mechanism for GMD requires oxidoreductase chemistry mediated by a dinucleotide cofactor [31]. Addition of exogenous NAD(P) was not required for catalytic activity of the P. aeruginosa enzyme, indicating that the cofactor had copurified with the protein. Some GMDs are bifunctional, being able to catalyze the subsequent reduction of their initial 4-ketosugar nucleotide products to produce 6-deoxysugar nucleotides [5,16,17]. Addition of NADPH to the incubation of the P. aeruginosa enzyme resulted in the gradual, His6-GMD-dependent reduction of GDP-6-deoxy-d-lyxo-hexos-4-ulose to GDP-d-Rha (Fig. 2C,D). Therefore, like its homologs from Klebsiella pneumonia, A. thermoaerophilus and PBCV-1, His6-GMD from P. aeruginosa is a bifunctional 4,6-dehydratase, and a stereospecific 4-reductase.
Fig. 2.
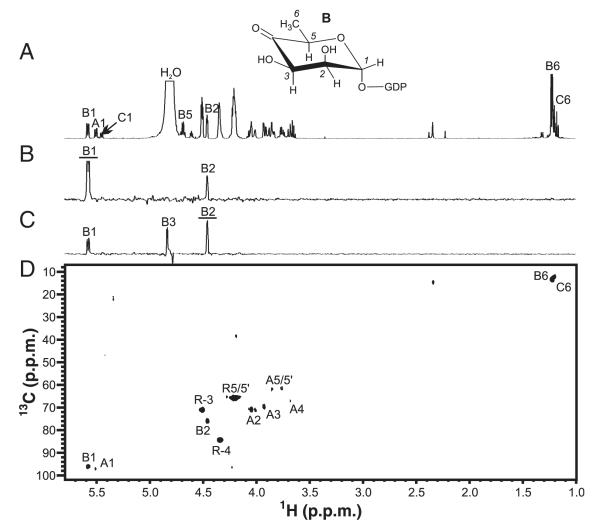
CE analysis of dehydratase and reductase activities exhibited by His6-GMD. Sugar nucleotide peaks were identified by NMR; other peaks were identified by comparison with standards. His6-GMD catalyzes the production of GDP-6-deoxy-d-lyxo-hexos-4-ulose from GDP-d-Man. When reduced cofactor (NADPH) is added, His6-GMD can catalyze the reduction of this intermediate to GDP-d-Rha, but the reaction is incomplete after 1 h. Traces: (A) standard, GDP-d-Man; (B) product of incubation of His6-GMD with GDP-d-Man; (C) products of incubation of His6-GMD with GDP-6-deoxy-d-lyxo-hexos-4-ulose (generated in situ) and NADPH for 1 h; (D) as in (C), but incubated for 2 h; (E) reaction in (D) spiked with GDP-d-Man. Spiking demonstrates that the final product is not the same as the starting material.
CE analysis of His6-RMDPa incubations and His6-RMDPa–His6-GMD coincubation
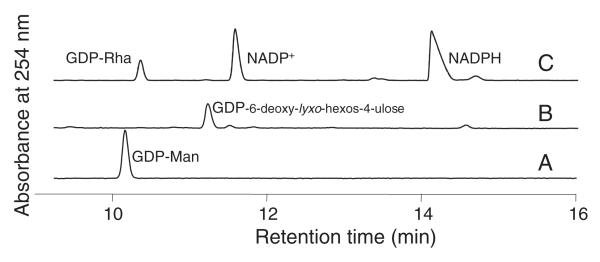
RMDs use the product of the GMD-catalyzed 4,6-dehydration reaction as substrate, and employ an NAD(P)H cofactor as an electron donor. We incubated His6-GMD with GDP-d-Man for 1 h, which was enough time for complete conversion of the GDP-d-Man, and then removed the enzyme by filtration. His6-RMDPa and NADPH were then added to the reaction mixture. The GDP-6-deoxy-d-lyxo-hexos-4-ulose substrate was generated in situ because it is unstable, and its purification is therefore impractical [5,17]. CE analysis of the reaction products (Fig. 3) showed that, in the presence of excess NADPH, His6-RMDPa catalyzed the conversion of GDP-6-deoxy-d-lyxo-hexos-4-ulose to GDP-d-Rha. When His6-GMD and His6-RMDPa were coincubated with GDP-d-Man and NADPH, however, no reaction was observed by CE (not shown).
Fig. 3.
CE analysis of His6-RMDPa reactions. His6-RMD converts the product of the His6-GMD-catalyzed reaction, GDP-6-deoxy-d-lyxo-hexos-4-ulose, to GDP-d-Rha, in the presence of NADPH. Traces: (A) standard, GDP-d-Man; (B) the product of His6-GMD incubation with GDP-d-Man after removal of His6-GMD by filtration; (C) the His6-GMD product shown in (B), after subsequent incubation with His6-RMDPa and NADPH.
Identification of reaction products by NMR spectroscopy
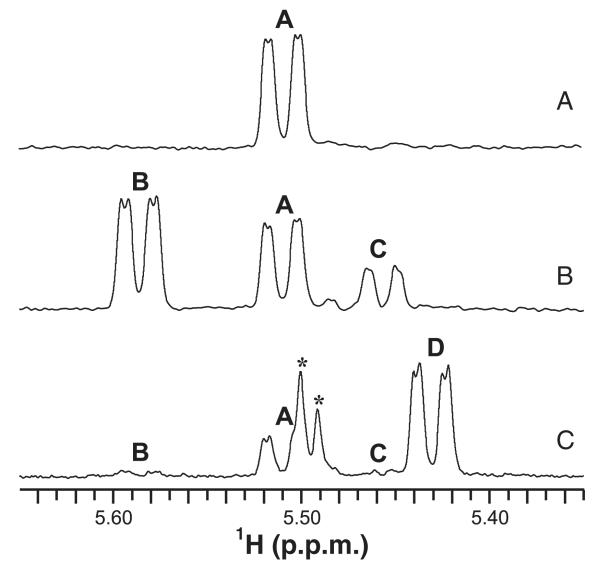
To precisely define the functions of His6-GMD and His6-RMDPa in vitro, we identified the products of these enzyme–substrate incubations by NMR spectroscopy. As it is not possible to purify the labile product of the GDP-d-Man 4,6-dehydration, we performed the enzyme incubation in an NMR spectrometer, and monitored the reaction directly in the tube (Figs 4 and 5). This technique was previously used for identification of labile 4-keto UDP-sugars [32]. Monitoring of the anomeric region of the 1D-1H-spectrum over the course of incubation with His6-GMD, without NADPH, showed progressive depletion of signals from GDP-d-Man (compound A) and the growth of peaks corresponding to the anomeric resonances of the 4-keto (compound B) and gem-diol (compound C) forms of GDP-6-deoxy-d-lyxo-hexos-4-ulose (Fig. 4B). On the basis of integration of the anomeric signals, the 4-keto and gem-diol forms of this compound coexist in equilibrium at an approximately 5 : 2 ratio. Full assignment of the NMR spectra of compounds B and C and measurement of coupling constants (Table 1) was achieved after removal of enzyme by filtration at the 16-h time point, using selective 1D-TOCSY and NOESY NMR experiments.
Fig. 4.
NMR spectroscopy of the active His6-GMD reaction directly in aqueous reaction buffer. The reaction buffer was: 5 mm GDP-α-d-mannose, 90 μg of His6-GMD, 25 mm NaPO4, 50 mm NaCl (pH 7.2), and 90% H2O/10% D2O. (A) 1H-NMR spectrum of the His6-GMD reaction buffer at the beginning of the reaction, showing the anomeric region. (B) 1H-NMR spectrum of the His6-GMD reaction buffer after 16 h. (C) 1H-NMR spectrum of the His6-GMD reaction buffer after 16 h following the addition of NADPH. A, GDP-d-Man; B, GDP-6-deoxy-d-lyxo-hexos-4-ulose; C, gem-diol form of compound B; D, GDP-d-Rha; *unknown impurities.
Fig. 5.
