Abstract
Hepatitis B virus (HBV) infection is major global issue, because chronic HBV infection is strongly associated with liver cancer. HBV spread worldwide with various mutations and variations. This variability, called quasispecies, is derived from no proof-reading capacity of viral reverse transcriptase. So far, thousands of studies reported that the variety of genome is closely related to the geographic distribution and clinical characteristics. Recent technological advances including capillary sequencer and next generation sequencer have made in easier to analyze mutations. The variety of HBV genome is related to not only antigenicity of HBs-antigen but also resistance to antiviral therapies. Understanding of these variations is important for the development of diagnostic tools and the appropriate therapy for chronic hepatitis B. In this review, recent publications in relation to HBV mutations and variations are updated and summarized.
Keywords: Hepatitis B virus, Mutation, Quasispecies
Core tip: Hepatitis B virus infection is major global issue. HBV spread worldwide with various mutations and variations. So far, thousands of studies reported that the variety of genome is closely related to the geographic distribution and clinical characteristics. Recent technological advances have made in easier to analyze mutations. Understanding of these variations is important for the development of diagnostic tools and the appropriate therapy for chronic hepatitis B.
INTRODUCTION
Hepatitis B virus (HBV) was first discovered by Blumberg et al[1] in 1965, and the relationship between HBV and acute hepatitis after blood transfusion was reported by Okochi in 1968[2]. At that time, most studies were based on immunological and serological methods. Molecular-based analyses progressed rapidly after the HBV particle was discovered[3] and the HBV genome cloned[4].
HBV infection is major global issue, and is a particular concern in Asia and Africa. Although HBV itself is not directly cytotoxic, the immune response to HBV infection causes liver damage and eventually leads to liver cirrhosis and hepatocellular carcinoma (HCC)[5]. More than 350 million people worldwide are thought to be chronically infected with HBV and 1-2 million people die every year from HBV-related cirrhosis and HCC[6]. The long-term outcomes of chronic hepatitis B (CHB) vary among different countries. The annual incidence of cirrhosis is estimated to range from 2% to 6% in hepatitis B e antigen (HBeAg)-positive patients and from 8% to 10% in HBeAg-negative patients. The annual incidence of HCC ranges from 2% to 3% in cirrhotic patients[7]. The goal of treating CHB is to suppress HBV replication before significant and irreversible liver damage occurs, such as end-stage decompensated cirrhosis and HCC. There are currently two main treatment options for chronic HBV infection, interferon (IFN) and nucleos(t)ide analogues.
HBV GENOME AND REPLICATION
HBV is approximately 42 nm in size. It is an incomplete double-stranded DNA virus from the genus Orthohepadnavirus and family Hepadnaviridae. Its genome consists of full-length coding minus strand DNA and incomplete noncoding plus strand DNA. The viral particle, a Dane particle, comprises an envelope and a core particle (Figure 1). The envelope is composed of a double lipid layer and three envelope proteins: L (large), M (medium), and S (small). The core particle (27 nm in size) consists of the core protein (HBc antigen) and the incomplete double-stranded DNA genome.
Figure 1.
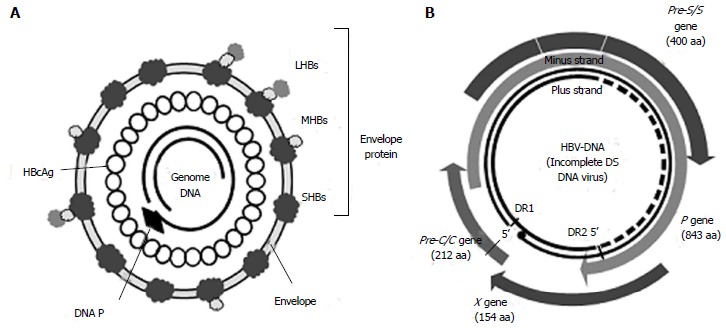
Structures of the hepatitis B virus (Dane particle) (A) and genome (B). aa: Amino acids; C: Core; DS: Double-stranded; LHBs: Hepatitis B large surface protein; HBV: Hepatitis B virus; MHBs: Hepatitis B medium surface protein; P: Polymerase; SHBs: Hepatitis B small surface protein; HBcAg: Hepatitis B c antigen.
The HBV genome is contained within the capsid. It is approximately 3200 bp long, with four overlapping open reading frames (ORFs), which encode the polymerase (P), core (C), surface antigen (S), and X protein (Figure 1B)[8]. Seven viral proteins (HBeAg, HBcAg, LHBs, MHBs, SHBs, polymerase, and HBx) are produced from transcripts (Table 1).
Table 1.
