Abstract
CarD is an essential and global transcriptional regulator in mycobacteria. While its biological role is unclear, CarD functions by interacting directly with RNA polymerase (RNAP) holoenzyme promoter complexes. Here, using a fluorescent reporter of open complex, we quantitate RPo formation in real time and show that Mycobacterium tuberculosis CarD has a dramatic effect on the energetics of RNAP bound complexes on the M. tuberculosis rrnAP3 ribosomal RNA promoter. The data reveal that Mycobacterium bovis RNAP exhibits an unstable RPo that is stabilized by CarD and suggest that CarD uses a two-tiered, concentration-dependent mechanism by associating with open and closed complexes with different affinities. Specifically, the kinetics of open-complex formation can be explained by a model where, at saturating concentrations of CarD, the rate of bubble collapse is slowed and the rate of opening is accelerated. The kinetics and open-complex stabilities of CarD mutants further clarify the roles played by the key residues W85, K90 and R25 previously shown to affect CarD-dependent gene regulation in vivo. In contrast to M. bovis RNAP, Escherichia coli RNAP efficiently forms RPo on rrnAP3, suggesting an important difference between the polymerases themselves and highlighting how transcriptional machinery can vary across bacterial genera.
INTRODUCTION
The regulation of gene expression via the control of DNA transcription allows all living organisms to adapt their cellular biochemistry to changes in their environment. In bacteria, the transcriptional machinery consists of RNA polymerase (RNAP) holoenzyme composed of the catalytic core enzyme (ββ′α2ω) and a dissociable sigma factor (σ) subunit that directs promoter recognition. Transcription initiates when RNAP holoenzyme recognizes a promoter sequence and forms the RNAP-promoter closed complex (RPc) where the DNA strands are still annealed in the duplex. Through a series of conformational changes the RNAP-promoter open complex (RPo) forms where the DNA strands are melted from the −10 to +2 positions and the active site is accessible to the initiating nucleotide. In Escherichia coli, the mechanism of open-complex formation includes multiple kinetic intermediates between the initially bound complex and the stable open complex (1). While the core machinery and major intermediate states are conserved across bacteria, the detailed kinetics of transcription likely vary between bacterial species to accommodate different physiologies and niches. Therefore, a minimal kinetic scheme describing promoter binding and opening by RNAP in two reversible steps (R + P ↔ RPc ↔ RPo) provides a useful starting point when investigating open-complex formation in a non-model system.
Regulation of transcription initiation is achieved through the modulation of the stabilities of intermediate states and/or the rates of exchange between these states on the pathway to promoter escape. Transcription factors can mediate this regulation by directly affecting the polymerase-promoter interaction, manipulating the equilibrium between RPc and RPo, or affecting rates of promoter escape (2,3). Much of what has been studied in terms of the mechanisms of transcription initiation and its regulation has used E. coli as a model system. However, it has become evident that some bacteria require specialized factors to allow for efficient gene transcription. For example, many of the sequences and proteins required to regulate transcription initiation in E. coli are absent in mycobacteria, including Fis (4), DksA (5), AT-rich upstream activating elements (6,7) and GC-rich discriminator sequences (8). The question remains how bacteria missing these factors confer efficient and regulable transcription.
CarD is a recently discovered RNAP-binding protein that is conserved in numerous bacterial species but not present in E. coli. CarD homologs are essential in Mycobacterium tuberculosis, Mycobacterium smegmatis (9) and Myxococcus xanthus (10) and knockouts were not attainable in Borrelia burgdorferi (11). Furthermore, CarD is essential for the response of M. tuberculosis to oxidative stress and certain antibiotics, as well as the acute and persistent infection of mice (9,12). Given the tremendous impact of M. tuberculosis on global health (1.3 million related deaths a year, WHO, 2013), it is of particular interest to understand unique biochemical pathways required by this pathogen. Yet, the reasons for CarD essentiality remain enigmatic primarily due to a lack of understanding of its molecular mechanisms.
CarD is associated with all RNAP-σA (mycobacterial house-keeping sigma, homologous to E. coli σ70) transcription initiation complexes within the M. smegmatis genome (13) and interacts directly with the β1 region of the RNAP β subunit through its N-terminal RNAP interaction domain (RID) (9–10,12). The current model for CarD activity is that this protein is directed to promoters via its interaction with RNAP β (13,14). At the promoter, a basic patch in the C-terminus of CarD contacts the DNA and the association of RNAP-bound CarD with the promoter DNA stabilizes the RNAP-promoter complex (13,14). Within this stabilized CarD-RNAP-promoter complex, a conserved tryptophan in CarD is required for proper transcriptional regulation, however its exact role is still unknown. Together, these three activities of CarD (RNAP binding, DNA binding and the conserved tryptophan) promote a gene expression profile that supports viability (9,12,14), but the mechanism by which CarD regulates transcription is unknown. Understanding how CarD stabilizes RNAP-promoter complexes and regulates transcriptional activity is crucial for developing models of transcriptional regulation in mycobacteria and for furthering our knowledge of the molecular physiology of M. tuberculosis.
Here, we perform a detailed mechanistic analysis of M. tuberculosis CarD in the context of open-complex formation using a real-time fluorescence assay on the rrnAP3 ribosomal RNA promoter from M. tuberculosis. CarD is known to regulate transcription from this promoter and is an activator of ribosomal RNA transcription (9). We show that CarD has dramatic effects on both the equilibrium concentration of open complex and on the kinetics of the approach to equilibrium. Analysis of CarD concentration-dependent data suggest that CarD stabilizes open complex via a two-tiered kinetic mechanism. First, CarD associates with RNAP open complexes with high affinity and slows the rate of DNA closing. Second, CarD associates with RNAP closed complexes with lower affinity and increases the rate of DNA opening. Thus, we propose that CarD regulates transcription by increasing the stability of RPo relative to RPc by affecting both the effective forward and reverse rates between these two states. Furthermore, comparisons between open-complex formation by E. coli and Mycobacterium bovis RNAPs suggest that E. coli does not posses a homolog of CarD because its polymerase is intrinsically more active in open-complex formation.
The work presented here reveals important details of the mechanism of CarD-dependent transcription initiation in mycobacteria and provides an example of the possible intrinsic differences between RNAPs from different classes of bacteria. This observation highlights the importance of research in prokaryotic transcription in organisms other than E. coli, especially with respect to understanding mechanisms of transcriptional regulation in pathogenic species such as M. tuberculosis. Lastly, our findings provide an important addition to the diverse paradigms of transcription initiation in prokaryotes.
MATERIALS AND METHODS
Protein purification
Mycobacterium bovis core RNAP and σA were over-expressed and purified using methods slightly modified from the literature (15). Mycobacterium bovis RNAP holoenzyme is identical to M. tuberculosis RNAP except for the 69th residue of β′ which is a proline in M. bovis and an arginine in M. tuberculosis. The M. bovis core RNAP subunits were co-over-expressed from plasmid pAC22 in BL21(λDE3) pRARE2. Cells were grown at 37°C until an OD600 of 0.8. Cells were induced with 250 μM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and grown 4 h at 20°C. Cell pellets were lysed and loaded onto a 10 mL Ni2+ affinity column (HP HiTrap, GE Healthcare) using a 5–1000 mM imidazole gradient. The eluted sample was dialyzed, concentrated and further purified by size exclusion chromatography (HiPrep Sephacryl 300, Pharmacia). The peak of core RNAP was dialyzed in storage buffer (50% glycerol, 10 mM Tris pH 7.9, 200 mM NaCl, 0.1 mM ethylenediaminetetraaceticacid (EDTA), 1 mM MgCl2, 20 μM ZnCl, 2 mM Dithiothreitol (DTT)), concentrated to 4 μM (Vivaspin 20, MWCO 100 kDa, GE Healthcare), flash frozen in liquid nitrogen and stored at −80°C.
Mycobacterium bovis σA was over-expressed from plasmid pAC27 BL21(λDE3) pRARE2. Cells were grown at 37°C until an OD600 of 0.8. Cells were induced with 250 μM IPTG and grown 4 h at 20°C. Cell pellets were lysed and σA was purified by Ni2+ affinity chromatography (HP HiTrap, Pharmacia) using a 5–1000 mM imidazole gradient. Eluted protein was dialyzed into storage buffer (10 mM Tris pH 7.9, 250 mM NaCl, 0.1 mM EDTA, 1 mM MgCl2, 20 μM ZnCl, 2 mM DTT), concentrated to 24 μM (Vivaspin 20, MWCO 30 kDa, GE Healthcare), flash frozen and stored at −80°C.
