Abstract
Stabilization of the ribosomal complexes plays an important role in translational control. Mechanisms of ribosome stabilization have been studied in detail for initiation and elongation of eukaryotic translation, but almost nothing is known about stabilization of eukaryotic termination ribosomal complexes. Here, we present one of the mechanisms of fine-tuning of the translation termination process in eukaryotes. We show that certain deacylated tRNAs, remaining in the E site of the ribosome at the end of the elongation cycle, increase the stability of the termination and posttermination complexes. Moreover, only the part of eRF1 recognizing the stop codon is stabilized in the A site of the ribosome, and the stabilization is not dependent on the hydrolysis of peptidyl-tRNA. The determinants, defining this property of the tRNA, reside in the acceptor stem. It was demonstrated by site-directed mutagenesis of tRNAVal and construction of a mini-helix structure identical to the acceptor stem of tRNA. The mechanism of this stabilization is different from the fixation of the unrotated state of the ribosome by CCA end of tRNA or by cycloheximide in the E site. Our data allow to reveal the possible functions of the isodecoder tRNAs in eukaryotes.
INTRODUCTION
Translation termination occurs with the participation of class-1 and class-2 release factors (RFs). Class-1 release factors RF1/2 in bacteria and eRF1 in eukaryotes recognize stop codons and hydrolyze peptidyl-tRNA (1). Class-2 release factors are GTPases and have slightly different functions. In bacteria RF3 promotes the release of the RF1/2 from the ribosome after hydrolysis of the peptidyl-tRNA whereas in eukaryotes eRF3 increases the efficiency of hydrolysis of the peptidyl-tRNA by eRF1 (2). eRF3 has a typical structure of GTPases involved in translation and consists of three domains, the G, II and III domains (3). eRF1 also consists of three domains, N, M and C, each of which has its independent function (4). The N domain recognizes stop codon in the A site of the ribosome (5), the M domain is involved in the hydrolysis of the peptidyl-tRNA in the peptidyl transferase center (PTC) (6), and interacts with the G domain of eRF3 (7). The C domain binds to the domain III of eRF3 (8).
It was shown that the complex of release factors eRF1–eRF3 changes its conformation during translation termination on the ribosome (8). Initially, eRF1 interacts with eRF3–GTP through its C and M domains, and its M domain is located distantly from the PTC and closely to the G-domain of eRF3. After GTP hydrolysis and a conformational change of eRF3 or, possibly, the dissociation of eRF3 from the ribosome, the M domain of eRF1 moves to the PTC where it stimulates peptidyl-tRNA hydrolysis. This model is confirmed by experiments using a non-hydrolysable analog of GTP (GMPPNP) to inhibit peptidyl-tRNA cleavage (9). A structural change in the ribosome was observed during binding of eRF1 to the stop codon, which is detected as a two nucleotide downshift of the ribosomal complex on the mRNA in primer extension experiments (9,10). This conformational change can be the result of the stop codon recognition by eRF1 and the following movement of the mRNA into the ribosome as in case of the RelE toxin (11,12) or the result of rearrangements in the mRNA channel (13).
eRF1 is able to hydrolyze peptidyl-tRNA independently of eRF3 but at a slow rate. In the presence of eRF3 the hydrolysis rate increases by at least two orders of magnitude (14). Furthermore, eRF3 increases the capacity of eRF1 to cause a conformational change in the ribosomal complex (10,11). Probably, eRF1 itself has low affinity to the ribosome or its association with the ribosome is unstable and eRF3 improves the efficiency of eRF1 binding with the ribosome. Apparently, binding of eRF3 to the ribosome and the resulting limitation in the mobility of the eRF1 in complex with eRF3 increase the affinity of the eRF1 to the ribosome (13).
To be recycled in another round of protein biosynthesis, ribosomal subunits, after termination of translation, need to be separated from each other. In prokaryotes, the dissociation of the ribosomal subunits is accomplished by ribosome recycling factor (RRF) and elongation factor G (EF-G) (15). In eukaryotes, ribosome recycling can be done in 2 ways (16). Addition of initiation factors eIF1, eIF1A and eIF3 leads to the dissociation of the ribosomal subunits at low concentrations of magnesium. This is a passive type of recycling and is based on the competition between initiation factors and the 60S ribosomal subunit for binding to the 40S subunit. The active pathway of ribosome recycling requires class-1 release factor eRF1 and special ATPase ABCE1 and operates at a high magnesium concentration (8,16).
As mentioned above, active ribosomal recycling requires eRF1. It is known that in the presence of GMPPNP, which does not allow eRF3 dissociation or a conformational change of the eRF1–eRF3 complex, ABCE1-dependent recycling is inhibited (16). This implies that, after GTP hydrolysis and dissociation of eRF3 from the ribosome and before joining ABCE1, eRF1 can stay alone in the A site of the ribosome. Taking into account the kinetic instability of the complex, it is reasonable to presume that additional factors stabilize the intermediate complexes following peptide release. In search of ligands capable of stabilizing eRF1 in the termination complex, it was found that tRNA in the E site of the ribosome is able to accomplish this function (17). Using toe-print analysis of the ribosomal complexes, it was shown that tRNALys and tRNAHis increase the intensity of the two nucleotide toe-print shift, which characterizes eRF1 binding, regardless of the codon in the E site of the ribosome.
Studying the mechanism of eukaryotic translation termination, we have found that not every deacylated tRNA is able to stabilize the termination complex. We have analyzed the impact of twelve different tRNAs on eukaryotic translation termination and provided evidence that only eight of those stabilized the eRF1-ribosome complex. Furthermore, stabilization occurred during binding of the eRF1 or its N domain to a stop codon, and was independent of peptidyl-tRNA hydrolysis. Mutagenesis of tRNAVal showed that several substitutions in its acceptor stem dramatically alter the ability of the tRNA to stabilize the ribosomal complex. The comparison of the stabilizing activities of deacylated tRNA and cycloheximide on the termination complexes allows us to propose the existence of an alternative mechanism of fixing ribosomal complexes, that is not associated with the inhibition of the rotated conformation of the ribosome.