NMR spectroscopy of GDP-6-deoxy-α-d-lyxo-hexos-4-ulose (B). These spectra were measured directly in aqueous reaction buffer (5 mm GDP-d-Man, 90 μg of His6-GMD, 25 mm NaPO4, 50 mm NaCl, pH 7.2, 90% H2O/10% D2O). (A) 1H-NMR spectrum. (B) 1D-TOCSY of compound B H1 (80 ms). (C) 1D-TOCSY of compound B H2 (80 ms). (D) 13C-HSQC spectrum (128 transients, 128 increments, 1JC,H = 140 Hz, 12 h). For selective 1D experiments, excited resonances are underlined. A, GDP-d-Man; C, gem-diol form of compound B; R, ribose.
Table 1.
NMR data for sugar nucleotide metabolites in the GDP-d-Rha pathway of P. aeruginosa. Resonances were referenced to an internal acetone standard at δH = 2.225 p.p.m. and δC = 31.07 p.p.m.
|
1H and 13C chemical shifts [δ (p.p.m.)], and proton coupling constants (JH,H (Hz)] |
|||||||
|---|---|---|---|---|---|---|---|
| Compound | H1 C1 J1,2 | H2 C2 J2,3 | H3 C3 J3,4 | H4 C4 J4,5 | H5 C5 J5,6 | H6/6′ C6 | |
| GDP-α-d-mannose (A) | δ H | 5.51 | 4.05 | 3.92 | 3.68 | 3.84 | 3.75/3.85 |
| δ C | 97.1 | 70.9 | 69.6 | 67.2 | 74.1 | 61.6 | |
| 3 J H,H | 1.8 | 3.4 | 10.3 | 10.0 | |||
| 3 J H,P | 7.9 | ||||||
| GDP-α-d-6-deoxy-d-lyxo-hexos-4-ulose (B) | δ H | 5.58 | 4.46 | 4.84 | 4.68 | 1.22 | |
| δ C | 96.3 | 76.0 | 73.1 | 208.8 | 71.9 | 13.4 | |
| 3 J H,H | 2.2 | 3.8 | 6.5 | 6.5 | |||
| 3 J H,P | 7.6 | ||||||
| gem-Diol form of GDP-α-d-6-deoxy-d-lyxo-hexos-4-ulose (C) | δ H | 5.45 | 4.01 | 3.93 | 4.06 | 1.20 | |
| δ C | 97.4 | 71.5 | 69.6 | 94.0 | 71.0 | 12.3 | |
| 3 J H,H | 1.8 | 3.5 | 6.5 | 6.5 | |||
| 3 J H,P | 7.6 | ||||||
| GDP-α-d-rhamnose (D) | δ H | 5.43 | 4.03 | 3.86 | 3.42 | 3.89 | 1.25 |
| δ C | 97.2 | 71.2 | 70.4 | 72.8 | 70.4 | 17.6 | |
| 3 J H,H | 1.2 | 3.5 | 9.7 | 9.8 | 6.1 | ||
| 3 J H,P | 7.6 | ||||||
One-dimensional TOCSY of compound A H1 revealed a single J-coupled signal corresponding to H2 (Fig. 5B). The small J1,2 coupling observed for compound B is consistent with a manno-configured sugar ring [33]. Owing to this small J1,2 coupling, a 1D-TOCSY experiment on compound B H2 was needed to assign H3 (Fig. 5C). Proton assignments for compound C were also made on the basis of the results of 1D-TOCSY experiments on H1 and H2 (not shown). Selective 1D-NOESY experiments revealed NOEs between H3 and H5 for compounds B and C, which indicated that these protons were in close spatial proximity, and thus occupied the trans position on the sugar ring (data not shown). These NOEs, along with the small J1,2-confirmed compounds B and C, had the manno configuration. Carbon assignments were made on the basis of the results of a 13C heteronuclear single quantum correlation (HSQC) experiment (Fig. 5D). Whereas 13C–1H correlations were readily observed for compounds A and B, signals for compound C were only visible at higher intensity. Three-bond 13C–1H correlations observed using a heteronuclear multiple bond correlation (HMBC) experiment were used to assign C4 of compounds B and C. It is of importance that a signal corresponding to C4 of compound B was observed at δC = 208.8 p.p.m. and was indicative of a carbonyl group, whereas that of compound C at δC = 94.0 p.p.m. was consistent with a diol form [34]. Together, these spectroscopy results for the ‘real-time’ enzyme–substrate reaction in the NMR tube containing the His6-GMD–substrate reaction mixture provided unambiguous identification of the structure of compound B as GDP-6-deoxy-α-d-lyxo-hexos-4-ulose and that of compound C as the gem-diol form of compound B.
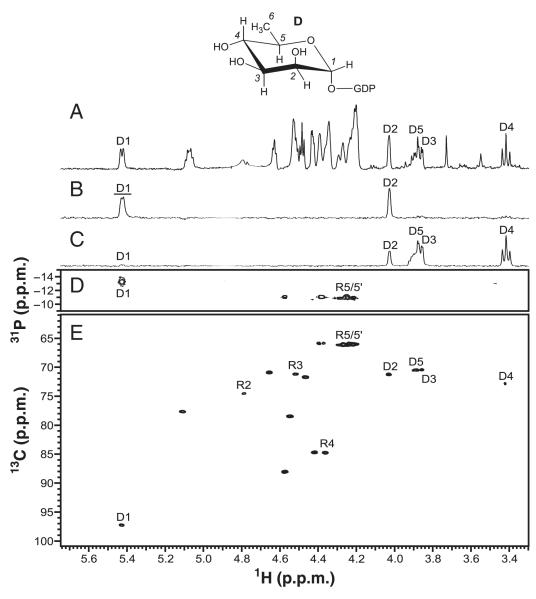
The product of the His6-RMD-catalyzed reaction (compound D) was purified by anion exchange chromatography before being analyzed by NMR. This sample contained NADP+ as a minor contaminant, but this did not prevent identification of the reaction product. Proton chemical shifts and JH,H coupling constants determined using 1D-TOCSY experiments agreed well with those previously reported for GDP-d-Rha [5] (Fig. 6A–C, Table 1). Results from a 31P-HMQC experiment showed a 1H–31P correlation for the anomeric signal of compound D at δP = −13.2 p.p.m., and another at δP = −10.8 p.p.m., corresponding to H5/5′ of ribose (Fig. 6D). Carbon chemical shifts and connectivities determined using 13C-HSQC (Fig. 6E) and HMBC were nearly identical to those reported for GDP-d-Rha [5]. On the basis of these NMR findings, compound D was concluded to be GDP-α-d-Rha. These results for His6-RMDPa therefore confirm that this enzyme is a GDP-6-deoxy-α-d-lyxo-hexos-4-ulose-4-reductase (GDP-d-Rha-forming).
Fig. 6.
NMR spectroscopy of the purified product from the His6-RMDPa-catalyzed reaction, GDP-α-d-rhamnose (D). (A) 1H-NMR spectrum. (B) 1D-TOCSY of compound D H1 (80 ms). (C) 1D-TOCSY of compound D H6 (80 ms). (D) 31P-HMQC spectrum (128 transients, 32 increments, 1JH,P = 8 Hz, 4 h). (E) 13C-HSQC spectrum (128 transients, 32 increments, 1JC,H = 150 Hz, 15 h). For selective 1D experiments, excited resonances are underlined. R represents ribose.