Position of hepatitis B virus regions and transcripts
| ORF | Protein | Position | Amino acids | Protein |
| Pre-S/S | Pre-S1 | 2854-3211 | 119 | LHBs |
| Pre-S2 | 3211-155 | 55 | MHBs/LHBs | |
| S | 155-835 | 226 | SHBs/MHBs/LHBs | |
| P | P | 2357-1623 | 843 | Polymerase |
| X | X | 1374-1838 | 154 | HBxAg |
| Pre-C/C | Pre-C | 1814-1901 | 29 | HBeAg, HBcAg |
| C | 1901-2458 | 183 |
ORF: Open reading frame; S: Surface protein; LHBs: Hepatitis B large surface protein; MHBs: Hepatitis B medium surface protein; SHBs: Hepatitis B small surface protein; P: Polymerase; HBxAg: Hepatitis B X antigen; C: Core; HBeAg: Hepatitis B e antigen; HBcAg: Hepatitis B c antigen.
The entry of HBV into human hepatocytes is the initial step of viral infection. It has been reported that the pre-S1 sequence at amino acids 2-48 mediates the attachment of the virus to its target cell[9]. After invading the target cells, HBV is transported to the nucleus where covalently closed circular DNA is constructed as the replication template of HBV.
Following infection, the HBV DNA is integrated into the host’s cellular DNA. HBV integration induces various kinds of secondary genetic alterations within the host’s genome, including deletions, translocations, and genomic instability[10]. It has been reported that the loss of chromosomal integrity in HCC is attributable to deletions in some chromosomes. In particular, losses in chromosomes 1p, 4q, 5q, 6q, 8p, 9p, 13q, 16p, 16q and 17p have been detected in 25%-45% of patients, whereas gains occur in chromosomes 1p, 6p, 8q, and 17q in 30%-55% of patients[11].
Unlike other DNA viruses, reverse transcriptase is necessary for HBV replication. Because reverse transcriptase has no proof-reading capacity, DNA mutations frequently occur during replication. In general, the mutation rate of the hepadonaviruses is estimated to be 2 × 104 base substitutions/site/year. This mutation rate is approximately 100 times higher than that of other DNA viruses, but 100-1000 times lower than that of RNA viruses[12]. The mutation rate of HBV is reported to be in the range of 1.4-3.2 × 10-5 base substitutions/site/year[13].
Mutations and variations that occur naturally or during antiviral therapy play important roles in viral latency, the pathogenesis of liver disease, immune escape, and resistance to antiviral therapies.
HBV GENOTYPES/SUBTYPES AND MUTATIONS
The major hepatitis B s antigen (HBsAg) protein carries a pair of mutually exclusive determinants, d or y and w or r, which are associated with variations in single amino acids at positions 122 and 160, respectively. Differences in the epitope result in four major serotypes (adr, adw, ayr and ayw) and ten subtypes (Figure 2)[14]. The serotypes and subtypes show differing geographic distributions and affect the antigenic characteristics of HBV[15,16].
Figure 2.
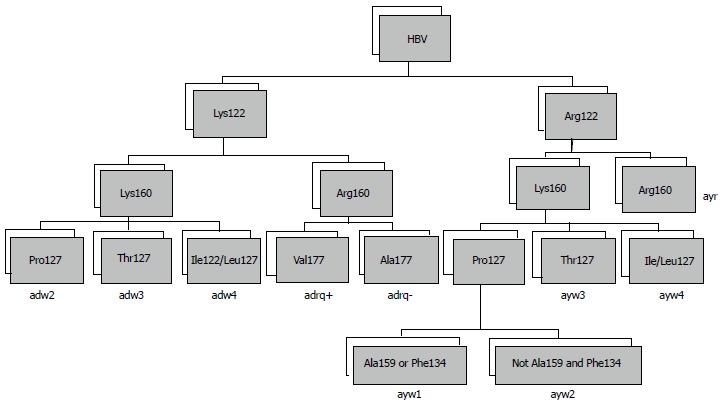
Algorithms for determining hepatitis B virus subtype from the primary structure of the S gene. HBV: Hepatitis B virus.
HBV has been classified into at least 10 genotypes (A-J) according to the divergence of their viral DNA sequences, with distinct geographic distributions (Table 2)[17-19]. In addition, many studies have revealed that the HBV genotype is strongly associated with disease progression and responses to antiviral therapies[20,21].
Table 2.