MATERIALS AND METHODS
Ribosomal subunits and recombinant proteins
The 40S and 60S ribosomal subunits, as well as eukaryotic translation factors eIF2, eIF3, eIF4F, eEF1H and eEF2, were purified from a rabbit reticulocyte lysate as described (9). The eukaryotic translation factors eIF1, eIF1A, eIF4A, eIF4B, eIF5B, ΔeIF5, wt eRF1, mutant eRF1s, and eRF3c lacking the N-terminal 138 amino acid residues were produced as recombinant proteins in Escherichia coli strain BL21 with subsequent protein purification on Ni-NTA agarose and ion-exchange chromatography (6,9).
Cloning and mutagenesis of tRNA
tRNAs sequences from the database tRNAdb (http://trna.bioinf.uni-leipzig.de/DataOutput/Search) were used for primers construction (Supplementary Table S1). For mutant tRNAVal(GC) primers contain substitution in respective positions. Direct primers contain T7 promoter sequence, reverse primers carry BstOI restriction site. PCR products were cloned into pUC18 by HindIII and XbaI restriction sites.
In vitro transcription of mRNA and tRNA
mRNA and tRNAs were transcribed by T7 RNA polymerase from MVHL-stop, MVVV-UAA and corresponding tRNA plasmids. mRNA plasmids contained T7 promoter, four CAA repeats, the β-globin 5′-untranslated region (UTR), and corresponding amino acid codons (MVHL, MVVV) followed by the stop codon and a 3′-UTR comprising the rest of the natural β-globin coding sequence (18). For run-off transcription mRNA plasmids were linearized with XhoI, tRNA plasmids with BstOI.
Pretermination complex assembly and purification
Pretermination complexes (preTC) were assembled as described (9). Briefly, 37 pmol of mRNA were incubated for 30 min in buffer A (20 mM Tris–acetate, pH 7.5, 100 mM KAc, 2.5 mM MgCl2, 2 mM DTT) supplemented with 400 U RNase inhibitor (RiboLock, Fermentas), 1 mM ATP, 0.25 mM spermidine, 0.2 mM GTP, 75 μg total tRNA (acylated with all or individual amino acids and [35S]Met), 75 pmol 40S and 60S purified ribosomal subunits, 125 pmol eIF2, eIF3, eIF4F, eIF4A, eIF4B, eIF1, eIF1A, eIF5, eIF5B each, 200 pmol eEF1H and 50 pmol eEF2 and then centrifuged in a Beckman SW55 rotor for 95 min at 4°C and 50 000 rpm in a 10–30% (w/w) linear sucrose density gradient prepared in buffer A with 5 mM MgCl2. Fractions corresponding to preTC complexes according to optical density and the presence of [35S]Met were combined, diluted 3-fold with buffer A containing 1.25 mM MgCl2 (to a final concentration of 2.5 mM Mg2+) and used in peptide release assay or conformational rearrangement analysis.
Peptide release assay
The peptide release assay was conducted as described (9) with minor modifications, as follows. Aliquots containing 0.15 pmol of the preTC assembled in the presence of [35S]Met-tRNA were incubated at 37°C for 3 min with 1 pmol of eRF1, 0.4 pmol of eRF3c and 10 pmol of different deacylated tRNAs. Ribosomes and tRNA were pelleted with ice-cold 5% TCA supplemented with 0.75% (w/v) casamino acids and centrifuged at 14 000 g at 4°C. The amount of released [35S]-containing peptide was determined by scintillation counting of supernatants using an Intertechnique SL-30 liquid scintillation spectrometer.
Conformational rearrangement analysis
Aliquots containing 0.2 pmol of the preTC were incubated with 1 pmol of eRF1/eRF1(AGQ) or domains of eRF1 (10–30 pmol), and 1 pmol of eRF3c with 0.2 mM GTP/GMPPNP and 10 pmol of different deacylated tRNAs (40 pmol of mini-helix tRNA) or 10 pmol cycloheximide for 20 min at 37°C and analyzed using a primer extension protocol, as described (10,19). Toe-printing analysis was performed with a 5′-FAM labeled primer 5′-FAM-GCATTTGCAGAGGACAGG-3′ complementary to β-globin mRNA nucleotides 197–214. cDNAs were separated by electrophoresis using standard GeneScan® conditions on an ABI Prism® Genetic Analyser 3100 (Applera) with ILS 600 molecular weight marker (Promega).
RelE assay
RelE analysis was performed as described (20). Aliquots containing 0.2 pmol of the preTC were incubated with 10 pmol of eRF1 and tRNAHis for 10 min at 37°C and then RelE (to final concentration of 2 μM) was added for 10 min at 37°C. The RNA was purified by phenol/chloroform extraction and precipitated with ethanol, and then reverse transcription was carried out with the same primer as for toe-printing.
RESULTS
Deacylated tRNA stabilizes eRF1 on the ribosome
We tested deacylated tRNAHis as a possible stabilizing factor of eRF1 association with the ribosome. Pretermination complexes (preTC) were reconstituted in an in vitro system composed of individual eukaryotic translation factors and purified on sucrose gradients. They contained MVHL-UAA mRNA with the leucine codon CUG in the P site of the ribosome, the histidine codon CAU in the E site and the UAA stop codon in the A site. The toe-print shift of two nucleotides caused by binding of eRF1 to the stop codon (9) was more pronounced in the presence of tRNAHis than in the absence of tRNA (Figure 1A). It was previously shown that the toe-print shift disappeared after dissociation of eRF1 from the ribosome during centrifugation of the termination complex (TC) in a sucrose gradient at a low magnesium concentration (16). Therefore, we interpreted the impact of the deacylated tRNA on the TC toe-print as a stabilization of the eRF1–ribosome complex. The stabilizing effect of tRNAHis was also observed in the presence of both eukaryotic release factors, eRF1 and eRF3c (Figure 1A). Binding of tRNAHis with the preTC without eRF1 did not change the position of the ribosome. According to cryoEM data, the centrifugation of reconstructed preTC leads to the dissociation of the tRNA from the E site. (13) and the RelE analysis of the ribosomal complexes has shown that the A site remains empty after incubation with deacylated tRNA (Supplementary Figure S1A). We therefore assumed that the deacylated tRNA binds to the free E site of the ribosome after preTC purification and stabilizes eRF1 binding during the termination reaction. Similar stabilization of the termination complexes has earlier been shown for tRNALys and tRNAHis (17).
Figure 1.