Time courses of His6-GMD and His6-RMDPa reactions determined by in-NMR-tube enzyme incubation
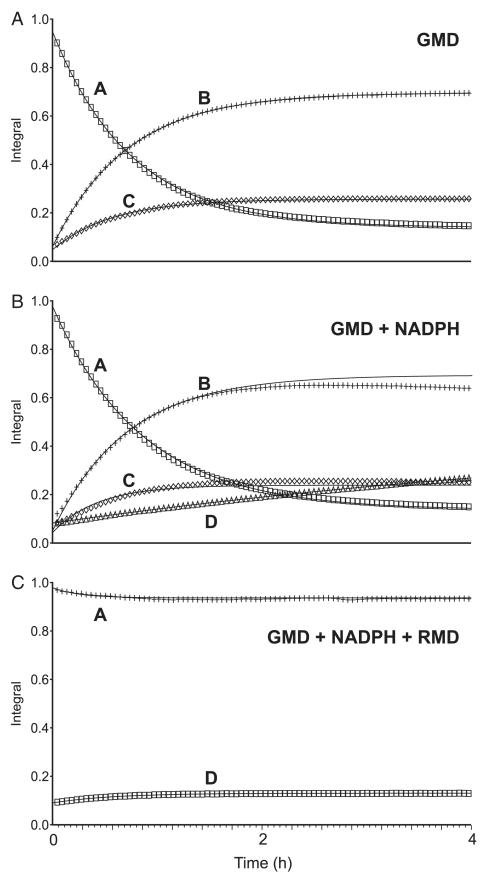
The NMR spectroscopic measurement of substrate and product concentrations during enzyme incubations in NMR tubes enables sensitive observation, in real time, of the course of enzyme-catalyzed reactions, and is particularly suitable when these reactions have labile starting materials or products [32,35]. To assess the relative reaction rates for the enzyme-catalyzed conversions described above, we performed His6-GMD incubations in an NMR tube. The progress of the reaction was monitored by acquiring a proton spectrum (1H) every 2.8 min. Time course graphs were created using vnmrj software (Varian, Palo Alto, CA, USA) by plotting the integrals for the anomeric signals for compounds A, B, C and D versus time. Lines of best fit (solid lines) were generated through the data points using vnmrj software. In the reaction containing His6-GMD and NADPH, build-up of compounds B and C in the reaction tube was observed, and these were slowly converted into compound D (Fig. 7B), indicating that the reductase activity of His6-GMD is much slower than its 4,6-dehydratase activity in these conditions. The 4,6-dehydration reaction creating compounds B and C proceeded at very similar rates in the presence and absence of NADPH (Fig. 7A,B).
Fig. 7.
NMR spectroscopy of active enzyme–substrate incubations directly in aqueous reaction buffer. The time course of reactions was monitored by 1H-NMR over a 4 h period. The changing relative concentrations of each sugar nucleotide are shown here in plots of their anomeric signal integrals versus time. GDP-d-Man was incubated with the enzyme(s), with or without NADPH. Coincubation of His6-RMDPa with His6-GMD and NADPH inhibits the 4,6-dehydratase activity of His6-GMD. A, GDP-d-Man; B, GDP-6-deoxy-d-lyxo-hexos-4-ulose, C, gem-diol form of compound B; D, GDP-d-Rha.
We also used this technique to corroborate our observation, by CE analysis, that His6-GMD–His6-RMD–NADPH coincubation inhibits the 4,6-dehydration reaction. We observed the same phenomenon (Fig. 7C). This more sensitive technique revealed that a small amount of compound D was produced but the majority of the GDP-d-Man starting material remained unchanged. Neither of the intermediate compounds, B or C, was detected, indicating that the ketone was converted to compound D faster than it was produced, probably by His6-RMD. The activity of the His6-RMDPa protein preparation used in this His6-GMD–His6-RMDPa–NADPH experiment was confirmed by incubation with the product of the His6-GMD incubation; all of the ~ 25 mm compound B plus compound C present was converted to compound D by His6-RMDPa within 4 min (data not shown).
Kinetic analysis of the His6-GMD GDP-d-Man 4,6-dehydratase activity
To compare the P. aeruginosa GMD with GMDs from other organisms, we determined its kinetic parameters. His6-GMD exhibits non-Michaelis–Menten kinetics producing typical curves corresponding to the substrate inhibition model with the following kinetic parameters: Km = 14.02 ± 6.05 μm; Vmax = 3.64 ± 1.37 μmol·min−1·mg−1; kcat = 8.82 s−1; Ki = 2.859 ± 1.31 μm; kcat/Km = 6.3 × 105 m−1·s−1.
Structural characterization of His6-RMD from A. thermoaerophilus
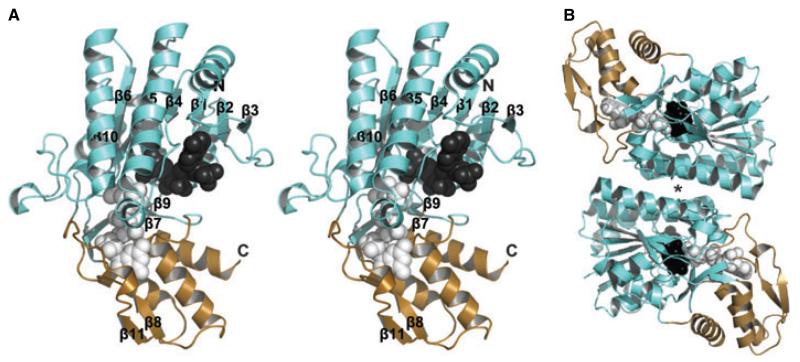
To gain further understanding of the second step in the GDP-d-Rha pathway, we set out to structurally characterize RMD. Attempts to obtain high-quality crystals of His6-RMDPa were unsuccessful, but we were able to determine the crystal structure of His6-RMDAt to 1.8 Å resolution, in complex with the product analog GDP-d-Man and a partially disordered NADP(H) cofactor. The nicotinamide ring was not resolved in the electron density (Fig. 8), so this molecule was modeled into the structure as adenine-phosphoribose-pyrophosphate-ribose (APPR). The protein has the typical architecture of the sugar nucleotide-modifying SDR family, folding into two domains: a Rossmann fold domain, which binds cofactor, and a mixed α/β domain, which binds substrate and confers substrate nucleotide specificity (Fig. 9A). The catalytic triad is located at the interface between these two domains. The structure also exhibits the typical dimer interface for this protein family, consisting of a four-helix bundle, where each monomer provides two helices (Fig. 9B).
Fig. 8.
The RMD active site. Stereoview of the RMD active site showing the refined 2Fo–Fc electron density map around GDP-d-Man and APPR, contoured at 1.0σ. Ser114, Tyr140 and Lys144 form the catalytic triad. No clear electron density is seen for the nicotinamide ring, and even the nicotinamide ribose shows some indications of disorder; in fact, the average B-factor for the nicotinamide ribose is significantly higher than for either the adenine or guanine riboses. An asterisk is placed in the expected position of the disordered nicotinamide moiety. His170 is positioned in part of this ‘open’ space, a change of about 3.4 Å as compared with the position of the equivalent residue, His180, in P. aeruginosa GMD.
Fig. 9.
Structure of RMD from A. thermoaerophilus. (A) Stereoview of the RMD monomer. The cofactor-binding domain and the substrate-binding domain are shown in aqua and light sand, respectively; the APPR portion of the cofactor (dark gray) and the ligand analog GDP-d-Man (light gray) are represented as space-filling models. Termini and secondary structural elements are labeled. (B) View of the RMD homodimeric structure; an asterisk highlights the four-helix bundle, the typical SDR enzyme dimerization mode.
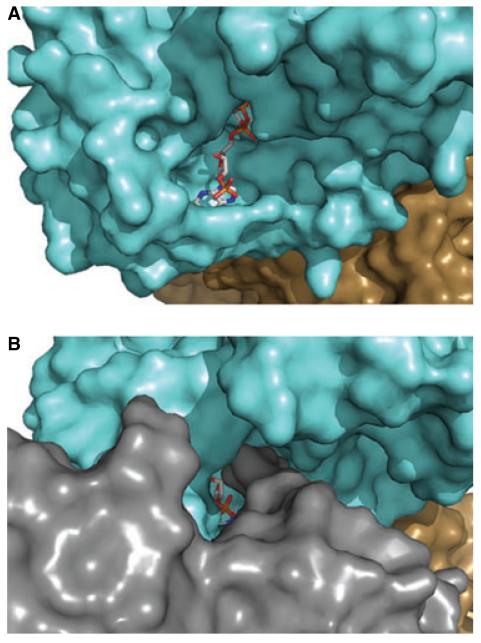
Comparison of the RMD structure with the structure of P. aeruginosa GMD [27] indicates that the dinucleotide cofactor-binding site is more open to solvent in RMD (Fig. 10). The active form of P. aeruginosa GMD is a tetramer, and has a structural feature called the ‘RR loop’ (comprising Arg35–Arg43). The RR loop stretches from each molecule into the adjacent monomer, and undergoes interactions with the neighboring protein and cofactor across the tetramer interface. In sequence alignments with GMD, both the P. aeruginosa and A. thermoaerophilus RMD sequences have gaps in the region of the RR loop, and in the His6-RMDAt structure, the β2–β3 loop is notably shorter than in GMD. The truncation of this structural feature probably explains why the RMD structure does not exhibit a tetramer-forming interaction like GMD, and why the RMD cofactor-binding site is less occluded than in GMD.
Fig. 10.