Mycobacterium tuberculosis CarD was over-expressed from a pETSUMO plasmid (13). Cells were grown at 37°C until OD600 of 0.8, induced with 1 mM IPTG, and grown 3 h at 37°C. Cell pellets were lysed and His-SUMO-CarD was purified by Ni2+ affinity chromatography (Ni-NTA Agarose, Qiagen). Elution at 250 mM imidazole was quantified and His-Ulp1 protease added to cleave the His-SUMO tag from CarD. This mix was dialyzed overnight in 3 l of 20 mM Tris pH 8.0, 150 mM NaCl and 1 mM β-ME and dialyzed again in fresh buffer for four more hours. Ni2+ resin was added to dialyzed proteins and, after incubating for 1 h at 4°C, CarD was collected in the flow through. Protein was concentrated to 100 μM (Vivaspin 20, MWCO 3 kDa, GE Healthcare), flash frozen and stored at −80°C.
Preparation of fluorescent promoter DNA fragments
The DNA template contained the −41 to +4 bases of the rrnAP3 ribosomal RNA promoter (16) of M. tuberculosis centered in a 150 bp template (Supplemental Information). Fluorescently labeled promoter DNA was prepared as previously described (17). In short, two 85-mer oligonucleotides with a 20 bp overlapping sequence were synthesized and high performance liquid chromatography (HPLC) purified (IDT). The 85-mer corresponding to the non-template strand was ordered with a C6 amine-modification on the +2 thymine for covalent attachment to Cy3-NHS (Lumiprobe). Labeled oligonucleotides were purified from unlabeled oligos and excess Cy3 using reverse-phase chromatography (C18 column, XTerra). Template and Cy3-non-template strands were annealed and extended with Taq polymerase (Invitrogen). The extension products were then purified by HPLC (DNA Swift Column, Dionex). Finally, the pure Cy3-labeled extension products were spin-concentrated and exchanged into Tris-EDTA buffer (Millipore Amicon Ultra, MWCO 30 kDa). Concentrations of dsDNA (A260) and Cy3 (A550) were measured by spectrophotometer (Nanodrop) and indicated a equimolar ratio of dye to DNA template demonstrating 100% labeling. Labeled DNA template was stored at −20°C.
Stopped-flow assay for real-time monitoring of open-complex formation
Stopped-flow experiments were performed as previously described (17,18) with minimal exceptions. All experiments were performed by mixing equal volumes of protein solution from one syringe with fluorescent promoter DNA from the other syringe. Accordingly, initial syringe concentrations of protein and DNA were prepared at twice the desired final concentration for the reaction. Including contributions from protein storage buffers, final reaction buffer conditions were as follows: 14 mM Tris pH 8.0, 120 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.1 mg/ml bovine serum albumin (BSA) and 10% glycerol by volume.
Protein solutions for stopped-flow experiments were prepared as follows. In the case of MboRNAP, core was mixed with saturating concentrations of σA and allowed to equilibrate for 15 min at room temperature. In the case of EcoRNAP, σ70-saturated holoenzyme was used (New England Biolabs). When required, CarD was added to RNAP holoenzyme and allowed to equilibrate for an additional 15 min at room temperature. Protein solutions were then diluted into a solution containing 10 mM Tris pH 8.0, 40 mM NaCl, 10 mM MgCl2, 1 mM DTT and 0.1 mg/ml BSA. Since MboRNAP core storage buffer contained 50% glycerol, the protein solution contained 10% glycerol by volume. For all experiments, any absence of protein volume was balanced by its appropriate storage buffer in order to preserve identical buffer conditions for different reactions.
Promoter DNA solutions were prepared for stopped-flow experiments as follows. Stock DNA was diluted to 20 nM into buffer containing 10 mM Tris pH 8.0, 40 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA and 10% glycerol by volume. Glycerol was added to the DNA solution to match the glycerol content of the protein solution, to minimize rapid mixing artifacts that can occur with asymmetric viscosities between syringes.
Experiments were performed on an SX-20 stopped-flow spectrophotometer (Applied Photophysics, dead-time < 2 ms) with a shot volume of 150 μl, using excitation at 515 nm (monochromator), and emission at 570+ nm (long-pass filter). Unless otherwise noted, data were collected for 20 min and at 25°C by sampling 10,000 points in a logarithmic decay over the time course of the experiment. A circulating water bath with active feedback was used to maintain temperature within 0.1°C. Multiple buffer shots as well as DNA only shots were performed before every experiment. At least two shots were collected for every protein condition, which were averaged before plotting as fold change (FC) where FC = (F − F0)/F0, where F = experimental signal − buffer signal in volts and F0 = DNA signal − buffer signal in volts. In all figures, error bars of fold change represent the standard error of the mean (SEM) for conditions that were repeated multiple times on different days. From these SEMs, average errors were then applied to conditions that were only repeated multiple times on the same day to better estimate the real error for these points.
Triple exponential fits of kinetic traces to extract observed rates
Fold changes as a function of time were fit to a sum of exponentials using the ProData Viewer software (Applied Photophysics). Single and double exponentials were insufficient to capture the entire shape of the curve without substantial systematic residual error and triple exponential fits of the entire time course yielded inconsistent assignment of phases (fast, medium and slow fits were interchangeable between a1, k1, a2, k2, a3 and k3). To fit the curves with three exponentials while preserving assignment of fast, medium and slow phases, 1–1200 s was fit to a double exponential and the amplitudes and observed rates for the medium (a2, k2) and slow (a3, k3) phases were fixed. Then, a third exponent was added and the time for fitting was expanded to include 0.1–1200 s in order to capture the fastest phase (a1, k1). In this manner, the curves could be fit with three exponentials from 0.1 to 1200 s, and assignments of amplitudes and observed rates for all phases between traces remained consistent. In all figures, error bars of observed rates represent the SEM for conditions that were repeated multiple times on different days. From these SEMs, average errors were then applied to conditions that were only repeated multiple times on the same day to better estimate the real error for these points.
Kinetic simulations
Simulation of the kinetic model of open-complex formation was carried out in MATLAB R2014b (Mathworks). Model parameters were determined by manually adjusting rate constants and comparing with the data. The model was adjusted until the trends of equilibrium fluorescence and observed rate as functions of RNAP and CarD concentrations were best fit. By assuming that the apparent bimolecular association rates were all equal, fixing the equilibrium between closed and open complex based on the MboRNAP data in the absence of CarD, and requiring that the model satisfied detailed balance, only three adjustable rate constants remained. This allowed for the manual exploration of parameter space and yielded a model that is able to capture the observed trends. The rate constants used to generate Figure 6 are listed in the Supplemental Information.
Figure 6.
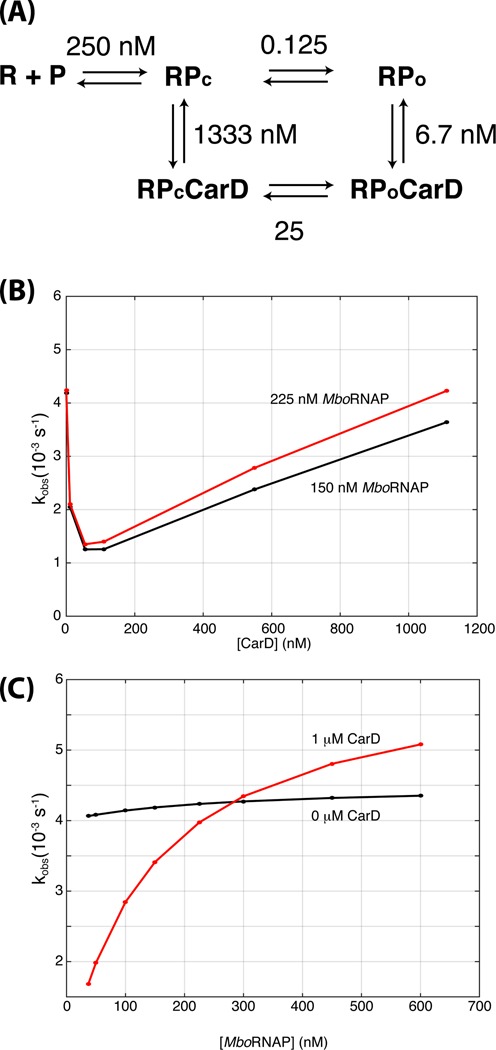
A CarD-dependent kinetic model of open-complex formation. (A) The model consists of five states: unbound DNA (R + P), closed complex (RPc), open complex (RPo), closed complex bound to CarD (RPcCarD) and open complex bound to CarD (RPoCarD). Values indicate the ratio of rate constants between each pair of states in the model. For example, at 250 nM, the forward and reverse rates between R + P and RPc are equal. Furthermore, the ratio of forward to reverse rates between RPc and RPo are 0.125 and 40 in the absence and presence of CarD, respectively. The rates used to generate the figures below can be found in Supplementary Information. (B) Simulations of the kinetic model titrating CarD concentration with 150 nM (black) and 225 nM (red) MboRNAP. (C) Simulations of the kinetic model titrating RNAP concentration with 0 nM (black) and 1 μM (red) CarD.