Hepatitis B virus subtypes and genotypes and their geographic distributions
| Genotype/ subgenotype | Geographic location |
| A | |
| A1 | Saharan Africa, India |
| A2 | Northern Europe |
| A3 | Western Africa |
| B | |
| B1 (Bj) | Japan |
| B2-5 | East and Southeast Asia |
| B6 | Alaska, Northern Canada |
| C | |
| C1-3 | Taiwan, China, South Korea, Southeast Asia |
| C4 | Australia |
| C5 | Philippines, Vietnam |
| D | |
| D1-5 | Africa, Europe, Mediterranean countries and India |
| E | West Africa |
| F | Central and South America |
| G | France, Germany, United States |
| H | Central America |
| I | Vietnam and Laos |
| J | Ryukyu, Japan (Kalimantan) |
HBV/A is mainly distributed in Africa (HBV/A1), the United States of America, and Europe (HBV/A2). Several reports from Africa, India, and Brazil have shown that HBV/A1 is associated with a high incidence of HCC in younger patients without cirrhosis[22]. By contrast, HBV/A2 is reported to be associated with a lower incidence of HCC than HBV/D and HBV/F[23,24]. HBV/A2 readily progresses to chronic infection after an acute infection, and is a major genotype in cases of vertical transmission[25]. HBV/B is mainly distributed in Asia, and is subclassified into HBV/B1/Bj and HBV/B2-5/Ba. HBV/B1 is found in Japan and is the most asymptomatic genotype. HBV/B2-5 is mainly detected in South-East Asia and has similar clinical characteristics to HBV/C. HBV/B and HBV/C are prevalent in the Far East and in South-East Asia. Several studies from Taiwan, Thailand, China, and Japan have shown that HBV/C is more aggressive and is associated with a greater risk of HCC than HBV/B[26-29]. HBV/D is detected worldwide, with HBV/D1 in Central Asia, HBV/D2 in Russia, HBV/D3 in Inner Mongolia, and HBV/D4 in Africa. HBV/D is reportedly associated with worse clinical outcomes than HBV/A[23,30].
The therapeutic efficacy of antiviral drugs is also related to the HBV genotype. HBV/A and HBV/B had better persistent response to IFN than HBV/C and HBV/D. A meta-analysis revealed that the response to IFN, including HBeAg seroconversion, loss of HBeAg and loss of HBV DNA, is better in HBV/A compared with HBV/D, and the response of HBV/B is better than that of HBV/C[31]. Whereas the HBeAg seroconversion rate by one-year treatment were respectively HBV/A 47%, HBV/B 44%, HBV/C 28%, and HBV/D 25%, HBsAg seroclearance rates were respectively HBV/A 14%, HBV/B 9%, HBV/C 3%, and HBV/D 2%[32]. It has also been reported that the HBs and HBe seroconversion rates are higher for HBV/A than for other genotypes[33]. A recent study reported that the response to IFN in HBV/E was worse than that in other genotypes[34]. However, the therapeutic responses to nucleotide analogue have been shown as mostly similar. Several meta-analyses revealed that HBV/B had no different response to lamivudine as HBV/C[35,36]. Though few studies were available, the therapeutic efficacy to other nucleos(t)ide analogues except lamivudine was same among genotypes[37]. It would be because the genomic variety in relation to antiviral resistance within polymerase region were mostly same among genotypes (Table 3).
Table 3.
Nucleotide alignment of polymerase region among different genotypes
| Position | 169 | 173 | 180 | 184 | 202 | 204 | 207 | 213 | 221 | 231 | 236 | 238 | 248 | 250 | |||||||||||||||||||
|
Consensus |
I1 |
V1 |
L1 |
A1 |
T |
- |
S1 |
Y |
M1 |
D |
D |
V |
- |
S |
V |
Q |
H |
L |
- |
F |
T |
A |
V |
- |
L |
- |
N1 |
P |
N |
- |
N |
F |
M1 |
| A1_AY233288_SouthAfrica | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | - | - | - | - | - | - | - | - | - | - | - | - | - |
| A2_AJ309370_France | - | - | - | - | - | - | - | - | - | - | - | - | - | T | - | - | - | R | - | Y | - | - | - | - | - | - | - | - | - | - | - | - | - |
| B1_D23679_Japan | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | A | - | - | - | - | - | - | - | H | - | - | - | - |
| B2_AB073639_Taiwan | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | A | - | - | - | - | - | - | - | H | - | - | - | - |
| B3_M54923_Indonesia | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | A | - | - | - | - | - | - | - | Q | - | - | - | - |
| C1_AB112471_Thailand | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | I | - | - | - | - | - | - | - | - | - | - |
| C2_AB014376_Japan | - | - | - | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | - | S | I | - | - | - | - | - | - | - | - | - | - |
| D1_FJ386590_China | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | H | - | - |
| D2_JF754597_Turkey | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | H | - | - |
| D3_EU594434_Estonia | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| E_X75664_Senegal | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | - | - | - | Y | - | S | - | - | - | - | - | - | - | - | - | - | - |
| F_AF223963_Argentina | - | - | - | - | - | - | - | - | - | - | - | L | - | - | - | - | - | - | - | Y | - | - | - | - | V | - | - | T | S | - | - | - | - |
| G_AB064312_United States | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | - | - | - | - | - | - | - | - | D | - | - | - | - |
| I_AB241408_Vietnam | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | - | - | - | - | - | - | - | - | - | - | - | - | - |
Indicated the known antiviral resistant against nucleos(t)ide analogue.