The RMD cofactor-binding site is readily accessible to solvent. Surface representations of (A) A. thermoaerophilus RMD and (B) P. aeruginosa GMD, looking into the cofactor-binding site. Corresponding monomers from RMD and GMD are colored the same. An additional monomer of the GMD tetramer (gray) significantly reduces the accessibility of cofactor to bulk solvent.
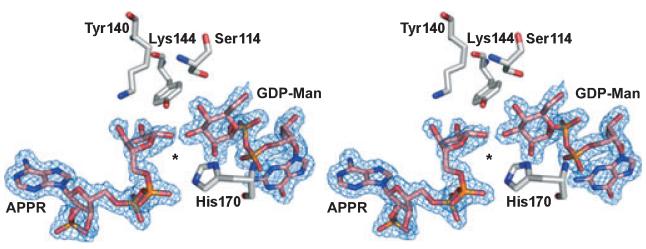
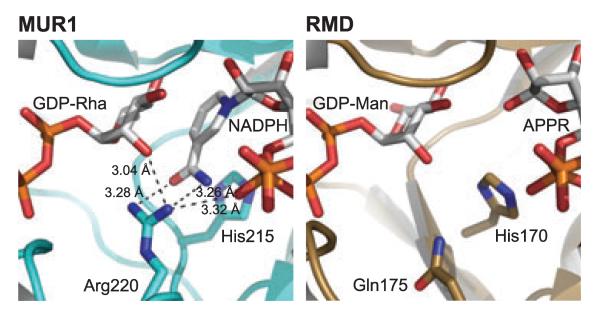
A curious feature of RMD, which was suggested by sequence alignments and confirmed by the RMD structure, is that the active site of this enzyme is very similar to that of GMD. The two protein structures superimpose quite well, with an rmsd of 1.2 Å over 281 equivalent Cα atoms. In addition, not only is the SDR catalytic triad present in the RMD structure (Ser105, Tyr131, and Lys135), but so is the conserved 4,6-dehydratase active site glutamate (Glu128 in P. aeruginosa GMD, and Glu116 in A. thermoaerophilus RMD). This glutamate is proposed to be the active site base that abstracts the C5 proton in the dehydration reaction [36]. Sequence alignments suggest, however, that this glutamate is not conserved in RMD from P. aeruginosa (Asp107 occupies this position). Comparison of other amino acid side chains lining the active sites of A. thermoaerophilus RMD and P. aeruginosa GMD shows that all residues are conserved, with the exception of RMD Gln175 (Arg185 in GMD). This arginine is conserved in all characterized GMD sequences, and in the Ar. thaliana MUR1 structure this side chain is close enough to suggest hydrogen-bonding interactions with a cofactor phosphate, the nicotinamide carboxyamide, and the rhamnosyl O2 hydroxyl of the substrate analog (Fig. 11). The degree of conservation for Gln175 among RMD enzymes is unclear, because so few bona fide RMDs have been identified and characterized. In a blast search (using the blastp algorithm [37]) of the P. aeruginosa RMD sequence, however, 89 of the top 100 hits had glutamine in this position; 10 of the others had arginine in its place, and the final variant had glutamate.
Fig. 11.
The potential hydrogen-bonding interactions of a conserved GMD arginine. The active sites of A. thermoaerophilus RMD and Ar. thaliana MUR1 are shown in equivalent orientations for comparison. MUR1 Arg220 is conserved in all GMDs, and during catalysis may coordinate with a cofactor phosphate, the substrate hexose, and the nicotinamide carboxyamide. The distances between these groups in the MUR1 crystal structure are indicated. In the RMD structure, the position of the MUR1 Arg220 is occupied by a glutamine, and this amino acid side chain is too short to mediate the same protein–ligand interactions. This may account for the disordering of the nicotinamide ring in the RMD crystal.
Discussion
We present the biochemical characterization of His6-GMD and His6-RMD from P. aeruginosa. Despite being the focus of some research interest in the past, including the publication of the P. aeruginosa GMD crystal structure [27], the enzymatic functions of these proteins have not previously been characterized using in vitro assays with purified proteins. Their functions had only been inferred from genetic experiments [23,24] and functional assays using cell lysates as enzyme source [23,25]. We have reconstituted the pathway in vitro using both P. aeruginosa proteins, and used NMR to unambiguously identify the reaction products (Fig. 1). We have also defined conditions for purification, long-term storage, and the performance of enzyme–substrate incubations, so that these enzymes can be used as synthetic tools to prepare GDP-d-Rha, or its 4-keto precursor. The stability of P. aeruginosa His6-GMD and His6-RMDPa makes them suitable for this application, and the kinetic parameters for P. aeruginosa GMD are comparable with those of GMD enzymes from other organisms [12,16,38–40].
Bifunctionality of P. aeruginosa GMD
We observed that P. aeruginosa GMD, like the enzymes from K. pneumoniae, A. thermoaerophilus, and PBCV-1 [5,16,17], is able to catalyze the reductase reaction leading to GDP-d-Rha. This is consistent with previous observations: when P. aeruginosa GMD was expressed from plasmid pFV39 (which contains the full-length gmd gene and a nonfunctional fragment of rmd that lacks the first 97 rmd codons), it was able to catalyze the conversion of GDP-d-Man to GDP-d-Rha [23], although this assay was conducted with E. coli cell lysates, and the reaction product was only identified at that time by paper chromatography. The ability of GMD to catalyze the reduction reaction indicates that exchange of cofactor with solution must be possible for this enzyme. In the current understanding of the mechanism, the 4,6-dehydratase reaction catalyzed by these enzymes involves an initial oxidation of the sugar nucleotide, followed by subsequent reduction, and the cofactor, presumed to be bound as NAD(P)+ in the resting state, is therefore regenerated in the catalytic cycle [31]. Conversely, reduction of GDP-6-deoxy-d-lyxo-hexos-4-ulose to GDP-d-Rha requires the formation of an initial enzyme–NAD(P)H complex, whereupon the cofactor is oxidized to NAD(P)+ during the reaction. Therefore, the reduced cofactor must be replaced from the solution before the next reaction cycle. Recent evidence suggests that several different GMDs contain a proportion or majority of cofactor bound in the reduced state, and that this is important for protein stability in solution [38]. Facile exchange of cofactor with bulk solution is sometimes reflected by binding of cofactor in a solvent-exposed groove (e.g. RmlD [41]). In contrast, the P. aeruginosa GMD structure [27] shows that the cofactor’s access to solvent is blocked in large part by interactions with the RR loop. Given that the dimer–dimer interface in P. aeruginosa GMD is apparently stabilized by interactions between cofactor and the neighboring monomer, it is conceivable that oxidation of the cofactor may alter the conformation of the RR loop or destabilize the dimer–dimer interface in a manner that allows cofactor exchange. In support of this hypothesis, the oligomerization state of PBCV-1 GMD is responsive to the oxidation state of bound NADP. In this viral enzyme, addition of NADPH, but not NADP+, induces dimerization of the apoenzyme, and oxidation of the bound NADPH results in dimer dissociation [38].
Unlike the case of PBCV-1, where the GMD has a higher specific activity as a reductase than as a 4,6-dehydratase, the bifunctionality of P. aeruginosa GMD is unlikely to be metabolically significant in vivo, at least in terms of biosynthesis, because P. aeruginosa expresses a dedicated reductase, RMD, to perform this synthetic step. It is still possible, however, that the GMD-catalyzed reductase reaction is functionally important, either in regulation of enzyme activity or as a mechanism to change the oxidation state of bound cofactor. There are properties of GMDs, e.g. stimulation of catalytic activity by addition of micromolar NADPH [39] and the exclusive presence of NADPH in GMD crystals, which are unexplained by the current mechanism [38].
Feedback inhibition
Our observation of strong inhibition of GDP-d-Man consumption by His6-GMD when incubated with NADPH and His6-RMD indicates that a feedback mechanism inhibits the 4,6-dehydratase activity of His6-GMD in these conditions. Such feedback inhibition, by which a sugar nucleotide controls the rate of its own synthesis, is not unusual, and is well documented for GMDs: there are multiple examples of organisms that incorporate l-fucose into oligosaccha-rides or polysaccharides, where GMD is inhibited by GDP-l-fucose [12,13,15,28,42,43]. Presumably, this feedback inhibition has evolved to prevent build-up of excess GDP-l-fucose and/or excessive consumption of the starting material. In the case of P. aeruginosa GMD, tight control of GDP-d-Man consumption may be important, because this sugar nucleotide is also an intermediate in the biosynthetic pathway for another virulence factor, alginate [44].