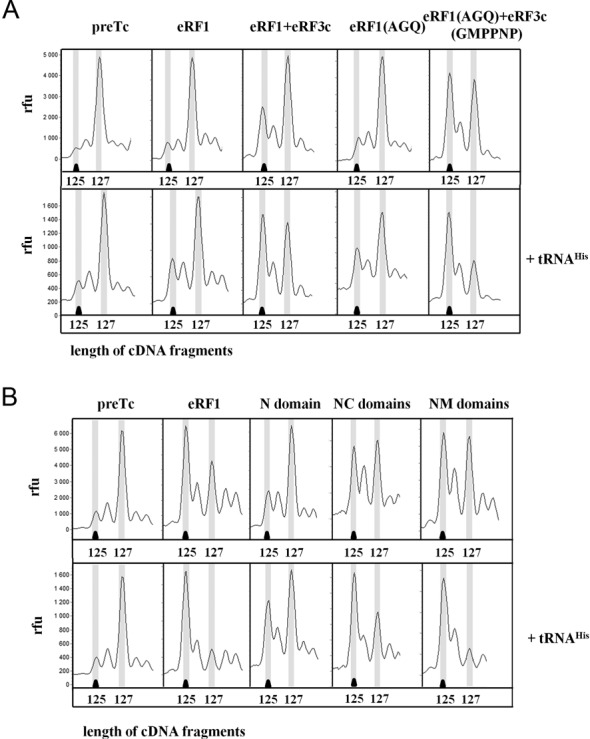
Stabilization of the termination complexes by tRNAHis. (A) Toe-prints with eRF1, eRF1 + eRF3c-GTP, eRF1(AGQ), eRF1(AGQ) + eRF3c-GMPPNP at UAA stop codon. (B) Toe-prints with eRF1 or its domains at UAA stop codon. cDNAs corresponding to preTC and TC have the 127 and 125 nt length, respectively. RFU, relative fluorescence unit. The experiments have been replicated at least three times.
It is known that the eRF1(AGQ) mutant is unable to hydrolyze the peptidyl-tRNA (6). The ribosomal complexes with the mutant protein in the presence or absence of eRF3 were stabilized by tRNAHis as observed with the wild type eRF1 (Figure 1A) and the addition of GMPPNP to the assembly reaction further increased the stabilizing effect (Figure 1A). It has been shown earlier that GMPPNP suppresses the hydrolysis of the peptidyl-tRNA by inhibiting the GTPase activity of eRF3 (9), and by keeping eRF1 in a catalytically inactive conformation (8,13). Thus, stabilization of the protein complex eRF1(AGQ) + eRF3c-GMPPNP on the ribosome by the deacylated tRNA showed that this stalling occurred before peptidyl-tRNA hydrolysis.
It is necessary to note that the positions of the M and C domains of eRF1 in the ribosome are significantly different with eRF3-GMPPNP present in the complex as compared to eRF1 alone (8,13). Since we observed an increase of the TC toe-print by the deacylated tRNA with and without the presence of eRF3–GMPPNP, we concluded that the deacylated tRNA stabilizes the eRF1–ribosome complex by affecting the binding of the eRF1 N domain to the ribosome. To test this hypothesis, we obtained TCs with the NM, NC and N domains of eRF1. All three proteins are able to recognize stop codons and to induce a conformational rearrangement in the ribosome (11). The conformational rearrangement induced by these proteins was strengthened in the presence of the deacylated tRNA (Figure 1B).
To test the effect of deacylated tRNA on the efficiency of peptidyl-tRNA hydrolysis induced by release factors, peptide release experiments were conducted during translation termination in the reconstituted system (Supplementary Figure S1C). The purified assembled preTC contained the peptidyl-tRNA (MVHL-tRNALeu) labeled by [35S]-methionine. Release factors eRF1 and eRF3 were added to this complex in the presence of the deacylated tRNAHis. Deacylated tRNA has a negligible or no effect on the reaction of peptidyl-tRNA hydrolysis in the ribosome.
Different tRNAs stabilize eRF1 on the ribosome with different efficiencies
We tested the influence of 10 different deacylated tRNAs on the stability of the TC reconstituted using MVHL-UAA mRNA, i.e. with the histidine codon CAU in the ribosomal E site, (Table 1, Figure 2A). We found that in addition to tRNAHis also tRNAAla, tRNALeu2, tRNAAsn, tRNALys and even bacterial tRNAiMet effectively stabilized eRF1 on this mRNA in the ribosome. On the other hand, tRNAVal1, tRNAVal2, tRNAMet and tRNALeu1 had no effect on the stability of TC. The anticodons of the tRNAs in each group are very different (Table 1) and the codon-anticodon interaction in the E site could therefore not play a significant role in the stabilization of the TC by tRNA. It should be noted that different tRNAs with the same amino acid specificity influenced the stability of the TC differently. Thus, tRNAiMet and tRNAMet, containing the same anticodon, exhibited various effects on TC. Moreover, we observed different stabilization activities for two tRNALeu with different anticodons.
Table 1. TC stabilization and elongation activities of tRNA transcripts.
| tRNA | Anticodon | Stabilization | Elongation | Sprinzl ID |
|---|---|---|---|---|
| ala | AGC | + | + | DA9380 |
| val1 | AAC | − | + | DV9990 |
| val2 | CAC | − | + | DV9991 |
| val (CG) | CAC | + | + | DV9994 |
| val (GC) | CAC | + | + | mut |
| leu1 | AAG | − | + | DL9991 |
| leu2 | CAG | + | + | DL9350 |
| his | GUG | + | + | DH9330 |
| asn | GUU | + | * | DN9240 |
| lys | UUU | + | + | DK9990 |
| met(ini) | CAU | + | + | DM7630 |
| met | CAU | − | * | DM1231 |
* Marks tRNAs with undetermined elongation activity.
Figure 2.