RESULTS
A real-time fluorescence-based assay of open complex
To detect the formation of open complexes, a Cy3 label was incorporated on the +2 nucleotide of the non-template strand of a M. tuberculosis rRNA promoter rrnAP3 DNA template via a NHS-C6 amide linkage (Figure 1A). Cy3 exhibits a 2-fold enhancement in fluorescence intensity in the open complex and has been used to monitor DNA opening and promoter escape in E. coli (17). The rrnAP3 promoter is the major ribosomal RNA promoter in M. tuberculosis (16), has been used previously in studies of CarD, and is thought to be regulated by CarD in vivo (13,14). Mixing of protein components with labeled DNA templates was performed via stopped-flow spectrophotometry and fluorescence was monitored for 20 min. The addition of 21 nM E. coli RNAP (EcoRNAP) sigma factor 70 (σ70) holoenzyme to rrnAP3 resulted in robust enhancement of the fluorescence intensity as compared to DNA alone (>60% increase or an enhancement of 0.6, Figure 1B). In contrast, very small (∼0.05) fluorescence enhancements are observed when the same concentration of EcoRNAP is added to templates lacking the rrnAP3 promoter sequence (Figure 1B), demonstrating that fluorescence enhancement is promoter dependent. Furthermore, an enhancement of approximately 0.3 is observed at low temperature where open-complex formation is inhibited (19–21) and the fluorescence enhancement increases with increasing temperature consistent with the known temperature-dependence of open-complex formation (Figure 1C). A possible structural rational for the specific dependence on open complex exhibited by the observed fluorescence enhancement is that the +2 position is positioned downstream of closed complex DNase I footprints, but is located within the footprint of open complex (22). Taken together, these control experiments confirm that fluorescence enhancement of a non-template +2 Cy3 in the M. tuberculosis rrnAP3 template DNA faithfully tracks the formation of the open initiation complex. Importantly, as with EcoRNAP, M. bovis RNAP (MboRNAP, identical to M. tuberculosis RNAP except β′ R69 is P69) also stimulates fluorescence enhancement in a promoter- and temperature-dependent manner (Figure 1D and E). The time traces obtained in this assay exhibit multiple phases consistent with the known kinetic complexity of transcription initiation (1) and provide insight into both the kinetics of open-complex formation and the equilibrium concentration of open complex.
Figure 1.
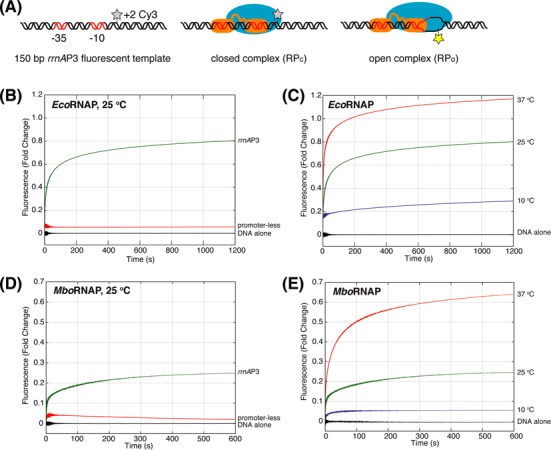
A real-time fluorescence assay for monitoring open-complex formation. (A) The assay utilizes a Cy3 dye at the +2 position of the non-template strand of a rrnAP3 ribosomal RNA promoter DNA template. The Cy3 has a baseline fluorescence in the free and closed complexes that is enhanced upon the formation of open complex. (B) EcoRNAP-dependent (21 nM) fluorescence enhancement in the presence (green) and absence (red) promoter sequence at 25°C. (C) EcoRNAP-dependent (21 nM) fluorescence enhancement at 10°C (blue), 25°C (green) and 37°C (red). (D) MboRNAP-dependent (100 nM) fluorescence enhancement in the presence (green) and absence (red) of promoter sequence at 25°C. (E) MboRNAP-dependent (100 nM) fluorescence enhancement at 10°C (blue), 25°C (green) and 37°C (red).
M. bovis RNAP forms open complex less efficiently than E. coli RNAP on rrnAP3
We first studied the concentration dependence of EcoRNAP (σ70) and MboRNAP (σA) holoenzymes on open-complex formation on the M. tuberculosis rrnAP3 promoter at 25°C. Fluorescence enhancements after the addition of 2–283 nM EcoRNAP (Figure 2A) or 37.5–600 nM MboRNAP (Figure 2B) were monitored for 20 min. Final fold enhancements were plotted versus RNAP holoenzyme concentration and fit to extract a concentration at which the enhancement is half-maximal (Keff, Figure 2C). As expected, the amount of open complex increases as concentration increases for both forms of RNAP, however EcoRNAP exhibits a Keff almost 10-fold smaller than MboRNAP (23 ± 5 nM versus 212 ± 43 nM, 95% confidence bounds). More strikingly, at saturating concentrations of the respective polymerases, the fluorescence enhancement for EcoRNAP is 1.44 ± 0.1 while MboRNAP only reaches an enhancement of 0.30 ± 0.1. The enhancement for EcoRNAP is comparable to that described previously for EcoRNAP on a consensus promoter (∼1.4) (17), suggesting that this signal is indicative of fully open DNA (i.e. 100% open complex) on rrnAP3. By comparison, MboRNAP σA holoenzyme, even when fully occupying the DNA template at saturating concentrations of polymerase, is not capable of opening a large percentage of the promoters leaving the majority of bound complexes in the closed state. This demonstrates that MboRNAP forms a significantly less stable open complex as compared to EcoRNAP, even on its own mycobacterial promoter.
Figure 2.
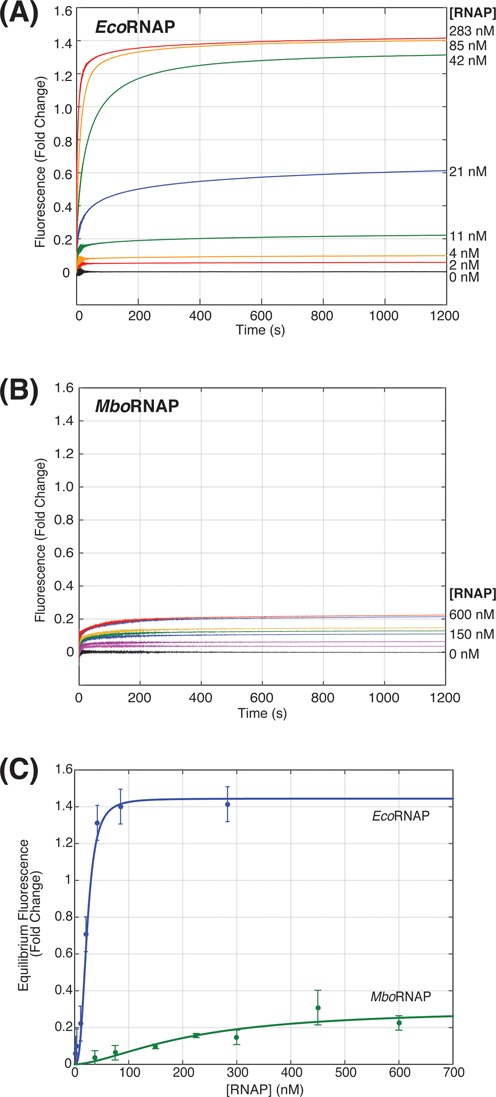
RNAP concentration dependence of promoter opening. (A) Fluorescence enhancement as a function of time for increasing concentrations of EcoRNAP (0–238 nM). (B) Fluorescence enhancement as a function of time for increasing concentrations of MboRNAP (0–600 nM). (C) Equilibrium fluorescence fold change for both EcoRNAP and MboRNAP as a function of polymerase concentration. Fits (solid lines) of the data allow the extraction of concentrations of half-maximal effect (23 ± 5 nM and 212 ± 43 nM) and saturated fluorescence enhancements (1.44 and 0.28) for EcoRNAP and MboRNAP, respectively.