In preliminary experiments to elucidate the inhibitory mechanism, we have observed that strong inhibition of the His6-GMD reaction only occurs in the presence of His6-RMD, raising the possibility that the mechanism of inhibition involves a GMD–RMD protein–protein interaction. The reaction was also strongly inhibited when His6-GMD was incubated with His6-RMD and NADP+, which rules out the possibility that GMD is inhibited simply by the exchange of bound NADP+ with NADPH preventing the first oxidative step of the 4,6-dehydratase reaction (data not shown). At the present time, the inhibitory mechanism remains unclear, but this will be an interesting subject for further study.
RMD structure
The similarity of the A. thermoaerophilus RMD structure to GMD structures is, in some respects, unsurprising. Where a bifunctional GMD enzyme is able to catalyze the same reaction as RMD, a close resemblance between the two active sites makes sense, at least as far as substrate binding and the SDR Ser/Thr-Tyr-Lys catalytic triad are concerned. What is more intriguing is that all of the amino acid side chains that have been proposed to function in acid–base catalysis of the 4,6-dehydratase reaction of GMD [27] are conserved in A. thermoaerophilus RMD. The conservation of these residues has been noted previously [45]; the RMD structure confirms that their orientation in space is also conserved. Why, then, is this RMD protein unable to catalyze the GDP-d-Man 4,6-dehydration reaction? Previously, the absence of such potential catalytic side chains has been used to rule out possible functions for SDRs [46]. The structure of RMD that we report here emphasizes that the inverse argument does not apply: the presence of such residues does not necessarily mean that the catalytic competence is likewise conserved. The disordered nature of the NADP nicotinamide ring in the RMD crystal indicated, however, an important difference between the two active sites. We propose that Arg185 in P. aeruginosa GMD is important for aligning NADP and GDP-d-Man in the active site for the dehydratase reaction. This role is suggested by the close relative positions of the corresponding side chain, Arg220, the NADPH cofactor and the substrate analog hexose in the MUR1 structure (Fig. 11). Some of the distances between these groups are rather long for classic hydrogen bonding, but relative motion of ligand molecules is expected during the catalytic cycle: In the GMD reaction, the nicotinamide must extract hydride from the substrate C4′, and later donate it back at C6′. In RMDAt, the amino acid occupying the position of the conserved GMD arginine is Gln175, and the side chain of this residue is too short to undergo these interactions. This may be the reason why a productive ternary RMD–NADP+–GDP-d-Man complex cannot assemble in the configuration necessary for this reaction. The bioinformatics analysis suggested that Arg185 is absolutely conserved among GMDs from diverse organisms, and that Gln175 is well conserved among close RMD homologs. The 10 RMD homolog sequences examined that had arginine in this position may, in fact, represent GMDs. We are currently working to test experimentally whether exchange of arginine and glutamine at this position can interconvert the catalytic functions of these GMD and RMD enzymes. Subject to this experimental verification, this current report may have helped to identify a diagnostic amino acid for distinction of GMD/RMD enzyme functions from sequence alone. As has been previously discussed [46], such indicators are important to make full use of the vast amount of sequence information available in the genome databases, and to provide useful indicators for the accurate annotation of this important class of enzymes.
Conclusions
We have verified, biochemically, the functions of GMD and RMD from P. aeruginosa, and showed that GMD from this organism is a bifunctional 4,6-dehydratase and a stereospecific 4-reductase. Reconstitution of the P. aeruginosa GDP-d-Rha pathway in vitro revealed a feedback mechanism inhibiting the first step that may be important for the regulation of GDP-d-Man consumption. Finally, structural analysis of RMD from A. thermoaerophilus identified an amino acid, Arg185, in P. aeruginosa GMD that may be critical for correct orientation of GDP-d-Man and NADP+ cofactor for the 4,6-dehydration reaction. The corresponding residue in RMDAt is Gln175. The amino acid in this position may be a key indicator of specificity among these closely related GMD/RMD enzymes.
Experimental procedures
Materials
Unless stated otherwise, all materials were purchased from Sigma-Aldrich (Oakville, Canada).
DNA methods
The rmd genes from P. aeruginosa and A. thermoaerophilus were amplified from chromosomal DNA templates, using the following primer pairs: P. aeruginosa, 5′-AGGCCCGCTTCCGGATCCACTCAGCGTCTG-3′ and 5′-AAAAAAGTCGACTTCTTCTCGTACCCGTGACTC-3′; and A. thermoaerophilus, 5′-TTGGATCCATGAGAGCCCTAATCACTGGA-3′ and 5′-TAGGTACCTTATGCTTGACGGTAACTTTGT-3′. Restriction enzyme sites included on the primers are underlined. The amplified P. aeruginosa gene was ligated into pQE-30 (Qiagen, Missisauga, Canada), using BamHI and SalI restriction sites. The A. thermoaerophilus gene was cloned into the BamHI–KpnI-digested pQE-80 (Qiagen). Both expression vectors encode N-terminal His6-tag fusions of their respective RMD proteins.
Protein expression and purification
His6-GMD was expressed from the pQE-30-gmd vector, which has been described previously [27]. His6-tagged RMD proteins (His6-RMDPa and His6-RMDAt) were expressed from the vectors described above, and E. coli M15(pREP4) or E. coli BL21(DE3) was used as the host strain. Cells from an overnight culture were used to inoculate 1 L of LB broth. When the attenuance at 600 nm (D600 nm) reached 0.5–0.6, protein expression was induced by addition of isopropyl-thio-β-d-galactoside to 0.25 mm, and the cultures were shaken for a further 16 h at room temperature. Cells were harvested by centrifugation (10 000 g, 10 min), and then suspended in 50 mm Hepes, 300 mm NaCl, and 5 mm imidazole (pH 8.0). Cell lysis was performed with ultrasonication. After centrifugation (10 000 g, 20 min), the supernatant was passed through a 4 mL column of Ni2+–nitrilotriacetic acid resin (Qiagen). The column was thoroughly washed (50 mm Hepes, 300 mm NaCl, 20 mm imidazole, pH 7.5), and then eluted by increasing the imidazole concentration to 200 mm in the same buffer. Proteins were then purified by anion exchange chromatography using a 1 mL HiTrap-Q column (GE Healthcare, Baie d’Urfé, Canada), pre-equilibrated with 20 mm Tris/HCl (pH 8.5), and eluted with a linear, 40 mL gradient from 0 to 1 m NaCl. For NMR analysis of enzyme incubations, proteins were buffer-exchanged using a PD-10 column (GE Healthcare), into 25 mm sodium phosphate and 50 mm NaCl (pH 7.5). For long-term storage, 25% glycerol was added to protein aliquots, which were then frozen (−80 °C).
Enzyme–substrate incubations
The enzymes used for in vitro assays were the N-terminal His6-tagged fusions of the P. aeruginosa proteins. Unless otherwise stated, enzyme–substrate incubations were performed in 40 mm Tris/HCl (pH 7.5) and 10 mm MgCl2 with 1.0 mm GDP-d-Man. In reactions that were to be analyzed by CE, NADP+ and/or NADPH was added at 0.01 or 0.1 mm to assist in alignment of CE traces. In reductase reactions requiring cofactor as a reagent, NADPH was added in molar excess with respect to the sugar nucleotide substrate. Reactions were started by the addition of enzyme, and were incubated at 37 °C. To determine enzyme catalytic activities with GDP-6-deoxy-d-lyxo-hexos-4-ulose, a GMD reaction was performed for 1 h, the enzyme was removed by filtration, and then enzyme and cofactor for the second reaction were added. To determine the optimal pH for enzyme-catalyzed reactions, the extent of substrate conversion was determined after 5 min in the standard reaction mixture, but with Mes (pH 5, 5.5, 6, and 6.5) or Bis/Tris propane (pH 7, 7.5, 8, 8.5, 9, 9.5, and 10) in place of Tris/HCl (pH 7.5).