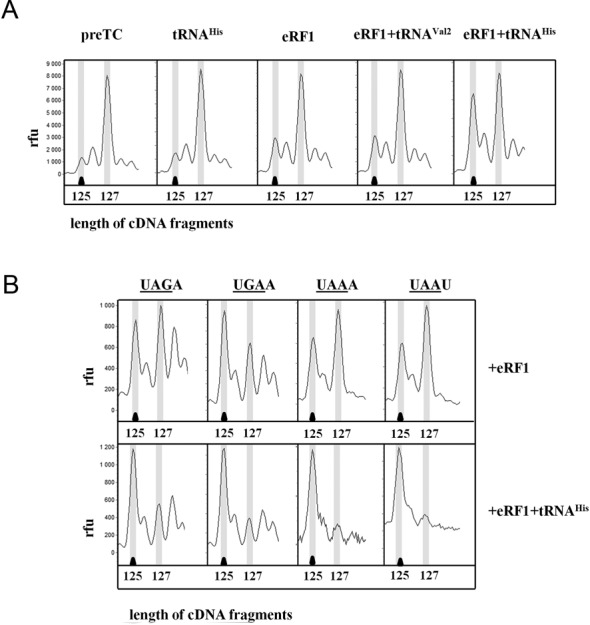
Influence of different tRNAs and sequence of the stop codons or their 3′-context on the stability of the termination complexes. (A) Toe-prints with eRF1 and tRNAHis or tRNAVal2 at UAA stop codon. (B) Toe-prints with eRF1 and tRNAHis on UAA, UAG, UGA codons in two 3′-contexts. cDNAs corresponding to preTC and TC have the 127 and 125 nt length, respectively. RFU, relative fluorescence unit. The experiments have been replicated at least three times.
Since the efficiencies of translation termination at various stop codons are significantly different (11) and can furthermore be affected by the nucleotide following the stop codon (21), we tested the effect of the deacylated tRNA on the stabilization of TCs assembled on mRNAs containing one of the three stop codons and two different nucleotides at position +4 after the stop codon. We found that the tRNAHis stabilized TCs reconstituted on all three stop codons UAA, UAG and UGA (Figure 2B). To study the effect of the nucleotide variation in the stop codon context, we changed the nucleotide at position +4 after UAA into A or U. No differences were detected in the stabilization of the TC by tRNAHis in the different 3′-contexts (Figure 2B). Thus, neither the sequence of the stop codon nor the nucleotide at position +4 affected the ability of the deacylated tRNA to stabilize the TC.
Mutations of human tRNAVal alter its ability to stabilize the TC
It is known that about half of the human tRNA genes are isodecoders, which may differ in any position other than the anticodon (22). Their function is still unknown. In the human genome, several tRNAVal genes were identified and they differ in primary structure to a small extent (22,23). It was shown that in placenta, in addition to the main form of tRNAVal DV9991 with anticodon CAC (tRNAVal2 here), another form of tRNAVal DV9994 with the same anticodon is transcribed (24). This isodecoder tRNAVal (tRNAVal(CG)), has a CG base pair instead of the TA base pair at positions 2–71, and UA instead of CG at positions 6–67 (Figure 3A). It should be noted that tRNAVal2 did not stabilize the TC in our experiments (Table 1). We constructed the isodecoder tRNAVal(CG) and its mutant with a GC base pair at the positions 2–71 tRNAVal(GC). We checked their abilities to stabilize the TC and observed that in the presence of eRF1 both tRNAVal(CG) and tRNAVal(GC) stimulated the two-nucleotide shift of the toe-print on the MVHL-UAA mRNA (Figure 3B). These data were also confirmed using the AGQ mutant of eRF1 (data not shown). Thus, just two pairs of nucleotides (2–71 and 6–67) in the acceptor stem of tRNA determined the ability of the tRNA to stabilize the TC.
Figure 3.
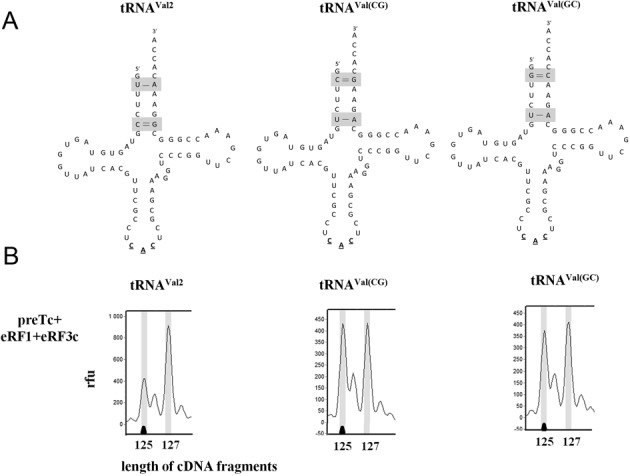
Influence of mutations in tRNAVal2 sequence on the stability of the termination complexes. (A) Positions where substitutions were inserted. (B) Toe-prints with eRF1+eRF3c+GTP and mutated tRNAVal2 at UAA stop codon. cDNAs corresponding to preTC and TC have the 127 and 125 nt length, respectively. RFU, relative fluorescence unit. The experiments have been replicated at least three times.
To confirm the correct folding and functional activity of all in vitro transcribed tRNAsVal, they were aminoacylated by the corresponding aminoacyl-tRNA synthetase and tested in the elongation reaction of protein synthesis. For this purpose, the model mRNA MVVV-UAA was used. We showed that all in vitro transcribed variations of the tRNAVal could be aminoacylated and valine was effectively inserted into the polypeptide chain in the reconstituted translation system (Supplementary Figure S2).
An analogue of the acceptor stem of tRNA stabilizes the TC
Since we found that substitutions of just two base pairs in the acceptor stem of the valine tRNA completely changed its effect on the TC, we supposed that determinants in the acceptor stem are responsible for stabilizing the TC. To test this hypothesis, we obtained a mini-helix analogue of the acceptor stem of tRNAVal(CG) (Figure 4A). It has previously been shown that some truncated forms of tRNA could also bind to the ribosomal E site (25). This analog was tested on purified preTC reconstituted on MVHL-UAA mRNA in the presence of eRF1 and eRF1-eRF3. The truncated tRNA acceptor stem in the same concentration as the full-length tRNA also stabilized the TC but rather weakly. This is probably due to the low affinity of the mini-helix to the ribosome, since it lacks additional ribosome binding determinants present in the native tRNA. At higher concentrations the mini-helix substantially increased the stability of the TC (Figure 4B). Thus, for stabilization of the preferred conformation of the termination ribosomes, only a small fragment of tRNA is sufficient and necessary.
Figure 4.