METHODS AVAILABLE FOR DETECTING MUTATIONS
Recent technological advances have made it easier to detect mutations in HBV DNA. Several approaches can be used to detect HBV genomic mutations. Polymerase chain reaction (PCR) amplification with direct Sanger sequencing is perhaps the most commonly used method, but it cannot detect variations in < 20% of viral quasispecies. By contrast, line probe assays can detect specific variants occurring in > 5% of viral quasispecies[38,39]. Other highly sensitive methods include restriction fragment length polymorphism analysis[40], clone-based sequencing[41], and real-time PCR. More recently, several next-generation sequencing methods have been developed, including ultra-deep pyrosequencing, which can detect thousands of clonally amplified regions[42-45]. However, this method also has some limitations and it is unclear whether the variants found in different positions are actually located in the same clones because the next-generation sequencing read is shorter than the Sanger sequence read. Furthermore, it is still difficult to analyze insertion and deletion variants.
CHARACTERISTICS OF THE PRE-S/S GENE
The S ORF (nt 2854-835) encodes three different translated genes: pre-S1, pre-S2, and S domain. The pre-S domain is essential for viral binding to hepatocyte receptors and contains several epitopes that are targeted by T and B cells. The S domain is also important in the production of HBsAg[9]. There are three forms of HBV surface proteins (S, M, and L), of different sizes[46]. The 24-kDa S protein, which contains 226 amino acids, is the major component of the envelope protein and is involved in particle budding. The 33-kDa M protein contains the S protein and an additional 55 amino acids encoded by the pre-S2 gene. Finally, the 39-kDa L protein contains the M protein and an additional 108 or 119 amino acids, the sequence of which depends on the genotype[47]. The three HBsAg types share a common region, consisting of the main antigenic loop (amino acids 124-147), which is called the “a” determinant region. The “a” determinant region is the main epitope to induce a protective immune response. It is located in the major hydrophilic region (MHR) of the S protein, which is between amino acids 103 and 173. The MHR forms a two-loop structure. Mutations and variations in the S gene have been reported in many countries[48]. Although these mutations occur naturally, they are also generated during immunoglobulin therapy or vaccine-induced immunity. Variations in the “a” determinant region cause changes in HBsAg antigenicity and may prevent the detection of HBsAg in HBsAg screening assays (Figure 3)[49,50]. These mutations also occur naturally in developing countries where nucleos(t)ide analogues are less frequently used than in developed countries[51,52].
Figure 3.
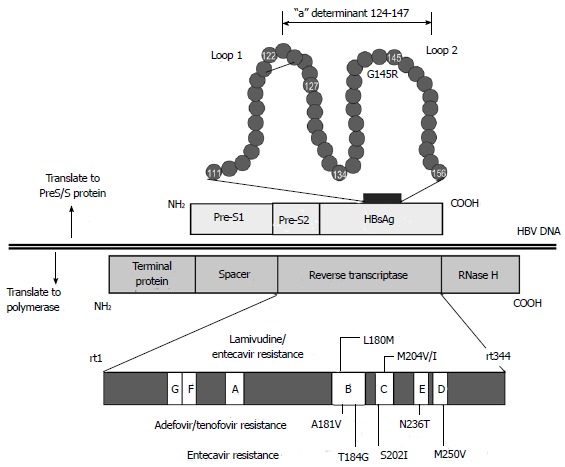
Structures of the overlapping P and S genes. HBsAg: Hepatitis B s antigen; P: Polymerase; S: Surface.
HBsAg is an important diagnostic marker and its expression level is related to the efficacy of an antiviral treatment. Recent studies have revealed that the HBsAg titer correlates with the level of HBV DNA and hepatocarcinogenesis. However, mutations in the pre-S/S region are strongly related to HBs antigenicity, and it is reported that the presence of pre-S/S variants correlates negatively with the HBsAg titer[53]. Mutations in the S region result in antigenic variations and may allow HBV to escape vaccination. Such mutations are known as “vaccine escape mutations”. A study of HBsAg-negative patients from Hong Kong revealed that a variety of mutations, including deletions in the promoter region, abolition of the pre-S2/S start codon, disruption of the pre-S2/S mRNA splice site, nucleotide duplications, and missense mutations in the “a” determinant region, contribute to defects in HBsAg production (Table 4)[54].