CE
CE analyses were performed using a P/ACE MDQ Glycoprotein System (Beckman Coulter, Fullerton, CA, USA), using a bare silica 75 μm × 57 cm capillary and a running buffer consisting of 25 mm sodium tetraborate (pH 9.5). Compound elution was monitored by measuring UV absorbance at 254 nm, with the UV detector positioned at 50 cm. The capillary was preconditioned before each run by washing with 0.2 m sodium hydroxide, water, and running buffer, each for 2 min. Samples were introduced by pressure injection for 8 s (for reaction composition analysis) or 24 s (for kinetic analysis), and separation was performed at 22 kV. Peak integration was performed using 32 karat software (Beckman).
Determination of kinetic parameters for GMD
Reactions were performed in triplicate, and contained 0.25 μg of protein and 0.5–40 μm GDP-d-Man in a total volume of 1 mL. Samples were incubated at 37 °C for 5 min, the reaction was stopped by flash freezing in a dry ice/ethanol bath, and this was followed by transfer to a boiling water bath for 10 min to denature the enzymes. Samples were refrozen, lyophilized and finally suspended in 25 μL of water prior to analysis by CE. Because of the instability of the ketone product in the enzyme-inactivation procedure, reaction product quantities were normalized by comparison with no-enzyme controls. Data were fitted to the substrate inhibition equation v = Vmax/(1 + Km/[S] + [S]/Ki), and the kinetic parameters were determined by nonlinear regression using the sigmaplot enzyme kinetics module. In the course of these experiments, we found that MgCl2 has a slight inhibitory effect on GMD activity (data not shown), and so all kinetic parameters were measured in the absence of MgCl2.
Purification of the product of the GMD/RMD sequential reaction (GDP-d-Rha)
In a large-scale reaction, 30 μmol of GDP-d-Man was completely converted to GDP-d-Rha in sequential reactions catalyzed by His6-GMD and then His6-RMD (1.5 mg of each enzyme) with a molar excess of NADPH. Protein was removed from the completed reaction by ultrafiltration through a Centriplus YM-3 cartridge (Millipore, Billerica, MA, USA). The reaction product was then purified as previously reported [47]. Briefly, the filtrate was subjected to anion exchange chromatography using an Econo-Pac High Q anion exchange column (Bio-Rad, Hercules, CA, USA) with a linear gradient of 0–500 mm triethylammonium bicarbonate (pH 8.0). Fractions were monitored by CE, and those containing the sugar nucleotide were pooled. Bicarbonate was removed (as CO2 gas) by addition of H+-charged AG 50W-X4 resin (Bio-Rad) until pH 4.5 was achieved. The resin was removed by filtration, and the solution was neutralized by the addition of triethylamine. Finally, water and triethylamine were removed by lyophilization.
NMR spectroscopy
NMR experiments were performed at 500 MHz (1H) in 10% D2O (90% H2O) or 99% D2O with a Varian Z-gradient 3 mm triple resonance (1H, 13C, 31P) probe (Varian). Standard homonuclear-correlated and heteronuclear-correlated 2D pulse sequences from Varian, such as COSY, TOCSY, HSQC, HMBC and 31P-HMQC, were used for general assignments. NMR experiments were typically performed at 25 °C with suppression of the deuterated H2O resonance. The methyl resonance of acetone was used as an internal reference at δH = 2.225 p.p.m. and δC = 31.07 p.p.m. Selective 1D-TOCSY experiments with a Z-filter, as well as 1D-NOESY experiments, were used for complete residue assignment and for the determination of JH,H coupling constants [48,49]. In initial experiments (data shown in Fig. 4) to observe the products of GMD-catalyzed, ‘in-NMR-tube’ reactions, the enzyme was first placed in a 3 mm NMR tube suspended in 200 μL of its reaction buffer (25 mm NaPO4, 50 mm NaCl, pH 7.2, 90% H2O/10% D2O). The reaction was started by the addition of 5 mm GDP-d-Man, ± 5 mm NADPH, to the reaction buffer, and the proton spectrum was taken at the start of the reaction and again 16 h later. To follow His6-GMD-catalyzed and His6-RMD-catalyzed reactions over time (for the data shown in Fig. 7), 25 mm GDP-d-Man was incubated with 7.5 lg·mL−1 each enzyme, ± 25 mm NADPH, in 25 mm NaPO4, 50 mm NaCl (pH 7.2), and 90% H2O/10% D2O. A proton spectrum was acquired every 2.8 min over a 4 h period.
Crystallography
Purified His6-RMDAt (i.e. the tagged A. thermoaerophilus protein) was concentrated to 10 mg·mL−1, and crystals were grown by the sitting drop vapor diffusion method in 35% pentaerythritol propoxylate (5/4 PO/OH; Hampton Research, Aliso Viejo, CA, USA), 100 mm Tris (pH 8.5), and 200 mm NaCl. As the natural substrate is unstable and the natural product is not available commercially, the product analog GDP-d-Man was added to the crystallization conditions, along with NADP or NADPH. The presence of GDP-d-Man was found to be absolutely necessary for crystal growth. The best crystals grew in a combination of 5 mm GDP-d-Man and NADPH. Prior to data collection, crystals were flash-frozen in liquid nitrogen directly from the crystallization drop.
X-ray diffraction data were collected at a wavelength of 1.0 Å on a MAR CCD detector at the Advanced Photon Source Beamline 5-ID (DND) (Argonne National Laboratory). Crystals were held at 100 K in a cryostream during data collection. Data were processed using xds software [50], and statistics are shown in Table 2.
Table 2.
Data collection and refinement statistics. R-factor = ∑∣Fo–Fc∣/∑Fo, where Fo and Fc are observed and calculated structure factors, respectively. R-free is the cross-validation R-factor computed for the test set of 3207 reflections (5% of the total unique reflections).
| Data collection statistics | |
| Space group | P1 |
| Unit cell parameters (Å , °) | a = 46.88, b = 55.74, c = 79.24, α = 72.54, β = 82.95, γ = 75.61 |
| Resolution range (Å ) | 30.0–1.8 |
| No. of observed reflections | 215 748 |
| No. of unique reflections | 64 129 |
| Completeness (%) | 96.5 (95.2)a |
| B-factor from Wilson plot | 20.35 |
| Rmerge (%) | 7.6 (43.0)a |
| Average I/σ (I) | 13.4 (3.7)a |
| Refinement statistics | |
| No. of residues | 618/618 |
| No. of water molecules | 350 |
| No. of heteromolecules | 4 |
| R factor | 16.5 |
| R free | 19.8 |
| rmsd bond lengths (Å ) | 0.014 |
| rmsd bond angles (°) | 1.47 |
| Average B-factor (Å2) | 25.6 |
| Ramachandran | |
| Most favored (%) | 93.8 |
| Allowed (%) | 6.2 |
Indicates statistic from highest-resolution shell.
The structure of RMD was determined by molecular replacement with mrbump (R. M. Keegan & M. D. Winn, unpublished data) in conjunction with the ccp4 suite [51], using a search model based on the known structure of GMD from P. aeruginosa (29% sequence identity; Protein Data Bank code: 1RPN [27]). arp/warp [52] was used for an initial round of automatic model building and refinement of the protein portion. The initial 2Fo–Fc electron density maps from arp/warp revealed clear density for a GDP-sugar and the APPR portion of the cofactor. Further refinement was carried out using the TLS option in ref-mac5 [53], alternated with manual model building in coot [54] using the 2Fo–Fc and Fo–Fc maps. Restraint libraries were constructed for APPR and GDP-d-Man using sketcher [51]. Water molecules were added using coot, and checked for accuracy by hand. The final model (Rfactor = 16.5%, Rfree = 19.8%) consists of an RMD dimer, 350 water molecules, and two molecules each of APPR and GDP-d-Man. Although the density is a little weak in one loop area (residues 33–36), there is clear density for residues 1–309 of 309 residues in each monomer. The model conforms ideally to the geometry defined by procheck [55]. The refinement statistics are presented in Table 2, and the final coordinates have been deposited in the Protein Data Bank under the accession number 2PK3.
Acknowledgements
We are grateful to E. F. Mulrooney for technical assistance. This work was supported by an operating grant to J. S. Lam from the Canadian Cystic Fibrosis Foundation (CCFF) and the Canadian Institutes of Health Research (grant no. MOP14687), a grant (no. P18013-B10) to P. Messner from the Austrian Science Fund, and a grant from the NIH (GM65501) to R. M. Garavito. K. K. H. Poon was a recipient of a postdoctoral fellowship from CCFF. J. S. Lam holds a Canada Research Chair in Cystic Fibrosis and Microbial Glycobiology funded jointly by the Canadian Foundation of Innovation and the Ontario Innovation Trust.