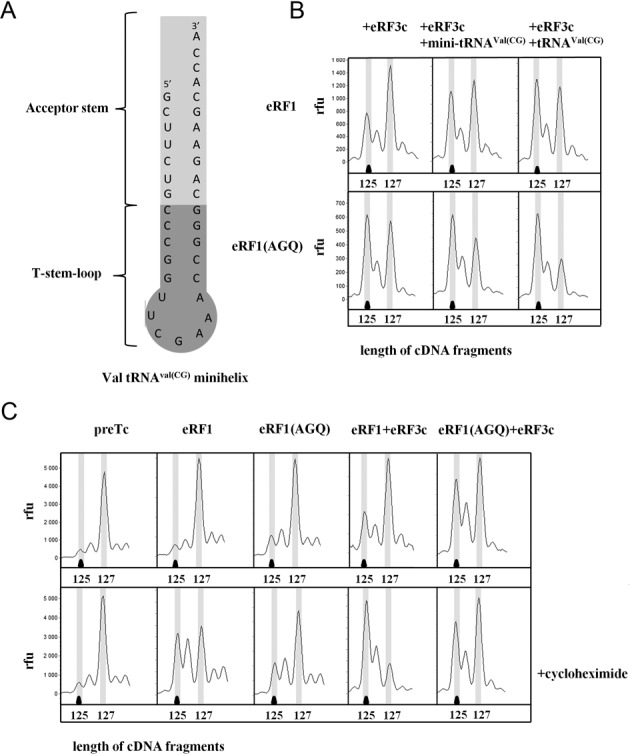
Influence of the analogs of the tRNA acceptor end on the stability of the termination complexes. (A) Structure of the mini-helix obtained from tRNAVal(CG). (B) Toe-prints with eRF1, eRF1(AGQ) and eRF3c+GTP and mini-helix obtained from tRNAVal(CG) at UAA stop codon. (C) Toe-prints with eRF1, eRF1(AGQ) and eRF3c + GTP and cycloheximide at UAA stop codon. cDNAs corresponding to preTC and TC have the 127 and 125 nt length, respectively. RFU, relative fluorescence unit. The experiments have been replicated at least three times.
Cycloheximide stabilizes the postTC
Inherent internal conformational mobility of the ribosome is essential for its functioning. Specifically, ribosomal subunits of eukaryotes and prokaryotes (26,27), are capable of mutual rotation, which is associated with a change in the positions of anticodon and acceptor ends of the tRNA during translocation (27,28). Thus, the rotated state of the ribosome is characterized by moving of the small subunit counterclockwise through 4–8° relative to the large subunit (27). The movement is consistent with the hybrid P/E position of deacylated tRNA, where the acceptor end is transferred to the E site of the large subunit whereas the anticodon loop remains in the P site (29). Such a hybrid position of the tRNA is only possible, if the E site of the large ribosomal subunit is vacant. Consequently, the backward movement of the ribosome can be suppressed, if deacylated tRNA is firmly bound to the E site.
Structural data of the ribosomal complexes with eRF1 and eRF3, and their homologs Dom34 and Hbs1 involved in rescuing of stalled ribosomes showed preferential binding of these factors with unrotated ribosome (13,30). Moreover, it was found that the deacylated tRNA and cycloheximide can revert the rotated state of the ribosome to an unrotated state (26). Cycloheximide is an antibiotic that inhibits translocation via binding to the E site of the large ribosomal subunit (31). Thus, the mechanism of action of cycloheximide on the rotational mobility of the ribosome may be similar to the effect of deacylated tRNA on TC.
To test this hypothesis, we added cycloheximide in a concentration of 0,6 mM to the purified preTC in the presence of eRF1/eRF3 and eRF1(AGQ)/eRF3 (Figure 4C). We found that cycloheximide stabilized the ribosomal complexes with eRF1 and eRF1–eRF3. On the contrary, the stability of the ribosomal complexes with eRF1(AGQ) and eRF1(AGQ)–eRF3 in the presence of cycloheximide were practically unchanged, in contrast to the action of deacylated tRNA. Thus, cycloheximide is only able to affect posttermination events in the ribosome, when the peptidyl-tRNA hydrolysis has already occurred and the deacylated tRNA from the P site can move to the E site. Since the eRF1(AGQ) mutant does not hydrolyze peptidyl-tRNA (5), tRNA is unable to adopt the hybrid P/E state and cycloheximide is unable to influence the ribosomal complex with eRF1(AGQ).
DISCUSSION
We have shown that the binding of eRF1 to the stop codon is improved by the presence of deacylated tRNA at the E site of the ribosome (Figure 1A). This does not require hydrolysis of the peptidyl-tRNA as the AGQ mutant of eRF1 and the complex of eRF1–eRF3–GMPPNP, unable to hydrolyze the peptidyl-tRNA, are also stabilized by the deacylated tRNA in the ribosome. Furthermore, the N domain of eRF1, containing only stop codon recognizing part, is also stabilized by the deacylated tRNA. We suggest that the binding of eRF1 to the stop codon can be improved by deacylated tRNA via fixing the conformation of the ribosome optimal for recognition.
Previously, it was suggested that deacylated tRNA by its CCA end or cycloheximide transfer ribosomes to an unrotated state by blocking the E site of the large subunit and the deacylated tRNA from the P site cannot adopt the hybrid P/E state (26). The deacylated tRNA in the P site may appear either as a result of transpeptidation during translation elongation or as a result of the peptidyl-tRNA hydrolysis in the translation termination. In available cryoelectronic structures of the ribosomal complexes with the eRF1–eRF3 (and their homologs Dom34-Hbs1) the ribosome is in an unrotated state (13,30). According to our data, both cycloheximide and deacylated tRNA stabilize eRF1 binding with the ribosome. One could assume that the mechanism of stabilization of eRF1 in the ribosome is similar in both cases. However, during termination of translation hybrid P/E state of tRNA is possible only after the peptidyl-tRNA hydrolysis, as the movement of the CCA end of tRNA to the E site of the large subunit is inhibited by the peptide. Cycloheximide does not stabilize the eRF1(AGQ) mutant in the ribosome, therefore fixation of the unrotated state is sensitive to the peptidyl-tRNA hydrolysis. On the contrary, the peptidyl-tRNA hydrolysis is not necessary for the stabilization of the TC complex by the deacylated tRNA, which indicates the existence of another mechanism of fixation preferred for binding of release factors conformation of the ribosomal A site.