CarD stabilizes the open complex of MboRNAP
We next looked at the effect of CarD on open-complex formation. Holoenzyme was incubated with CarD for 15 min prior to mixing with the DNA template and recording changes in fluorescence intensity over time. While CarD shows no fluorescence enhancement when added to DNA alone and has little effect on the fluorescence signal from EcoRNAP (Supplementary Figure S1), its presence leads to a dramatic increase in the magnitude of fluorescence enhancements observed when incubated with MboRNAP (Figure 3A). As fluorescence enhancement serves as a reporter of open complex, we conclude that CarD significantly stabilizes open complex specifically for MboRNAP. This is consistent with known differences in the β1 lobe of EcoRNAP and MboRNAP on the CarD-binding interface that would predict tighter binding of M. tuberculosis CarD to MboRNAP than to EcoRNAP (23). In the presence of 225 nM MboRNAP, a rise in fluorescence fold change can be seen as the concentration of CarD is increased from 0 - 1.1 μM. The concentration of CarD that exhibits half-maximal effect (77 ± 35 nM) and the fold fluorescence enhancement over 0 μM CarD at saturation (6.2-fold) was determined by normalizing the equilibrium (final) fluorescence enhancements relative to 0 μM CarD and fitting to a binding isotherm (Figure 3B). Assuming that EcoRNAP generates 100% open complex at saturating concentrations (283 nM, Figure 2A) and that MboRNAP generates the same enhancement of signal per open complex, at saturating concentrations of CarD (≥ 1 μM) the percent of DNA that are in open complexes at equilibrium in the presence of 225 nM MboRNAP increases from approximately 15% to 93% (Figure 3A).
Figure 3.
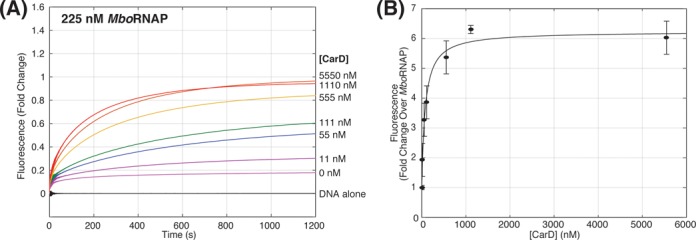
CarD increases open-complex formation with MboRNAP. (A) Fluorescence enhancement as a function of time for 225 nM MboRNAP with increasing concentrations of CarD (0–5.55 μM) at 25°C. (B) Equilibrium values of fluorescence enhancement are plotted as a function of CarD concentration. A fit (solid line) of the data reveals a concentration of half-maximal effect Keff = 77 ± 35 nM and an amplitude of 6.2-fold over 225 nM MboRNAP alone.
To compare the MboRNAP holoenzyme concentration dependence in the presence and absence of CarD, a holoenzyme titration (37.5–600 nM) was performed in the presence of saturating CarD (1 μM) at 25°C (Figure 4A). Comparing RNAP titrations in the presence and absence of CarD shows that 1 μM CarD reduces Keff from 212 ± 43 nM to 106 ± 3 nM and increases the amount of open complex 4.3-fold at saturation (Figure 4B).
Figure 4.
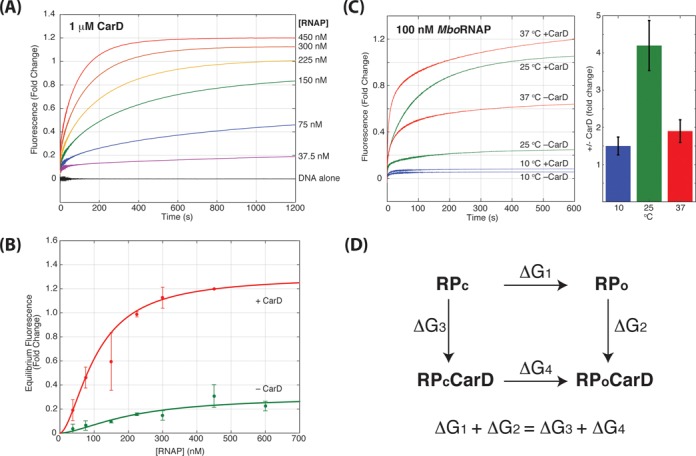
MboRNAP titration at saturating CarD. (A) Fluorescence enhancement as a function of time for 1 μM CarD with increasing concentrations of MboRNAP (0–450 nM) at 25°C. (B) Equilibrium values of fluorescence enhancement fold change are plotted in the presence (red) and absence (green) of 1 μM CarD. The data were best fit with an amplitudes of 1.3 and 0.28 and Keff of 106 ± 3 nM and 212 ± 43 nM in the presence and absence of CarD, respectively. (C) Observed traces for 100 nM MboRNAP in the presence or absence of 1 μM CarD at 10°C (blue), 25°C (green) and 37°C (red). The bar graph shows the fold increase in equilibrium fluorescence enhancement for each temperature. (D) The thermodynamic cycle linking closed complex (RPc), open complex (RPo), closed complex bound to CarD (RPcCarD) and open complex bound to CarD (RPoCarD). The equality shown below follows since the sum of the free energy differences along the two paths from RPc to RPoCarD must be equal.
The ability of CarD to stabilize open complexes is also temperature dependent. In experiments with 100 nM MboRNAP in the presence and absence of 1 μM CarD, traces at different temperatures show different amounts of CarD-dependent fold change (Figure 4C). Specifically, the CarD effect is significantly larger at 25°C (4.2-fold) than it is at either 10°C (1.5-fold) or 37°C (1.9-fold). These observations fit well with the known temperature-dependence of open complex (19–21). At low temperature, opening is severely inhibited and the binding energy from CarD is not sufficient to tilt the landscape enough toward opening to have a large effect. At 25°C, the energy landscape is more balanced between closed and open and the extra binding energy supplied by CarD is able to significantly affect the equilibria between the two states. Lastly, at 37°C, open complex forms more readily than at lower temperatures and thus there is an appreciable amount of open complex to begin with (i.e. 50%). In this case, even if CarD is capable of stabilizing all bound complexes in the open form, it will exhibit a lower fold change in open complex equilibrium concentration (i.e. 2-fold).
CarD pushes the equilibria of polymerase-bound states toward open complex
In the presence of a saturating concentration of MboRNAP (450 nM) where the DNA templates are fully occupied by holoenzyme, the amount of open complex observed in the presence of CarD is dramatically increased in the presence of 1 μM CarD (Figures 2B and 4A). This can only be the case if CarD stabilizes the open complex relative to closed complex either by increasing the effective rate of opening or decreasing the effective rate of closing or both.
Furthermore, the observation that CarD stabilizes open complexes relative to closed complexes suggests that CarD has a higher affinity to open complex than it does to closed complex. In a thermodynamic cycle (Figure 4D) where binding of CarD is coupled to the open/closed equilibrium, the sum of free energies around the cycle must be zero. The data show that RPo is more stable relative to RPc in the presence of CarD (ΔG4 < ΔG1). Since the sum of free energies along paths that begin and end in the same state must be the same, ΔG1 + ΔG2 = ΔG3 + ΔG4. This then requires the free energy differences between RP and RP·CarD in the open and closed states to satisfy ΔG2 < ΔG3. Thus, as the free energy difference determines the affinity (Kd = e−ΔG/RT), we predict that CarD binds more tightly to open complex than to closed complex.
The kinetics of open-complex formation can be monitored via the slowest observed rate
The time-dependent traces allow for an analysis of the kinetics of open-complex formation. The traces are well fit by a triple exponential resulting in three observed rates, consistent with the well-known multi-state kinetic complexity of open-complex formation (1) (Figure 5A, Materials and Methods). The three observed rates differ by orders of magnitude with a fast (k1obs ∼ 1 s−1), intermediate (k2obs ∼ 0.1 s−1) and slow (k3obs ∼ 0.005 s−1) observed rates. In traces exhibiting robust opening (i.e. high polymerase or high CarD concentrations), the amplitude of the curve is dominated by the amplitude of the slowest observed rate (Supplementary Figure S2). Furthermore, although promoter-less control traces (Figure 1B and D) do exhibit a modest amount of fluorescence enhancement, this enhancement occurs very rapidly (< 1 s) and the slow phase (k3obs) is not observed. Therefore, we reason that, to a first-approximation, the fastest observed rates report on a protein–DNA interaction not related to opening (i.e. binding) and the slowest observed rate (k3obs) specifically reports on the approach to equilibration of open complex for MboRNAP.