Table 4.
Representative mutations and clinical characteristics
| Region | Mutation | Clinical characteristics |
| Pre-S/S | P120S/E, K122R, T126A, P127T, Q129H/R, L134S, K141E, P142S, D144A/E/V, G145R/A | OBI/HBsAg decrease |
| Pre-S deletion | HCC | |
| Pre-C/C | A1896T, G1899A | HBe seroconversion HCC |
| X | C1653T, T1753C, A1762T, G1764A | HCC |
| P | L180M, A181V, T184G, S202I, M204V/I N236T, M250V | NA resistance |
S: Surface; OBI: Occult hepatitis B virus infection; HCC: Hepatocellular carcinoma; HBe: Hepatitis B e; HBsAg: Hepatitis B s antigen.
The HBs antigen was discovered pathologically as ground glass hepatocytes (GGH) in 1973[55]. Different types of GGHs are associated with the expression patterns of surface/core antigens and the stage of virus replication. Type I GGHs express an inclusion-like pattern of HBsAg and carry mutants with deletions in the pre-S1 region. By contrast, type II GGHs are distributed in clusters, emerge in the late replicative phase, and contain mutants with deletions in the pre-S2 region. Because the pre-S2 region includes an epitope targeted by cytotoxic T lymphocytes, type II GGHs may represent an immune escape mutant[56]. It has also been reported that pre-S mutants could induce endoplasmic reticulum stress, followed by oxidative DNA damage and genomic instability[57,58]. Recent studies have also shown that the pre-S2 region upregulates human telomerase reverse transcriptase expression and transactivates forkhead box P3 expression, which may promote the development of HCC[59]. Clinical studies have revealed that pre-S deletions, pre-S2 start codon mutations, and the T53C mutation in the pre-S2 region are related to the development of HCC[60].
CHARACTERISTICS OF THE P GENE
The P gene encodes the 843-amino-acid virus-specific DNA polymerase and partially overlaps the other three genes. The DNA polymerase is located in the core of the virus. It acts as the DNA primer and exhibits reverse transcriptase, RNaseH, and DNA-dependent DNA polymerase activities.
A tyrosine (Y)-methionine (M)-aspartic acid (D)-aspartic acid (D) motif (YMDD) starting at codon 203 forms the enzyme activity center of the polymerase There are several well-known hot spots in the HBV DNA where mutations lead to the emergence of antiviral drug resistance. In particular, long-term treatment with lamivudine sometimes leads to the emergence of YMDD variants and breakthrough hepatitis[61]. Nucleos(t)ide analogues approved for the treatment of CHB include lamivudine, adefovir, entecavir, telbivudine, and tenofovir. Nucleos(t)ide analogues have a similar structure to natural nucleotides and compete with natural nucleotides for binding sites on the polymerase during DNA synthesis. Incorporation of nucleos(t)ide analogues instead of natural nucleotides disrupts DNA synthesis and suppresses viral replication.
M204M/I is a well-known mutation that confers resistance to l-nucleosides, including lamivudine and telbivudine. The M204V/I mutation is also associated with compensatory mutations, such as L80V/I, I169T, V173L, L180M, T184S/G, S202I, and Q215S[62]. Mutations A181T and N236T, which are located outside the YMDD motif, are major mutations that confer resistance to alkyl-phosphonates, such as adefovir and tenofovir[63]. The mutations T184G/S, S202I/G, and M250V in combination with L180M and M204V confer resistance to d-cyclopentanes, including entecavir (Tables 4 and 5)[64].
Table 5.
Mutations associated with resistance to nucleos(t)ide analogues
| Nucleos(t)ide analogue | Resistance-conferring mutations |
| Lamivudine | I169T, V173L, L180M, T184G, S202G, M204V/I, M250V |
| Adefovir | A181V/T, N236T |
| Entecavir | I169T, V173L, L180M, T184G, S202G, M204V/I, M250V |
| Telbivudine | I169T, V173L, L180M, T184G, S202G, M204V/I, M250V |
| Tenofovir | N236T |
The overlapping region of the P and S genes is important for drug resistance and HBs antigenicity (Table 3). A triple mutation in the P protein (V173L + L180M + M204V) is accompanied by a double mutation in the S protein (E164D + I195M). This mutant may confer antiviral resistance and promotes vaccine escape[65,66]. Furthermore, the introduction of an rtA181T (A181T in reverse transcriptase) surface nonsense mutation (rtA181T/sW172*) reduced viral replication and increased drug resistance compared with the introduction of an rtA181T surface missense mutation (rtA181T/sW172S)[67].