Interestingly, our results lead to the paradoxical conclusion that cycloheximide can stabilize eRF1 in the posttermination state of the ribosome. It will result in increased efficiency of ribosome recycling induced by ABCE1, as this protein requires eRF1 in the A site for binding to the ribosome. Thus, cycloheximide, a well known and widely used inhibitor of polypeptide chain elongation, can stimulate protein synthesis.
It was recently shown that the large ribosomal subunit protein RPL36AL interacts with the CCA end of tRNA in the P site and eRF1 in the PTC and it can also move with the tRNA to the E site of the ribosome (32). It is reasonable to assume that this protein could serve as a bridge between the E site tRNA and eRF1, stabilizing its binding. For this interaction the physical presence of eRF1 in the ribosomal PTC is required. But we have shown that deacylated tRNA stabilizes even the N-terminal domain of eRF1, which binds with the ribosome near stop codon and is unable to reach the PTC. Therefore there is probably another component of the ribosome stabilizing the TC via interaction with the deacylated tRNA in the E site.
For such stabilization, the ability of the tRNA to bind the E site is very important. It has previously been shown that the strength of binding to the ribosomal E site between different tRNAs may vary by up to 10 times (33,34). We found that different deacylated tRNAs have different effects on the eukaryotic translation termination (Figure 2A, Table 1). From twelve tRNAs studied, eight stabilize the TC, and the remaining four have no effect on it.
We observed that several substitutions in the acceptor stem of tRNA change its properties (Figure 3). Thus, substitutions of nucleotides UA to CG in positions 2–71 and nucleotides CG to UA in positions 6–67 of tRNAVal2 altered its ability to stabilize eRF1 in the ribosome. It should be noted that these mutations cannot affect the process of transcription of mRNA by polymerase III, because tRNA transcription initiation sequence, box A, is located in the positions 8–19 of tRNA gene (35), which was not mutated. In addition, mutated nucleotides are not included in the area of recognition of valyl-tRNA synthetase (36,37), and should not affect the efficiency of aminoacylation. This was also confirmed in experiments on the synthesis of the MVVV peptide with different tRNAsVal (Supplementary Figure S2). Consequently, all the effects, which have been obtained by the mutagenesis of tRNA, appear at the ribosome binding. Different stabilizing TC activity of two isodecoders of tRNAVal may reflect a functional role of the isodecoder tRNAs in cells. In the human genome there are 440 isodecoders of approximately 600 tRNA genes (22) with unknown functional activity. There are multiple evidences of tissue-specific expression of different isodecoder tRNAs (24,38,39) and of heterogeneity of tRNA pool at different stages of the organism development (39–41), though the composition of tRNA pool in tissues correlates with the frequency of codon usage (42,43). Moreover, some isodecoder tRNAs have different affinity to the ribosome. For example, efficiencies of competition of some suppressor tRNAs isodecoders with release factors differ by 20-fold (44). Our data suppose that the difference in the affinity of isodecoder tRNAs to the ribosome, in particular to the ribosomal E site, is an element of fine-tuning of eukaryotic translation.
Besides binding with anticodon, deacylated tRNA in the E site has contacts of the elbow with the L1 ribosomal protein and in the regions of the acceptor stem and deacylated CCA terminus (45,46). It was shown that the codon-anticodon interaction plays a minor role in its affinity to the E site (34), which is consistent with our data (Table 1). So, such stabilization of the ribosomal complexes is not the function of maintaining the reading frame during termination of translation. Contacts of the elbow and CCA-end are conservative and should be very similar for different tRNAs (25,46). We found that the nucleotides in the acceptor stem affect the stabilization of eRF1 in the ribosome (Figures 3B and 4B). Mini-helix structure that is composed of the acceptor stem of tRNA, also stabilized the TC. It is likely that the acceptor stem of tRNA interacts with some regions of the ribosomal RNA or ribosomal proteins, which are responsible for the fixation of conformation of the ribosomal A site required for termination.
In bacteria, the interaction of H68 helix of large ribosomal RNA with the acceptor stem of tRNA includes minor interactions: ribose zipper with the ribose base 70, and possible A-minor interaction with the pair 2–71 (46,47). In the currently available structures of eukaryotic ribosomes helix H68 is also adjacent to the acceptor end of tRNA in the E site (26,48). It becomes clear that the stabilization of the TC is accomplished via interactions of tRNA with the large ribosomal subunit. The minimum stabilized by the tRNA part of eRF1 is the N domain. In our model of stop codon recognition (11), which correlates with the cryoEM data, the N domain contacts only with the residues 2253–2259 of 25S yeast rRNA (Supplementary Figure S3). These residues belong to the helix H69, which has a common base with the helix H68 and which is subjected to certain conformational changes in the 80S ribosome (48,49). It has been identified that the interaction of tRNA with the helix H69 of the 50S subunit plays a crucial role in the formation and stabilization of the A/P, the A, and the P states of tRNA (47,50,51). For prokaryotic ribosome was also shown coordinated motion of these helices during adoption of P/E state by tRNA (49). Moreover, it has recently been shown that the position of H69 in the yeast ribosome is different from that in the mammalian ribosome structure (48). Authors suppose that such a difference was observed perhaps because of the occupancy of the A site of the 40S subunit, which is located near H69 and this helix could be involved in A site tRNA repositioning in the course of translocation.
Combining our data with the information about structures of the ribosomal complexes we propose a model of the TC stabilization by deacylated tRNA (Figure 5). The acceptor stem of deacylated tRNA in the E site possibly interacts with the helix H68 and fixes its conformation and conformation of the helix H69, resulting in stabilization of the N domain of eRF1 in the A site of the ribosome (Figure 5B). After peptide release intersubunit rotation is suppressed by the CCA end of deacylated tRNA in the E site and eRF1 remains in the ribosome. If deacylated tRNA dissociates from the E site, the rearrangement of the helixes H68 and H69 changes the A site structure and destabilizes association of the eRF1-eRF3-GTP complex with the ribosome (Figure 5A). Empty E site after peptide release allows movement of the CCA end of deacylated tRNA from the P site and intersubunit rotation. Structure of the A site changes and eRF1 dissociates from the ribosome.
Figure 5.