Figure 5.
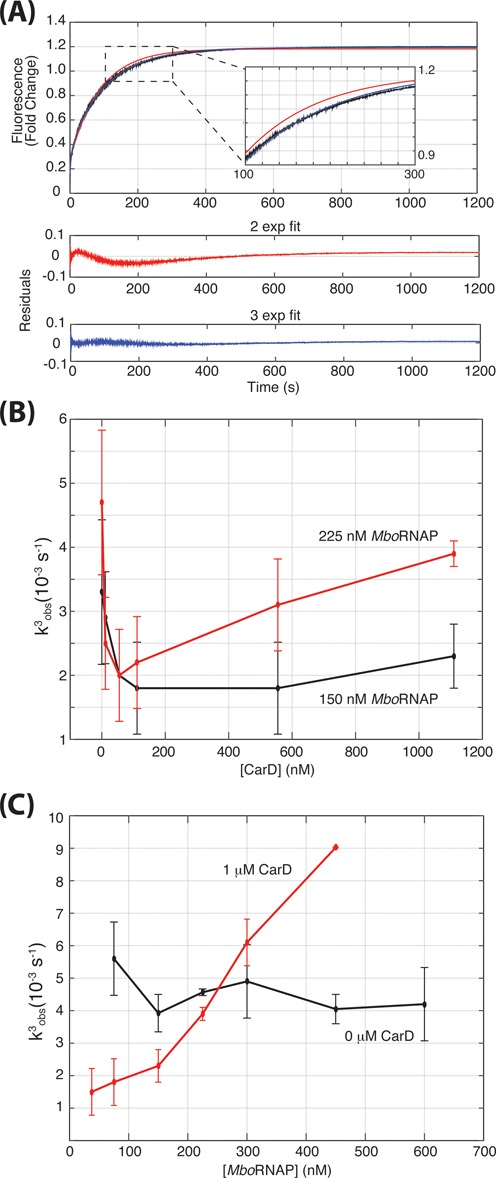
Kinetics of open-complex formation. (A) Triple exponential fits are required to obtain fits with good residuals for traces where DNA opening is occurring. A typical trace, the fits, and the resulting residuals are shown for with double (red) and triple exponential (blue) fits. (B) The CarD concentration dependence of k3obs in the presence of 150 nM (black) or 225 nM (red) MboRNAP. (C) The MboRNAP concentration dependence of k3obs in the presence (black) and absence (red) of 1 μM CarD.
Observed rates typically represent combinations of microscopic rate constants. In the simplest case, the two states A and B are connected by a forward and a reverse rate constant. In this case, if one starts with the system entirely in A and monitors the approach to equilibrium, the observed rate will simply be the sum of the two rate constants. There are two mechanisms by which B could be stabilized relative to A: (i) the forward rate constant could be accelerated or (ii) the reverse rate constant could be slowed. In the first case, B would be stabilized and the observed rate would increase while in the second case, B would be stabilized and the observed rate would decrease. In more complicated kinetic mechanisms, the interpretation may not be so straightforward, but nonetheless, these two limiting cases serve as a useful backdrop for extracting mechanistic information from trends in an observed rate (Supplementary Figure S3).
CarD exhibits a concentration-dependent effect on the kinetics of open-complex formation
To extract information regarding the effect of CarD on the kinetics of open-complex formation, we first analyzed the CarD-dependence of k3obs using data collected at 150 nM MboRNAP (Figure 5B). Here, k3obs decreases with increasing CarD concentration suggesting that CarD slows a reverse rate in order to stabilize open complex. This interpretation is consistent with recent results showing that CarD increases the lifetime of open complexes in the presence of competitor DNA (14) and suggests that CarD functions by inhibiting bubble collapse.
Interestingly, when the above analysis is repeated with a higher concentration of MboRNAP (225 nM), k3obs exhibits a markedly different dependence on the concentration of CarD (Figure 5B). At low concentrations of CarD (0–100 nM), k3obs decreases with increasing concentration as observed with 150 nM MboRNAP. However, at concentrations higher than 100 nM CarD, k3obs accelerates as a function of CarD concentration and almost reaches the magnitude observed in the absence of CarD. This biphasic dependence of the observed rate suggests that CarD stabilizes open complex via a more complicated mechanism and points to models where CarD modulates more than one rate constant in the kinetic mechanism. It further suggests that it does so with different concentration dependencies. Therefore, we propose a model, discussed in detail below, where CarD slows bubble collapse in the open complex at low concentrations and can also associate with closed complex at high concentrations and accelerate DNA opening. Importantly, CarD is one of the highest expressed proteins in M. tuberculosis (24) and quantitative western analysis shows that the concentrations of CarD in the cell are similar to the concentrations used for these assays (Supplementary Figure S4) suggesting that CarD both accelerates DNA opening and inhibits bubble collapse in vivo.
Since k3obs depends on RNAP concentration at 1 μM CarD (Figure 3), we analyzed this dependence further by titrating MboRNAP concentration in the presence of 1 μM CarD. At this concentration of CarD, k3obs increases as a function of polymerase concentration (Figure 5C). As increasing RNAP concentration increases the rate at which bound complexes will form, this result suggests that the rate of CarD-stimulated open-complex formation also depends on the rate of polymerase binding to the DNA template. Importantly, in the absence of CarD, k3obs does not show this dependency. Consistent with the rate limiting steps of CarD-independent bubble formation taking place between two bound complexes, k3obs is independent of MboRNAP in this concentration range in the absence of CarD (Figure 5C). These observations provide further constraints to models of the kinetic mechanism of CarD-dependent regulation of open-complex formation.
A kinetic model with CarD binding to both open and closed complexes
To understand the concentration-dependent effects of CarD, a kinetic model of open-complex formation in the presence of CarD was constructed (Figure 6A). The model consists of five states including unbound DNA (R + P), closed complex (RPc), open complex (RPo), CarD-bound closed complex (RPcCarD) and CarD-bound open complex (RPoCarD). In principle, CarD may interact both with DNA alone and unbound RNAP. However, CarD has been found by ChIP-seq only in genomic positions where RNAP holoenzyme is also found (13). In addition, the interaction between CarD and free RNAP is likely much weaker than the association of CarD to DNA-bound RNAP complexes where it may interact with both polymerase and DNA cooperatively. For these reasons, CarD-DNA and CarD-RNAP states are left out of the current model.
Initially models where CarD only associated with open complex were attempted, but these models are unable to capture the biphasic dependence of k3obs on CarD concentration. However, a model where CarD also interacts with closed complex is able to account for the concentration dependencies of both the polymerase and CarD with a single set of rate constants (Figure 6, Supplemental Methods). The model has three key features: (i) CarD has a lower affinity to closed complex than it does to open complex; (ii) in the open complex, CarD inhibits the rate of bubble collapse; and (3) in the closed complex, CarD accelerates the rate of DNA opening. Importantly, the model was required to satisfy detailed balance so that the energetics linking the affinities of CarD to the equilibria between open and closed complexes do not violate thermodynamics (Supplemental Methods).
The model was used to simulate time-dependent traces of open-complex formation and the resulting curves were fit to extract an observed rate of the approach to equilibrium (kobs). The model captures the dependence of the observed rate on CarD concentration at different fixed MboRNAP concentration (Figures 5B and 6B). Specifically, the observed rate decreases initially at both polymerase concentrations and then increases more at 225 nM RNAP than it does 150 nM RNAP. The model also captures the dependences of the observed rate on RNAP concentration in the presence and absence of 1 μM CarD (Figures 5C and 6C). Specifically, at 1 μM CarD, the observed rate increases with increasing RNAP concentration whereas in the absence of CarD, the observed rate is RNAP-concentration independent.