Abbreviations
- APPR
adenine-phosphoribose-pyrophosphate-ribose
- CE
capillary electrophoresis
- D-Man
α-D-mannose
- D-Rha
α-D-rhamnose
- GMD
GDP-D-mannose-4,6-dehydratase
- HMBC
heteronuclear multiple bond correlation
- HSQC
heteronuclear single-quantum correlation
- LPS
lipopolysaccharide
- PBCV
Paramecium bursaria chlorella virus
- RMD
GDP-6-deoxy-D-lyxo-hexos-4-ulose-4-reductase (GDP-D-rhamnose forming)
- SDR
short-chain dehydrogenase/reductase
Footnotes
Database Protein structure model data are available in the Protein Data Bank database under the accession number 2PK3
References
- 1.Kocharova NA, Knirel YA, Widmalm G, Jansson PE, Moran AP. Structure of an atypical O-antigen polysaccharide of Helicobacter pylori containing a novel monosaccharide 3-C-methyl-d-mannose. Biochemistry. 2000;39:4755–4760. doi: 10.1021/bi992635k. [DOI] [PubMed] [Google Scholar]
- 2.Senchenkova SN, Shashkov AS, Knirel YA, McGovern JJ, Moran AP. The O-specific polysaccha-ride chain of Campylobacter fetus serotype B lipopolysaccharide is a d-rhamnan terminated with 3-O-methyl-d-rhamnose (d-acofriose) Eur J Biochem. 1996;239:434–438. doi: 10.1111/j.1432-1033.1996.0434u.x. [DOI] [PubMed] [Google Scholar]
- 3.Molicaro A, Silip A, Lanzetta R, Newman MA, Dow JM, Parrilli M. Structural elucidation of the O-chain of the lipopolysaccharide from Xanthomonas campestris strain 8004. Carbohydr Res. 2003;338:277–281. doi: 10.1016/s0008-6215(02)00433-0. [DOI] [PubMed] [Google Scholar]
- 4.Wang IN, Li Y, Que Q, Bhattacharya M, Lane LC, Chaney WG, Van Etten JL. Evidence for virus-encoded glycosylation specificity. Proc Natl Acad Sci USA. 1993;90:3840–3844. doi: 10.1073/pnas.90.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kneidinger B, Graninger M, Adam G, Puchberger M, Kosma P, Zayni S, Messner P. Identification of two GDP-6-deoxy-d-lyxo-4-hexulose reductases synthesizing GDP-d-rhamnose in Aneurinibacillus thermoaerophilus L420-91T. J Biol Chem. 2001;276:5577–5583. doi: 10.1074/jbc.M010027200. [DOI] [PubMed] [Google Scholar]
- 6.Persson B, Kallberg Y, Oppermann U, Jornvall H. Coenzyme-based functional assignments of short-chain dehydrogenases/reductases (SDRs) Chem Biol Interact. 2003:143–144. 271–278. doi: 10.1016/s0009-2797(02)00223-5. [DOI] [PubMed] [Google Scholar]
- 7.Chang S, Duerr B, Serif G. An epimerasereductase in l-fucose synthesis. J Biol Chem. 1988;263:1693–1697. [PubMed] [Google Scholar]
- 8.Maki M, Jarvinen N, Rabina J, Maaheimo H, Mattila P, Renkonen R. Cloning and functional expression of a novel GDP-6-deoxy-d-talose synthetase from Actinobacillus actinomycetemcomitans. Glycobiology. 2003;13:295–303. doi: 10.1093/glycob/cwg035. [DOI] [PubMed] [Google Scholar]
- 9.Markovitz A. Biosynthesis of guanosine diphosphate d-rhamnose and guanosine diphosphate d-talomethylose from guanosine diphosphate alpha-d-mannose. J Biol Chem. 1964;239:2091–2098. [PubMed] [Google Scholar]
- 10.Albermann C, Piepersberg W. Expression and identification of the RfbE protein from Vibrio cholerae O1 and its use for the enzymatic synthesis of GDP-d-perosamine. Glycobiology. 2001;11:655–661. doi: 10.1093/glycob/11.8.655. [DOI] [PubMed] [Google Scholar]
- 11.Bonin CP, Potter I, Vanzin GF, Reiter WD. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-d-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-l-fucose. Proc Natl Acad Sci USA. 1997;94:2085–2090. doi: 10.1073/pnas.94.5.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bisso A, Sturla L, Zanardi D, De Flora A, Tonetti M. Structural and enzymatic characterization of human recombinant GDP-d-mannose-4,6-dehydratase. FEBS Lett. 1999;456:370–374. doi: 10.1016/s0014-5793(99)00982-5. [DOI] [PubMed] [Google Scholar]
- 13.Sturla L, Bisso A, Zanardi D, Benatti U, De Flora A, Tonetti M. Expression, purification and characterization of GDP-d-mannose 4,6-dehydratase from Escherichia coli. FEBS Lett. 1997;412:126–130. doi: 10.1016/s0014-5793(97)00762-x. [DOI] [PubMed] [Google Scholar]
- 14.Broschat KO, Chang S, Serif G. Purification and characterization of GDP-d-mannose 4,6-dehydratase from porcine thyroid. Eur J Biochem. 1985;153:397–401. doi: 10.1111/j.1432-1033.1985.tb09315.x. [DOI] [PubMed] [Google Scholar]
- 15.Rhomberg S, Fuchsluger C, Rendic D, Paschinger K, Jantsch V, Kosma P, Wilson IB. Reconstitution in vitro of the GDP-fucose biosynthetic pathways of Caenorhabditis elegans and Drosophila melanogaster. FEBS J. 2006;273:2244–2256. doi: 10.1111/j.1742-4658.2006.05239.x. [DOI] [PubMed] [Google Scholar]
- 16.Tonetti M, Zanardi D, Gurnon JR, Fruscione F, Armirotti A, Damonte G, Sturla L, De Flora A, Van Etten JL. Paramecium bursaria chlorella virus 1 encodes two enzymes involved in the biosynthesis of GDP-l-fucose and GDP-d-rhamnose. J Biol Chem. 2003;278:21559–21565. doi: 10.1074/jbc.M301543200. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto K, Katayama I, Onoda Y, Inami M, Kumagai H, Tochikura T. Evidence that the enzyme catalyzing the conversion of guanosine diphosphate d-mannose to a 4-keto sugar nucleotide intermediate requires nicotinamide adenine dinucleotide phosphate. Arch Biochem Biophys. 1993;300:694–698. doi: 10.1006/abbi.1993.1096. [DOI] [PubMed] [Google Scholar]
- 18.Emori TG, Gaynes RP. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivera M, McGroarty EJ. Analysis of a common-antigen lipopolysaccharide from Pseudomonas aeruginosa. J Bacteriol. 1989;171:2244–2248. doi: 10.1128/jb.171.4.2244-2248.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam MY, McGroarty EJ, Kropinski AM, MacDonald LA, Pedersen SS, Høiby N, Lam JS. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J Clin Microbiol. 1989;27:962–967. doi: 10.1128/jcm.27.5.962-967.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rocchetta HL, Burrows LL, Pacan JC, Lam JS. Three rhamnosyltransferases responsible for assembly of the A-band d-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol Microbiol. 1998;28:1103–1119. doi: 10.1046/j.1365-2958.1998.00871.x. [DOI] [PubMed] [Google Scholar]
- 22.Rocchetta HL, Lam JS. Identification and functional characterization of an ABC transport system involved in polysaccharide export of A-band lipopolysaccharide in Pseudomonas aeruginosa. J Bacteriol. 1997;179:4713–4724. doi: 10.1128/jb.179.15.4713-4724.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lightfoot J, Lam JS. Chromosomal mapping, expression and synthesis of lipopolysaccharide in Pseudomonas aeruginosa: a role for guanosine diphospho (GDP)-D-mannose. Mol Microbiol. 1993;8:771–782. doi: 10.1111/j.1365-2958.1993.tb01620.x. [DOI] [PubMed] [Google Scholar]
- 24.Rocchetta HL, Pacan JC, Lam JS. Synthesis of the A-band polysaccharide sugar d-rhamnose requires Rmd and WbpW: identification of multiple AlgA homologues, WbpW and ORF488, in Pseudomonas aeruginosa. Mol Microbiol. 1998;29:1419–1434. doi: 10.1046/j.1365-2958.1998.01024.x. [DOI] [PubMed] [Google Scholar]