CHARACTERISTICS OF THE X GENE
The X ORF (nt 1374-1838) encodes HBx, a 154-amino-acid 16.5 kDa protein. HBx is a multifunctional protein that modulates transcription, signal transduction, cell-cycle progression, protein degradation pathways, apoptosis, and genetic stability by interacting with a variety of host factors[68,69].
HBx protein is strongly associated with the development of HCC. HBx activates cAMP and several transcription factors, including nuclear factor κB and activating transcription factor 2. It also stimulates RAS, SRC, and c-JUN, resulting in activation of the RAS–RAF oncogenic pathways[70].
Deletion of the basal core promoter (BCP) causes a frame shift in the X gene, leading to the production of a truncated X protein. The truncated X protein is frequently detected in HCC, and is thought to contribute to hepatocarcinogenesis by upregulating RAS and MYC. Despite the deletions of nt 1637-1667, which regulate p53-dependent transcription, and nt 1733-1754, corresponding to the SP1-binding region in the CP domain, truncated X protein is still capable of regulating various transcription factors and competes with protein p53. Moreover, because amino-acid mutations at positions 130 and 131 of the X protein overlap the core promoter region, these mutations are associated with the progression of CHB and hepatocarcinogenesis.
Several mutations in the X gene are reported to be are associated with hepatocarcinogenesis. Liao et al[71] reviewed 85 case–control studies and reported that G1896A (OR = 1.46), G1899A (OR = 3.02), the pre-S1 deletion (OR = 2.94), and pre-S2 deletion (OR = 3.02) were significantly associated with the development of HCC. The A1762T/G1764A double mutant, T1753V and C1653T in the BCP were also associated with HCC. Similar results have reported in another meta-analysis of case–control studies[72], and several other mutations in the X gene are associated with hepatocarcinogenesis[73,74].
CHARACTERISTICS OF THE PRE-CORE/CORE GENOME
The pre-C/core region contains two regions; pre-C (nt 1814-1901) and core (nt 1901-2452). The core ORF encodes an 183-amino-acid core protein (HBcAg) and the pre-C ORF encodes the 29-amino-acid protein that connects to the N-terminal tail of the core protein. Although the pre-C/C gene produces HBcAg and HBeAg, only the cleaved form of HBeAg is released from infected cells into the blood, together with the HBV particle. Although the function of HBeAg is not completely understood, it may act as an immune “tolerogen”, contributing to the establishment of chronic infection[75].
The BCP and the adjacent pre-C region are crucial for the replication of HBV. The BCP binds to various liver factors and pre-C forms a pregenomic RNA structure that acts as the encapsidation signal[8]. Changes in viral replication may influence the progression of liver diseases[76]. In particular, nucleotide mutation G1896A, which replaces tryptophan with a stop codon at codon 28, is the most common and important factor responsible for the inhibition of HBeAg production[77,78].
A relatively common double mutation (A1762T and G1764A) in the BCP is responsible for reduced pre-C mRNA synthesis[79].
CONCLUSION
The treatment of CHB has changed dramatically in recent years. However, there is increasing evidence that viral variations and mutations that allow the virus to escape antiviral therapies are clinically important. HBV mutations are also closely related to the serological status of the patients. Understanding the viral mutations and their associations with the clinical characteristics of HBV infection should contribute to improvements in diagnostic procedures and therapeutic guidelines. Recent technological advances have made it easier to assess the HBV genome and detect possible variations or mutations in it. We believe it is important to discuss and implement generalized methods that are suitable for use worldwide.
Footnotes
Supported by A Grant-in-Aid from the Japan Initiative for Global Research Network on Infectious Disease (J-GRID) Program of the Ministry of Education, Culture, Sports, Science and Technology, Japan; a SATREPS Grant from the Japan Science and Technology Agency and the Japan International Cooperation Agency; and by the Ministry of Health, Labour, and Welfare of Japan, No. H25-general-008.