Lastly, the identical model parameters also reproduce the trends in the equilibrium concentration of open complex as a function of MboRNAP and CarD (Supplementary Figure S5). Although the model does not explicitly include a CarD-dependent change in the rates connecting free polymerase to closed complex, the approximately 2-fold reduction in Keff in the presence of 1 μM CarD (Figure 4B) is captured by the model (Supplementary Figure S5). This illustrates that a lower Keff is not necessarily due to a change in the energetics of free polymerase relative to the closed complex. Since open complexes cannot dissociate from the DNA without closing first, any stabilization of open complex relative to closed results in an increased apparent affinity of the polymerase for DNA.
CarD mutants are deficient in open-complex stabilization
Single amino acid mutants of CarD have previously been used in vivo and in vitro to understand the roles played by different domains or regions of the protein (12,14). Three groups of mutants have been identified according to their distinct phenotypic effects in vivo, namely those with DNA-binding defects, RNAP interaction defects and with an alanine substituted for a conserved tryptophan residue (W85A). Although W85 is part of the DNA-binding domain, it appears to play a more specific role in the stabilization of open complex, perhaps by interacting with the upstream end of the DNA bubble (13,14).
CarD mutants that weaken the interaction with RNAP (R25E, (12)), weaken the interaction with DNA (K90A, (14)) and W85A (13,14) were used as representative mutants from the three classes and were tested in the open-complex formation assay. Fluorescence enhancement traces using MboRNAP in the presence of 1 μM CarD (Figure 7A) show that all three mutants are partially deficient in stabilizing open complex relative to WT, as evidenced by their lower equilibrium (final) fluorescence enhancements. Increasing the concentration of mutant CarDs showed more enhancement, but the final enhancements at the highest protein levels tested were still less than 50% that of wild type (WT) (Figure 7B, Supplementary Figure S6). Fits of the mutant CarD titrations result in estimates of open-complex stabilization and concentrations of half-maximal effect. K90A and W85A both reach a maximum of 3-fold fluorescence enhancement as opposed to the 6.2-fold enhancement seen with WT CarD. The half-maximal concentrations of K90A and W85A are 400 ± 48 nM and 381 ± 118 nM respectively as compared to the 77 ± 35 nM observed with WT CarD. The data for R25E were unable to be fit as we were unable to reach saturation due to limitations of protein concentration, but the activity of the RNAP-interacting domain mutant is even further decreased relative to the other mutants. Taken together, the data demonstrate that all three residues, and by extension all three activities (polymerase binding, DNA binding and W85 activity), are required for full CarD activity.
Figure 7.
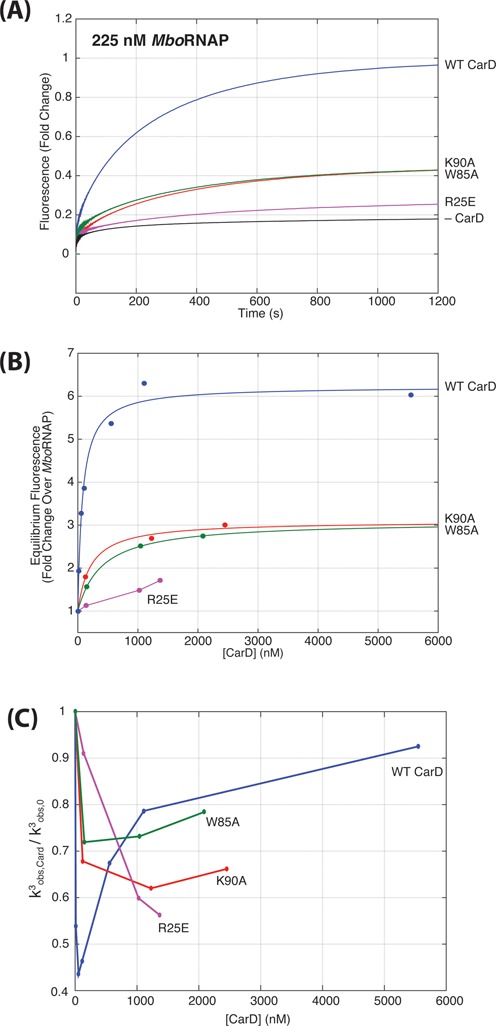
Mutants of CarD diminish the degree of open-complex stabilization. (A) Fluorescence enhancement as a function of time are shown for 225 nM RNAP + 1 μM WT and mutant CarDs at 25°C. W85A (green) and K90A (red) traces generate less than half the open complex at equilibrium as compared to WT (blue) and R25E (purple) is even further compromised. (B) Equilibrium fluorescence enhancement as a function of CarD concentration taken from titrations of mutant CarDs (Supplementary Figure S6). (C) The CarD concentration dependence of k3obs for WT (blue), W85A (green), K90A (red) and R25E (purple) mutant proteins.
Fits of the time-dependent fluorescence enhancements again showed three phases and the slowest rate (k3obs) in the presence of CarD mutants depended on CarD concentration as with WT CarD (Figure 7C, Supplementary Figure S6). However, the degree to which k3obs was decreased was less in all the mutants compared to WT. Based on the model that the decrease in k3obs is due to CarD's ability to inhibit DNA closing, this suggests that the mutants are unable to prevent bubble collapse to the same extent as WT. More specifically, the initial decrease of k3obs observed as a function of CarD concentration showed a similar concentration dependence as WT for the W85A and K90A mutants, but the observed rate decreased less (Figure 7C). In contrast, R25E exhibited a much slower decrease suggesting a lower affinity to open complex (Figure 7C) consistent with previous work showing that this mutant associates less strongly with MboRNAP (12). At higher concentrations, W85A and K90A show signs of an increasing k3obs suggesting that they are able to interact with closed complex to accelerate DNA opening, but not to the same extent of WT. Even though the observed rate does not decrease as much as with WT CarD at low concentrations, it is not as fast at higher CarD concentrations either (i.e. at 1 – 2 μM CarD). In contrast, R25E shows no signs of the increasing phase of k3obs suggesting, perhaps, that it is unable to effectively interact with closed complex at these concentrations. These observations coupled with the reduced fluorescence enhancement observed at saturating levels of mutant CarD, suggest that W85A and K90A mutants are able to associate with both open and closed complexes as in WT, but are deficient in both inhibiting the rate of bubble collapse and increasing rate of opening. In contrast, it appears that the association of R25E with the complexes occurs with a much lower affinity and that it may only be able to interact weakly with open complex.
DISCUSSION
A better understanding of the role of M. tuberculosis CarD in transcription initiation will not only be relevant to mycobacteria, but also to the diverse bacterial species that encode CarD homologs (9,13). The result that MboRNAP is less able to form rrnAP3 open complexes as compared to EcoRNAP rationalizes the essentiality of CarD in M. tuberculosis (and its absence from E. coli) and suggests that polymerases from organisms in which CarD is found may also be deficient in open-complex formation. This observation also stresses the importance of directly studying transcription systems in a wide spectrum of bacteria, especially in pathogens like M. tuberculosis, that may diverge from traditional model systems.
Previous studies of the effect of CarD on transcription initiation focused on open complex lifetimes in the presence of competitor DNA (13–14,25). Specifically, proteins and DNA were allowed to come to equilibration and open-complex stability was assayed by the time-dependent decay of reporters of open complex (i.e. the production of transcripts upon the addition of nucleoside triphosphate (NTPs)). These valuable assays have led to the model where CarD stabilizes open complex. However, in the cell, there is no initial equilibration time and transcriptional regulation must be enacted in real time. That is to say, the flux of transcript production must be attenuated or augmented. Therefore, both the rate at which open complexes are formed and the rate at which they decay are crucial for gene regulation in vivo. With this mindset, the rates at which DNA opens and closes in DNA-bound holoenzyme complexes and their dependence on CarD likely have important regulatory consequences.
In this study, we adapt a real-time, fluorescence-based assay to gain insights on the mechanisms used by M. tuberculosis CarD to stabilize mycobacterial open complexes. We show that CarD stabilizes RPo for MboRNAP on rrnAP3 in a concentration-dependent manner that saturates at 1 μM and approaches the level of stability displayed by EcoRNAP on the same promoter. The fact that CarD increases the concentration of open complex on RNAP-saturated DNA templates demonstrates that CarD specifically affects the equilibrium between promoter-bound complexes and stabilizes RPo with respect to RPc. This result, in combination with a thermodynamic cycle coupling CarD binding and the equilibrium between RPo and RPc, shows that CarD has a higher affinity to open complex as compared to closed complex. The temperature dependence of CarD activity also matches well with the known temperature dependence of open complex (Figure 4C) (19–21). Furthermore, the kinetics of the approach to equilibrium display interesting trends as a function of both RNAP and CarD concentrations. Specifically, the biphasic behavior of the slowest observed rate (k3obs) as a function of CarD concentration at high RNAP concentrations suggests that CarD uses more than one mechanism to stabilize open complex (Figure 5B). Lastly, we have constructed a detailed kinetic model of the formation of open complex in the presence of CarD that captures all of the experimental trends in the slowest observed rate with a single set of parameters.