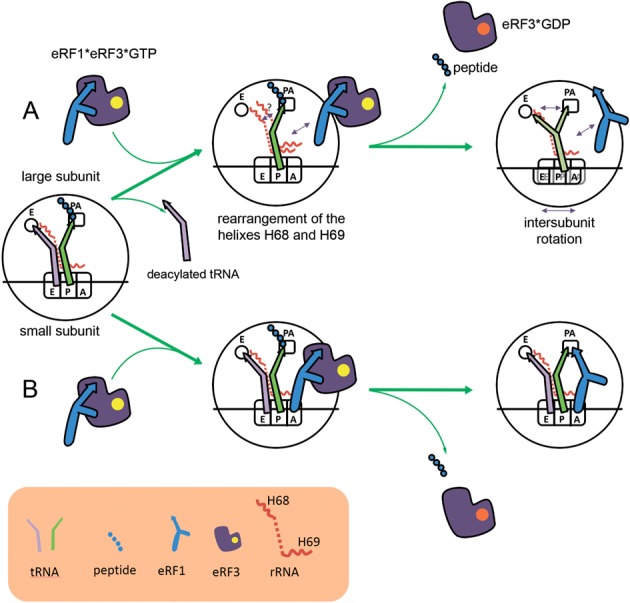
The model of stabilization of eRF1 in the A site of the ribosome by deacylated tRNA. During last round of elongation deacylated tRNA can dissociate from the E site (A). Rearrangement of the helixes H68 and H69 influences the stability of association of the eRF1–eRF3–GTP complex with the ribosome. After peptide and eRF3-GDP release intersubunit rotation is possible which destabilizes the eRF1 binding with the ribosome. When deacylated tRNA remains in the E site after elongation (B), it stabilizes H68 and H69 helixes and promote binding of the eRF1–eRF3–GTP complex with the ribosome. In this case intersubunit rotation after peptide release is suppressed and eRF1 remains in the ribosome.
What is the functional role of the TC stabilization? It may be necessary for proofreading during termination or for the binding of TC to proteins involved in the other steps of translation, for example to ABCE1 or NMD protein complex. Since the binding of eRF1 to the ribosome is required not only for the translation termination but also the further disruption of the 80S ribosomal complex by the ABCE1, the deacylated tRNA can influence this process. Indeed it was previously shown that the addition of tRNALys to translation termination in the presence of ABCE1 decreases stability of TC (17). But we found that deacylated tRNA stabilizes eRF1 in the ribosome before peptide release. Therefore, such stabilization is important for stop codon recognition step during termination of translation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful to A. Grishin for help with 3D structure analysis, K. Strub for critical reading of the manuscript, V. Hauryliuk for kindly providing RelE toxin, L. Frolova for providing plasmids encoding release factors and to T. Pestova and C. Hellen who providing us with recombinant plasmids encoding initiation factors. Sequencing of mutant tRNAs and cDNA fragment analyses were performed by the Center of the collective use ‘Genome’ of EIMB RAS.
FUNDING
Russian Scientific Fund [14-14-00487]. Funding for open access charge: Personal funds of the authors.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kisselev L., Ehrenberg M., Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 2003;33:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kisselev L.L., Buckingham R.H. Translational termination comes of age. Trends Biochem. Sci. 2000;25:561–566. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 3.Kong C., Ito K., Walsh M.A., Wada M., Liu Y., Kumar S., Barford D., Nakamura Y., Song H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 4.Song H., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F., Hemmings B.A., Barford D. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 5.Seit-Nebi A., Frolova L., Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frolova L.Y., Tsivkovskii R.Y., Sivolobova G.F., Oparina N.Y., Serpinsky O.I., Blinov V.M., Tatkov S.I., Kisselev L.L. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kononenko A.V., Mitkevich V.A., Dubovaya V.I., Kolosov P.M., Makarov A.A., Kisselev L.L. Role of the individual domains of translation termination factor eRF1 in GTP binding to eRF3. Proteins. 2008;70:388–393. doi: 10.1002/prot.21544. [DOI] [PubMed] [Google Scholar]
- 8.Preis A., Heuer A., Barrio-Garcia C., Hauser A., Eyler D.E., Berninghausen O., Green R., Becker T., Beckmann R. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 2014;8:59–65. doi: 10.1016/j.celrep.2014.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 10.Shirokikh N.E., Alkalaeva E.Z., Vassilenko K.S., Afonina Z.A., Alekhina O.M., Kisselev L.L., Spirin A.S. Quantitative analysis of ribosome-mRNA complexes at different translation stages. Nucleic Acids Res. 2010;38:e15. doi: 10.1093/nar/gkp1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kryuchkova P., Grishin A., Eliseev B., Karyagina A., Frolova L., Alkalaeva E. Two-step model of stop codon recognition by eukaryotic release factor eRF1. Nucleic Acids Res. 2013;41:4573–4586. doi: 10.1093/nar/gkt113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neubauer C., Gao Y.G., Andersen K.R., Dunham C.M., Kelley A.C., Hentschel J., Gerdes K., Ramakrishnan V., Brodersen D.E. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor D., Unbehaun A., Li W., Das S., Lei J., Liao H.Y., Grassucci R.A., Pestova T.V., Frank J. Cryo-EM structure of the mammalian eukaryotic release factor eRF1-eRF3-associated termination complex. Proc. Natl. Acad. Sci. U.S.A. 2012;109:18413–18418. doi: 10.1073/pnas.1216730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eyler D.E., Wehner K.A., Green R. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J. Biol. Chem. 2013;288:29530–29538. doi: 10.1074/jbc.M113.487090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weixlbaumer A., Petry S., Dunham C.M., Selmer M., Kelley A.C., Ramakrishnan V. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat. Struct. Mol. Biol. 2007;14:733–737. doi: 10.1038/nsmb1282. [DOI] [PubMed] [Google Scholar]
- 16.Pisarev A.V., Skabkin M.A., Pisareva V.P., Skabkina O.V., Rakotondrafara A.M., Hentze M.W., Hellen C.U., Pestova T.V. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skabkin M.A., Skabkina O.V., Hellen C.U., Pestova T.V. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol. Cell. 2013;51:249–264. doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alkalaeva E., Eliseev B., Ambrogelly A., Vlasov P., Kondrashov F.A., Gundllapalli S., Frolova L., Soll D., Kisselev L. Translation termination in pyrrolysine-utilizing archaea. FEBS Lett. 2009;583:3455–3460. doi: 10.1016/j.febslet.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gould P.S., Bird H., Easton A.J. Translation toeprinting assays using fluorescently labeled primers and capillary electrophoresis. Biotechniques. 2005;38:397–400. doi: 10.2144/05383ST02. [DOI] [PubMed] [Google Scholar]