Conflict-of-interest: None.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: August 28, 2014
First decision: November 14, 2014
Article in press: December 29, 2014
P- Reviewer: Al-Shamma S, Amarapurkar DN, Betrosian AP, El-Bendary M S- Editor: Tian YL L- Editor: A E- Editor: Wu HL
References
- 1.Blumberg BS, Alter HJ, Visnich S. A “new” antigen in leukemia sera. JAMA. 1965;191:541–546. doi: 10.1001/jama.1965.03080070025007. [DOI] [PubMed] [Google Scholar]
- 2.Okochi K, Murakami S. Observations on Australia antigen in Japanese. Vox Sang. 1968;15:374–385. doi: 10.1111/j.1423-0410.1993.tb04532.x. [DOI] [PubMed] [Google Scholar]
- 3.Dane DS, Cameron CH, Briggs M. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet. 1970;1:695–698. doi: 10.1016/s0140-6736(70)90926-8. [DOI] [PubMed] [Google Scholar]
- 4.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 5.Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 6.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 7.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–S50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glebe D, Urban S, Knoop EV, Cag N, Krass P, Grün S, Bulavaite A, Sasnauskas K, Gerlich WH. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology. 2005;129:234–245. doi: 10.1053/j.gastro.2005.03.090. [DOI] [PubMed] [Google Scholar]
- 10.Bréchot C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology. 2004;127:S56–S61. doi: 10.1053/j.gastro.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 11.Zhang SH, Cong WM, Xian ZH, Wu MC. Clinicopathological significance of loss of heterozygosity and microsatellite instability in hepatocellular carcinoma in China. World J Gastroenterol. 2005;11:3034–3039. doi: 10.3748/wjg.v11.i20.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buti M, Rodriguez-Frias F, Jardi R, Esteban R. Hepatitis B virus genome variability and disease progression: the impact of pre-core mutants and HBV genotypes. J Clin Virol. 2005;34 Suppl 1:S79–S82. doi: 10.1016/s1386-6532(05)80015-0. [DOI] [PubMed] [Google Scholar]
- 13.Orito E, Mizokami M, Ina Y, Moriyama EN, Kameshima N, Yamamoto M, Gojobori T. Host-independent evolution and a genetic classification of the hepadnavirus family based on nucleotide sequences. Proc Natl Acad Sci USA. 1989;86:7059–7062. doi: 10.1073/pnas.86.18.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Couroucé-Pauty AM, Plançon A, Soulier JP. Distribution of HBsAg subtypes in the world. Vox Sang. 1983;44:197–211. doi: 10.1111/j.1423-0410.1983.tb01885.x. [DOI] [PubMed] [Google Scholar]
- 15.Le Bouvier GL, McCollum RW, Hierholzer WJ, Irwin GR, Krugman S, Giles JP. Subtypes of Australia antigen and hepatitis-B virus. JAMA. 1972;222:928–930. doi: 10.1001/jama.222.8.928. [DOI] [PubMed] [Google Scholar]
- 16.Norder H, Couroucé AM, Magnius LO. Molecular basis of hepatitis B virus serotype variations within the four major subtypes. J Gen Virol. 1992;73(Pt 12):3141–3145. doi: 10.1099/0022-1317-73-12-3141. [DOI] [PubMed] [Google Scholar]
- 17.Norder H, Couroucé AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 18.McMahon BJ. The influence of hepatitis B virus genotype and subgenotype on the natural history of chronic hepatitis B. Hepatol Int. 2009;3:334–342. doi: 10.1007/s12072-008-9112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurbanov F, Tanaka Y, Mizokami M. Geographical and genetic diversity of the human hepatitis B virus. Hepatol Res. 2010;40:14–30. doi: 10.1111/j.1872-034X.2009.00601.x. [DOI] [PubMed] [Google Scholar]
- 20.Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554–559. doi: 10.1016/s0016-5085(00)70261-7. [DOI] [PubMed] [Google Scholar]
- 21.Kramvis A, Kew M, François G. Hepatitis B virus genotypes. Vaccine. 2005;23:2409–2423. doi: 10.1016/j.vaccine.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 22.Kew MC, Kramvis A, Yu MC, Arakawa K, Hodkinson J. Increased hepatocarcinogenic potential of hepatitis B virus genotype A in Bantu-speaking sub-saharan Africans. J Med Virol. 2005;75:513–521. doi: 10.1002/jmv.20311. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez-Tapias JM, Costa J, Mas A, Bruguera M, Rodés J. Influence of hepatitis B virus genotype on the long-term outcome of chronic hepatitis B in western patients. Gastroenterology. 2002;123:1848–1856. doi: 10.1053/gast.2002.37041. [DOI] [PubMed] [Google Scholar]