Importantly, k3obs also depends on RNAP concentration at high concentrations of CarD (Figure 5C). Since the concentration of the polymerase affects the rate at which it encounters and binds the template, this suggests that the effect of CarD on the kinetics of open-complex formation depends on the rate of association of RNAP. One possibility is that CarD interacts with free polymerase and accelerates its association with DNA. However, while we do not exclude this interaction as part of CarD's overall mechanism, it is unlikely to play a major role as k3obs shows no RNAP concentration dependence in the absence of CarD (Figure 5C, black) suggesting that only increasing the rate of association of holoenzyme has no effect. Instead, to account for the above observations, we hypothesize that CarD is able to associate with closed complex (Figure 6A). The addition of this state to the mechanism has the potential to account for all the data as the formation of a CarD-bound closed complex will naturally depend on the concentrations of both RNAP and CarD.
The full model postulates that at low concentrations, CarD associates with pre-formed open complex and stabilizes it, thus inhibiting its isomerization to closed complex. At higher concentrations, CarD associates with closed complex as well and destabilizes it, thus promoting its isomerization to open complex (Figures 6 and 8). The model satisfies detailed balance and is also consistent with the observation that CarD must have a higher affinity to RPo as compared to RPc. We caution that we do not interpret the presented model parameters as measurements of affinities or equilibria. These parameters will need to be determined via future experiments, however the ability of the model to capture all the trends we observe with a single parameter set suggests that the model topology is correct and is able to capture the critical interactions between MboRNAP, CarD and DNA. At saturating concentrations of both RNAP and CarD, the model predicts that CarD interacts with both open and closed complexes and tilts the equilibrium toward open complexes. As both proteins are present in high concentrations in the cell (> 1 μM, Supplemental Methods, Supplementary Figure S4 and (24)), both activities of CarD are apt to play important roles in transcriptional regulation in vivo.
Figure 8.
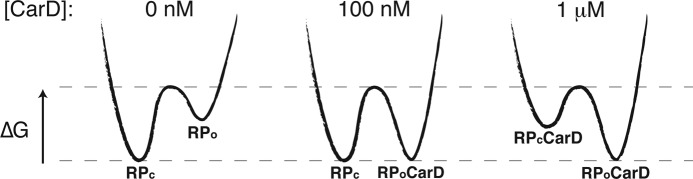
Energy diagram. Schematic energy diagrams showing the relative free energies of closed (RPc) and open (RPo) complexes. In the proposed model, in the absence of CarD (0 nM), RPc is more stable than RPo. In the presence of low concentrations (100 nM), CarD binds to and stabilizes RPo, effectively increasing the energy barrier for closing and inhibiting bubble collapse. In the presence of high concentrations (1 μM), CarD additionally interacts with and destabilizes RPc, effectively decreasing the energy barrier for opening and accelerating the formation of RPo.
Recently, DNA footprinting and transcription assays were used to measure the effect of CarD on the stability of the open complex and transcript production (25). This work nicely shows that although mycobacterial RNAP makes the same contacts with the promoter as E. coli RNAP, it forms an unstable open complex on the rrnAP3 promoter. Furthermore, the addition of CarD at high concentrations led to the stabilization of the open complex and stimulated single rounds of transcript production. Our results are consistent with these data with regard to CarD's ability to inhibit bubble collapse in the open complex. Additionally, the concentration-dependent kinetic analysis of the formation of open complex significantly expands our understanding of the mechanism of CarD and shows that it also functions by accelerating bubble formation.
The two-step binding and opening model is able to capture all of the trends in observed rates presented in this work. The model has served as a useful tool for analyzing open-complex formation of E. coli RNAP on a range of promoters (18,26) and to analyze open-complex formation for mycobacterial RNAP in the absence of transcription factors (27). Given that the mechanism of initiation in E. coli is known to be significantly more complex (1), we expect that the two-step model will serve as a starting point for more detailed studies of CarD's kinetic mechanism of regulating transcription initiation in mycobacteria. Promoter-specific initiation in E. coli involves transitions through multiple closed and open complexes of varying stabilities which are defined by conformational changes in both the DNA as well as RNAP (1). One subtle possibility with respect to the interpretation of the data presented is that early open intermediates in the E. coli mechanism (i.e. I2) also exist in the M. bovis mechanism and do not generate fluorescence enhancement. In this case, the data could be explained via the CarD-dependent acceleration of the formation of RPo via its association with these intermediates instead of with RPc. However, as transitions between intermediates prior to melting of the transcription bubble are thought to be rate limiting, transcription factors which act on these intermediates (i.e. CRP (28) and DksA (29)) can have more dramatic effects on open-complex kinetics. Thus, while we favor the model presented where CarD interacts directly with RPc, we exclude neither the possibility that multiple closed and open intermediate states exist in the mechanism of mycobacterial open-complex formation, nor the possibility that CarD may act on more than one of these states.
To dissect the roles played by different domains of CarD within the context of the proposed kinetic mechanism, real-time fluorescence enhancements were acquired using point-mutants of CarD. Mutants of CarD that weaken its interactions with DNA or the polymerase, or lack the critical tryptophan W85 all show defects in open-complex stabilization. The equilibrium concentration of open complex and the relative changes of the slowest observed rate with the DNA-binding (K90A) and W85A mutants are consistent with a model where they are able to associate with RNAP-bound complexes, but are unable to generate full stabilization (Figure 7B). This interpretation suggests that K90 and W85 are more important for the activity of CarD once it has bound than they are for the recruitment of CarD to the initiation complex. Furthermore, the trends in the observed rates for these mutants suggest that they are defective in both the inhibition of bubble collapse and the acceleration of DNA opening (Figure 7C).
In contrast, in the case of the R25E mutant, the CarD concentration dependence of both open-complex stability and observed rate show significant shifts to higher concentrations (Figure 7B and C). This result is consistent with the RID domain playing a role in the overall recruitment of CarD to RNAP-DNA complexes. Furthermore, the effect of 1 μM R25E CarD is similar to the effect of WT CarD on EcoRNAP (Supplementary Figure S7). This corroborates previous immunoprecipitation data that M. tuberculosis CarD associates with E. coli RNAP with low affinity (9), possibly due to the fact that EcoRNAP lacks glutamate 138 which is thought to form a salt bridge with R25 that is critical for CarD binding to RNAP β (12,23). Lastly, at the highest concentration of R25E, the observed rate is slower than the other mutants, but the equilibrium stabilization of open complex is less. Based on this result, we speculate that the dramatically reduced affinity of R25E completely prevents it from interacting with closed complex at the concentrations tested.
Structural modeling has positioned W85 in a position to interact with the upstream edge of the DNA bubble present in open complex (13). Given this location and its chemical nature, predictions of its role have included promoting formation of open complexes, in a way analogous to the tryptophan residues in σ factors (14,30), or stabilizing the open complex by intercalating into the distorted backbone of the DNA at the upstream edge of the open complex (13,14). The data presented here are consistent with a model in which W85 performs both of these roles. While it is not clear what interactions would lead to a different affinity between open and closed complex in WT CarD, one possibility is the hypothesized steric clash between W85 and closed complex DNA (13). This clash would result in non-optimal binding between the C-terminal domain of CarD and DNA and could lead to a destabilization of CarD-bound closed complex. This destabilization would naturally lead to an increase in all rates leading out of the CarD-bound closed complex. This would increase the rate of CarD dissociation, explaining the decreased affinity of CarD to closed complex and would also lead to an increase in the rate of isomerization to open complex resulting in an acceleration of DNA opening.