- 20.Andreev D., Hauryliuk V., Terenin I., Dmitriev S., Ehrenberg M., Shatsky I. The bacterial toxin RelE induces specific mRNA cleavage in the A site of the eukaryote ribosome. RNA. 2008;14:233–239. doi: 10.1261/rna.693208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eliseev B., Kryuchkova P., Alkalaeva E., Frolova L. A single amino acid change of translation termination factor eRF1 switches between bipotent and omnipotent stop-codon specificity. Nucleic Acids Res. 2011;39:599–608. doi: 10.1093/nar/gkq759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parisien M., Wang X., Pan T. Diversity of human tRNA genes from the 1000-genomes project. RNA Biol. 2013;10:1853–1867. doi: 10.4161/rna.27361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomann H.U., Schmutzler C., Hudepohl U., Blow M., Gross H.J. Genes, variant genes and pseudogenes of the human tRNA(Val) gene family. Expression and pre-tRNA maturation in vitro. J. Mol. Biol. 1989;209:505–523. doi: 10.1016/0022-2836(89)90590-1. [DOI] [PubMed] [Google Scholar]
- 24.Schmutzler C., Gross H.J. Genes, variant genes, and pseudogenes of the human tRNA(Val) gene family are differentially expressed in HeLa cells and in human placenta. Nucleic Acids Res. 1990;18:5001–5008. doi: 10.1093/nar/18.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmeing T.M., Moore P.B., Steitz T.A. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Budkevich T., Giesebrecht J., Altman R.B., Munro J.B., Mielke T., Nierhaus K.H., Blanchard S.C., Spahn C.M. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol. Cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank J., Agrawal R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 28.Moazed D., Noller H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 29.Agirrezabala X., Liao H.Y., Schreiner E., Fu J., Ortiz-Meoz R.F., Schulten K., Green R., Frank J. Structural characterization of mRNA-tRNA translocation intermediates. Proc. Natl. Acad. Sci. U.S.A. 2012;109:6094–6099. doi: 10.1073/pnas.1201288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker T., Armache J.P., Jarasch A., Anger A.M., Villa E., Sieber H., Motaal B.A., Mielke T., Berninghausen O., Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 31.Garreau de Loubresse N., Prokhorova I., Holtkamp W., Rodnina M.V., Yusupova G., Yusupov M. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014;513:517–522. doi: 10.1038/nature13737. [DOI] [PubMed] [Google Scholar]
- 32.Hountondji C., Bulygin K., Crechet J.B., Woisard A., Tuffery P., Nakayama J., Frolova L., Nierhaus K.H., Karpova G., Baouz S. The CCA-end of P-tRNA contacts both the human RPL36AL and the A-site bound translation termination factor eRF1 at the peptidyl transferase center of the human 80S ribosome. Open Biochem. J. 2014;8:52–67. doi: 10.2174/1874091X01408010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson J.M., Wintermeyer W. Mechanism of ribosomal translocation. tRNA binds transiently to an exit site before leaving the ribosome during translocation. J. Mol. Biol. 1987;196:525–540. doi: 10.1016/0022-2836(87)90030-1. [DOI] [PubMed] [Google Scholar]
- 34.Lill R., Wintermeyer W. Destabilization of codon-anticodon interaction in the ribosomal exit site. J. Mol. Biol. 1987;196:137–148. doi: 10.1016/0022-2836(87)90516-x. [DOI] [PubMed] [Google Scholar]
- 35.Goodenbour J.M., Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–6146. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K., Himeno H., Asahara H., Hasegawa T., Shimizu M. Identity determinants of E. coli tRNA(Val) Biochem. Biophys. Res. Commun. 1991;177:619–623. doi: 10.1016/0006-291x(91)91833-x. [DOI] [PubMed] [Google Scholar]
- 37.Florentz C., Dreher T.W., Rudinger J., Giege R. Specific valylation identity of turnip yellow mosaic virus RNA by yeast valyl-tRNA synthetase is directed by the anticodon in a kinetic rather than affinity-based discrimination. Eur. J. Biochem. 1991;195:229–234. doi: 10.1111/j.1432-1033.1991.tb15698.x. [DOI] [PubMed] [Google Scholar]
- 38.Dittmar K.A., Goodenbour J.M., Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagenbuchle O., Larson D., Hall G.I., Sprague K.U. The primary transcription product of a silkworm alanine tRNA gene: identification of in vitro sites of initiation, termination and processing. Cell. 1979;18:1217–1229. doi: 10.1016/0092-8674(79)90234-4. [DOI] [PubMed] [Google Scholar]
- 40.Kutter C., Brown G.D., Goncalves A., Wilson M.D., Watt S., Brazma A., White R.J., Odom D.T. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat. Genet. 2011;43:948–955. doi: 10.1038/ng.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouyang C., Martinez M.J., Young L.S., Sprague K.U. TATA-binding protein-TATA interaction is a key determinant of differential transcription of silkworm constitutive and silk gland-specific tRNA(Ala) genes. Mol. Cell. Biol. 2000;20:1329–1343. doi: 10.1128/mcb.20.4.1329-1343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong H., Nilsson L., Kurland C.G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 43.Duret L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–289. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- 44.Geslain R., Pan T. Functional analysis of human tRNA isodecoders. J. Mol. Biol. 2010;396:821–831. doi: 10.1016/j.jmb.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng S., Chen Y., Gao Y.G. Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex. PLoS One. 2013;8:e58829. doi: 10.1371/journal.pone.0058829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korostelev A., Trakhanov S., Laurberg M., Noller H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 47.Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 48.Svidritskiy E., Brilot A.F., Koh C.S., Grigorieff N., Korostelev A.A. Structures of yeast 80S ribosome-tRNA complexes in the rotated and nonrotated conformations. Structure. 2014;22:1210–1218. doi: 10.1016/j.str.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W., Frank J. Transfer RNA in the hybrid P/E state: correlating molecular dynamics simulations with cryo-EM data. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16540–16545. doi: 10.1073/pnas.0708094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valle M., Zavialov A., Li W., Stagg S.M., Sengupta J., Nielsen R.C., Nissen P., Harvey S.C., Ehrenberg M., Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 51.Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