- 25.Maki M, Jarvinen N, Rabina J, Roos C, Maaheimo H, Renkonen R. Functional expression of Pseudomonas aeruginosa GDP-4-keto-6-deoxy-d-mannose reductase which synthesizes GDP-rhamnose. Eur J Biochem. 2002;269:593–601. doi: 10.1046/j.0014-2956.2001.02688.x. [DOI] [PubMed] [Google Scholar]
- 26.Markovitz A. Isolation of d-talomethylose (6-deoxy-d-talose) and d-rhamnose (6-deoxy-d-man-nose) from capsular polysaccharide of a Gram-negative bacterium. J Biol Chem. 1962;237:1767–1771. [PubMed] [Google Scholar]
- 27.Webb NA, Mulichak AM, Lam JS, Rocchetta HL, Garavito RM. Crystal structure of a tetrameric GDP-d-mannose 4,6-dehydratase from a bacterial GDP-d-rhamnose biosynthetic pathway. Protein Sci. 2004;13:529–539. doi: 10.1110/ps.03393904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somoza JR, Menon S, Schmidt H, Joseph-McCarthy D, Dessen A, Stahl ML, Somers WS, Sullivan FX. Structural and kinetic analysis of Escherichia coli GDP-mannose 4,6 dehydratase provides insights into the enzyme’s catalytic mechanism and regulation by GDP-fucose. Structure. 2000;8:123–135. doi: 10.1016/s0969-2126(00)00088-5. [DOI] [PubMed] [Google Scholar]
- 29.Mulichak AM, Bonin CP, Reiter WD, Garavito RM. Structure of the MUR1 GDP-mannose 4,6-dehydratase from Arabidopsis thaliana: implications for ligand binding and specificity. Biochemistry. 2002;41:15578–15589. doi: 10.1021/bi0266683. [DOI] [PubMed] [Google Scholar]
- 30.Rosano C, Zuccotti S, Sturla L, Fruscione F, Tonetti M, Bolognesi M. Quaternary assembly and crystal structure of GDP-d-mannose 4,6-dehydratase from Paramecium bursaria chlorella virus. Biochem Biophys Res Commun. 2006;339:191–195. doi: 10.1016/j.bbrc.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Oths PJ, Mayer RM, Floss HG. Stereochemistry and mechanism of the GDP-mannose dehydratase reaction. Carbohydr Res. 1990;198:91–100. doi: 10.1016/0008-6215(90)84279-4. [DOI] [PubMed] [Google Scholar]
- 32.McNally DJ, Schoenhofen IC, Mulrooney EF, Whitfield DM, Vinogradov E, Lam JS, Logan SM, Brisson JR. Identification of labile UDP-ketosugars in Helicobacter pylori, Campylobacter jejuni and Pseudomonas aeruginosa: key metabolites used to make glycan virulence factors. Chembiochem. 2006;7:1865–1868. doi: 10.1002/cbic.200600298. [DOI] [PubMed] [Google Scholar]
- 33.Kanipes MI, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. A mannosyl transferase required for lipopolysaccharide inner core assembly in Rhizobium leguminosarum. Purification, substrate specificity, and expression in Salmonella waaC mutants. J Biol Chem. 2003;278:16356–16364. doi: 10.1074/jbc.M301255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenhofen IC, McNally DJ, Vinogradov E, Whitfield D, Young NM, Dick S, Wakarchuk WW, Brisson JR, Logan SM. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J Biol Chem. 2006;281:723–732. doi: 10.1074/jbc.M511021200. [DOI] [PubMed] [Google Scholar]
- 35.Pfoestl A, Hofinger A, Kosma P, Messner P. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-d-galactose in Aneurinibacillus thermoaerophilus L420-91T. J Biol Chem. 2003;278:26410–26417. doi: 10.1074/jbc.M300858200. [DOI] [PubMed] [Google Scholar]
- 36.Gross JW, Hegeman AD, Gerratana B, Frey PA. Dehydration is catalyzed by glutamate-136 and aspartic acid-135 active site residues in Escherichia coli dTDP-glucose 4,6-dehydratase. Biochemistry. 2001;40:12497–12504. doi: 10.1021/bi011138c. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fruscione F, Sturla L, Duncan G, Van Etten JL, Valbuzzi P, De Flora A, Di Zanni E, Tonetti M. Differential role of NADP+ and NADPH in the activity and structure of GDP-d-mannose 4,6-dehydratase from two chlorella viruses. J Biol Chem. 2008;283:184–193. doi: 10.1074/jbc.M706614200. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan FX, Kumar R, Kriz R, Stahl M, Xu GY, Rouse J, Chang XJ, Boodhoo A, Potvin B, Cumming DA. Molecular cloning of human GDP-mannose 4,6-dehydratase and reconstitution of GDP-fucose bio-synthesis in vitro. J Biol Chem. 1998;273:8193–8202. doi: 10.1074/jbc.273.14.8193. [DOI] [PubMed] [Google Scholar]
- 40.Wu B, Zhang Y, Wang PG. Identification and characterization of GDP-d-mannose 4,6-dehydratase and GDP-l-fucose synthetase in a GDP-l-fucose bio-synthetic gene cluster from Helicobacter pylori. Biochem Biophys Res Commun. 2001;285:364–371. doi: 10.1006/bbrc.2001.5137. [DOI] [PubMed] [Google Scholar]
- 41.Blankenfeldt W, Kerr ID, Giraud MF, McMiken HJ, Leonard G, Whitfield C, Messner P, Graninger M, Naismith JH. Variation on a theme of SDR. dTDP-6-deoxy-l-lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent dimerization mode. Structure. 2002;10:773–786. doi: 10.1016/s0969-2126(02)00770-0. [DOI] [PubMed] [Google Scholar]
- 42.Albermann C, Distler J, Piepersberg W. Preparative synthesis of GDP-beta-l-fucose by recombinant enzymes from enterobacterial sources. Glycobiology. 2000;10:875–881. doi: 10.1093/glycob/10.9.875. [DOI] [PubMed] [Google Scholar]
- 43.Kornfeld RH, Ginsburg V. Control of synthesis of guanosine 5′-diphosphate d-mannose and guanosine 5′-diphosphate l-fucose in bacteria. Biochim Biophys Acta. 1966;117:79–87. doi: 10.1016/0304-4165(66)90154-1. [DOI] [PubMed] [Google Scholar]
- 44.Deretic V, Gill JF, Chakrabarty AM. Gene algD coding for GDP-mannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987;169:351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maki M, Renkonen R. Biosynthesis of 6-deoxyhexose glycans in bacteria. Glycobiology. 2004;14:1R–15R. doi: 10.1093/glycob/cwh040. [DOI] [PubMed] [Google Scholar]
- 46.King JD, Harmer NJ, Preston A, Palmer CM, Rejzek M, Field RA, Blundell TL, Maskell DJ. Predicting protein function from structure – the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis. J Mol Biol. 2007;374:749–763. doi: 10.1016/j.jmb.2007.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulrooney EF, Poon KK, McNally DJ, Brisson JR, Lam JS. Biosynthesis of UDP-N-acetyl-l-fucosamine, a precursor to the biosynthesis of lipopolysaccharide in Pseudomonas aeruginosa serotype O11. J Biol Chem. 2005;280:19535–19542. doi: 10.1074/jbc.M500612200. [DOI] [PubMed] [Google Scholar]
- 48.Brisson JR, Sue SC, Wu WG, McManus G, Nghia PT, Uhrin D. NMR Spectroscopy of Glycoconjugates. Wiley-VCH; Weinhem: 2002. [Google Scholar]
- 49.Uhrin D, Brisson JR. Horizon Scientific Press. Wymondham, UK: 2000. Structure Determination of Microbial Polysaccharide by High Resolution NMR Spectroscopy. [Google Scholar]
- 50.Kabsch WJ. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 51.Collaborative Computational Project N The CCP4 Suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 52.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 53.Winn M, Isupov M, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]