In conclusion, the kinetic model of CarD-dependent open-complex stabilization proposed here might have important consequences in terms of the gene-specific effects of CarD. While CarD is localized in vivo at every σA-dependent promoter (13), its role at each promoter is unclear. The ability of CarD to accelerate opening and inhibit bubble collapse will likely lead to complex dependencies on both promoter sequence and the presence of additional transcriptional regulators. More specifically, CarD could either inhibit or promote transcription, depending on the basal kinetics of a given promoter and the basal rates of DNA opening, bubble collapse and promoter escape. Future research along these lines will be required to elucidate the role of CarD across the genome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors would like to thank Dr Bob Landick for assistance with protocols and reagents related to the isolation of MboRNAP. We also thank Dr Tim Lohman for training on and the use of his stopped-flow spectrophotometer and Tom Ellenberger for training on and the use of the HPLC and FPLC in his laboratory. We also thank Dr Lohman, Dr Roberto Galletto and Dr Tom Ellenberger for their scientific input. Lastly, we thank Dr Tomasz Heyduk for his assistance with setting up the fluorescence assay and for critically reading the manuscript while in preparation.
Author contributions: J.R. performed the experiments. E.A.G. and C.L.S. conceived the research. E.A.G. and J.R. planned the experiments, analyzed the data and interpreted the results. A.R.M. purified MboRNAP components and WT CarD, A.L.G. constructed and purified CarD mutants and measured protein levels in the cell. E.A.G., J.R. and C.L.S. wrote the paper.
FUNDING
National Institutes of Health (NIH) [R01GM107544 to E.A.G. and C.L.S.]; Sigma-Aldrich Predoctoral Fellowship [to J.R.]; NIGMS Cell and Molecular Biology Training [GM007067 to A.L.G.]; Stephen I. Morse Graduate Fellowship [to A.L.G.]. Funding for open access charge: NIH [R01GM107544].
Conflict of interest statement. None declared.
REFERENCES
- 1.Saecker R.M., Record M.T., Jr, deHaseth P.L. Mechanism of bacterial transcription initiation: promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rojo F. Mechanisms of transcriptional repression. Curr. Opin. Microbiol. 2001;4:145–151. doi: 10.1016/s1369-5274(00)00180-6. [DOI] [PubMed] [Google Scholar]
- 3.Lee D.J., Minchin S.D., Busby S.J.W. Activating transcription in bacteria. Annu. Rev. Microbiol. 2012;66:125–152. doi: 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 4.Kahmann R., Rudt F., Koch C., Mertens G. G inversion in bacteriophage Mu DNA is stimulated by a site within the invertase gene and a host factor. Cell. 1985;41:771–780. doi: 10.1016/s0092-8674(85)80058-1. [DOI] [PubMed] [Google Scholar]
- 5.Kang P.J., Craig E.A. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao L., Ross W., Appleman J.A., Gaal T., Leirmo S., Schlax P.J., Record M.T., Gourse R.L. Factor independent activation of rrnB P1. An ‘extended’ promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 7.Newlands J.T., Josaitis C.A., Ross W., Gourse R.L. Both fis-dependent and factor-independent upstream activation of the rrnB P1 promoter are face of the helix dependent. Nucleic Acids Res. 1992;20:719–726. doi: 10.1093/nar/20.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Travers A.A. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J. Bacteriol. 1980;141:973–976. doi: 10.1128/jb.141.2.973-976.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stallings C.L., Stephanou N.C., Chu L., Hochschild A., Nickels B.E., Glickman M.S. CarD Is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-Moreno D., Abellón-Ruiz J., García-Heras F., Murillo F.J., Padmanabhan S., Elías-Arnanz M. CdnL, a member of the large CarD-like family of bacterial proteins, is vital for Myxococcus xanthus and differs functionally from the global transcriptional regulator CarD. Nucleic Acids Res. 2010;38:4586–4598. doi: 10.1093/nar/gkq214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X.F., Goldberg M.S., He M., Xu H., Blevins J.S., Norgard M.V. Differential expression of a putative CarD-like transcriptional regulator, LtpA, in Borrelia burgdorferi. Infect. Immun. 2008;76:4439–4444. doi: 10.1128/IAI.00740-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss L.A., Harrison P.G., Nickels B.E., Glickman M.S., Campbell E.A., Darst S.A., Stallings C.L. Interaction of CarD with RNA polymerase mediates Mycobacterium tuberculosis viability, rifampin resistance, and pathogenesis. J. Bacteriol. 2012;194:5621–5631. doi: 10.1128/JB.00879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srivastava D.B., Leon K., Osmundson J., Garner A.L., Weiss L.A., Westblade L.F., Glickman M.S., Landick R., Darst S.A., Stallings C.L., et al. Structure and function of CarD, an essential mycobacterial transcription factor. Proc. Natl. Acad. Sci. USA. 2013;110:12619–12612. doi: 10.1073/pnas.1308270110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garner A.L., Weiss L.A., Manzano A.R., Galburt E.A., Stallings C.L. CarD integrates three functional modules to promote efficient transcription, antibiotic tolerance, and pathogenesis in mycobacteria. Mol. Microbiol. 2014;93:682–697. doi: 10.1111/mmi.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czyz A., Mooney R.A., Iaconi A., Landick R. Mycobacterial RNA polymerase requires a U-tract at intrinsic terminators and Is aided by NusG at suboptimal terminators. mBio. 2014;5:e00931. doi: 10.1128/mBio.00931-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-y-Merchand J.A., Colston M.J., Cox R.A. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology. 1996;142:667–674. doi: 10.1099/13500872-142-3-667. [DOI] [PubMed] [Google Scholar]
- 17.Ko J., Heyduk T. Kinetics of promoter escape by bacterial RNA polymerase: effects of promoter contacts and transcription bubble collapse. Biochem. J. 2014;463:135–144. doi: 10.1042/BJ20140179. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan J.J., Bjornson K.P., Sowers L.C., deHaseth P.L. Spectroscopic determination of open complex formation at promoters for Escherichia coli RNA polymerase. Biochemistry. 1997;36:8005–8012. doi: 10.1021/bi970363k. [DOI] [PubMed] [Google Scholar]
- 19.Kovacic R.T. The 0 degree C closed complexes between Escherichia coli RNA polymerase and two promoters, T7-A3 and lacUV5. J. Biol. Chem. 1987;262:13654–13661. [PubMed] [Google Scholar]
- 20.Cowing D.W., Mecsas J., Record M.T., Gross C.A. Intermediates in the formation of the open complex by RNA polymerase holoenzyme containing the sigma factor sigma 32 at the groE promoter. J. Mol. Biol. 1989;210:521–530. doi: 10.1016/0022-2836(89)90128-9. [DOI] [PubMed] [Google Scholar]
- 21.Schickor P., Metzger W., Werel W., Lederer H., Heumann H. Topography of intermediates in transcription initiation of E. coli. EMBO J. 1990;9:2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cook V.M., deHaseth P.L. Strand opening-deficient Escherichia coli RNA polymerase facilitates investigation of closed complexes with promoter DNA: effects of DNA sequence and temperature. J. Biol. Chem. 2007;282:21319–21326. doi: 10.1074/jbc.M702232200. [DOI] [PubMed] [Google Scholar]
- 23.Westblade L.F., Campbell E.A., Pukhrambam C., Padovan J.C., Nickels B.E., Lamour V., Darst S.A. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010;38:8357–8369. doi: 10.1093/nar/gkq692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu L.M., Fu-Liu C.S. The gene expression data of Mycobacterium tuberculosis based on Affymetrix gene chips provide insight into regulatory and hypothetical genes. BMC Microbiol. 2007;7:37–48. doi: 10.1186/1471-2180-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis E., Chen J., Leon K., Darst S.A., Campbell E.A. Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by CarD. Nucleic Acids Res. 2015;43:433–445. doi: 10.1093/nar/gku1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClure W.R. Rate-limiting steps in RNA chain initiation. Proc. Natl. Acad. Sci. USA. 1980;77:5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tare P., China A., Nagaraja V. Distinct and contrasting transcription initiation patterns at Mycobacterium tuberculosis promoters. PLoS ONE. 2012;7:e43900. doi: 10.1371/journal.pone.0043900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy S., Lim H.M., Liu M., Adhya S. Asynchronous basepair openings in transcription initiation: CRP enhances the rate-limiting step. EMBO J. 2004;23:869–875. doi: 10.1038/sj.emboj.7600098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutherford S.T., Villers C.L., Lee J.-H., Ross W., Gourse R.L. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panaghie G., Aiyar S.E., Bobb K.L., Hayward R.S., de Haseth P.L. Aromatic amino acids in region 2.3 of Escherichia coli sigma 70 participate collectively in the formation of an RNA polymerase-promoter open complex. J. Mol. Biol. 2000;299:1217–1230. doi: 10.1006/jmbi.2000.3808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