- 24.Livingston SE, Simonetti JP, McMahon BJ, Bulkow LR, Hurlburt KJ, Homan CE, Snowball MM, Cagle HH, Williams JL, Chulanov VP. Hepatitis B virus genotypes in Alaska Native people with hepatocellular carcinoma: preponderance of genotype F. J Infect Dis. 2007;195:5–11. doi: 10.1086/509894. [DOI] [PubMed] [Google Scholar]
- 25.Matsuura K, Tanaka Y, Hige S, Yamada G, Murawaki Y, Komatsu M, Kuramitsu T, Kawata S, Tanaka E, Izumi N, et al. Distribution of hepatitis B virus genotypes among patients with chronic infection in Japan shifting toward an increase of genotype A. J Clin Microbiol. 2009;47:1476–1483. doi: 10.1128/JCM.02081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CM, Chen CH, Lu SN, Tung HD, Chou WJ, Wang JH, Chen TM, Hung CH, Huang CC, Chen WJ. Prevalence and clinical implications of hepatitis B virus genotypes in southern Taiwan. Scand J Gastroenterol. 2003;38:95–101. doi: 10.1080/00365520310000500. [DOI] [PubMed] [Google Scholar]
- 27.Tangkijvanich P, Mahachai V, Komolmit P, Fongsarun J, Theamboonlers A, Poovorawan Y. Hepatitis B virus genotypes and hepatocellular carcinoma in Thailand. World J Gastroenterol. 2005;11:2238–2243. doi: 10.3748/wjg.v11.i15.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, Okanoue T, Yotsuyanagi H, Iino S. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33:218–223. doi: 10.1053/jhep.2001.20532. [DOI] [PubMed] [Google Scholar]
- 29.Zhang AM, Wang HF, Wang HB, Hu JH, He WP, Su HB, Chen J, Du N, Duan XZ. Association between HBV genotype and chronic/severe liver disease with HBV infection in Chinese patients. Zhonghua Shiyan He Linchuang Bingduxue Zazhi. 2010;24:178–180. [PubMed] [Google Scholar]
- 30.Thakur V, Guptan RC, Kazim SN, Malhotra V, Sarin SK. Profile, spectrum and significance of HBV genotypes in chronic liver disease patients in the Indian subcontinent. J Gastroenterol Hepatol. 2002;17:165–170. [Google Scholar]
- 31.Wiegand J, Hasenclever D, Tillmann HL. Should treatment of hepatitis B depend on hepatitis B virus genotypes? A hypothesis generated from an explorative analysis of published evidence. Antivir Ther. 2008;13:211–220. [PubMed] [Google Scholar]
- 32.Buster EH, Flink HJ, Cakaloglu Y, Simon K, Trojan J, Tabak F, So TM, Feinman SV, Mach T, Akarca US, Schutten M, Tielemans W, van Vuuren AJ, Hansen BE, Janssen HL. Sustained HBeAg and HBsAg loss after long-term follow-up of HBeAg-positive patients treated with peginterferon alpha-2b. Gastroenterology. 2008;135:459–467. doi: 10.1053/j.gastro.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 33.Boglione L, Cusato J, Cariti G, Di Perri G, D’Avolio A. The E genotype of hepatitis B: clinical and virological characteristics, and response to interferon. J Infect. 2014;69:81–87. doi: 10.1016/j.jinf.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 34.Chen XL, Li M, Zhang XL. HBV genotype B/C and response to lamivudine therapy: a systematic review. Biomed Res Int. 2013;2013:672614. doi: 10.1155/2013/672614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palumbo E. Hepatitis B genotypes and response to antiviral therapy: a review. Am J Ther. 2007;14:306–309. doi: 10.1097/01.pap.0000249927.67907.eb. [DOI] [PubMed] [Google Scholar]
- 36.Enomoto M, Tamori A, Nishiguchi S. Hepatitis B virus genotypes and response to antiviral therapy. Clin Lab. 2006;52:43–47. [PubMed] [Google Scholar]
- 37.Wen Z, Zhang H, Zhang M, Tan D, Li Q, Zhang H, Wu P, Deng L. Effect of hepatitis B virus genotypes on the efficacy of adefovir dipivoxil antiviral therapy. Hepat Mon. 2014;14:e10813. doi: 10.5812/hepatmon.10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Y, Guindon S, Rodrigo A, Wee LY, Inoue M, Thompson AJ, Locarnini S, Lim SG. Cumulative viral evolutionary changes in chronic hepatitis B virus infection precedes hepatitis B e antigen seroconversion. Gut. 2013;62:1347–1355. doi: 10.1136/gutjnl-2012-302408. [DOI] [PubMed] [Google Scholar]
- 39.Ghabeshi S, Sharifi Z, Hosseini SM, Mahmoodian Shooshtari M. Correlation between viral load of HBV in chronic hepatitis B patients and precore and Basal core promoter mutations. Hepat Mon. 2013;13:e7415. doi: 10.5812/hepatmon.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizokami M, Nakano T, Orito E, Tanaka Y, Sakugawa H, Mukaide M, Robertson BH. Hepatitis B virus genotype assignment using restriction fragment length polymorphism patterns. FEBS Lett. 1999;450:66–71. doi: 10.1016/s0014-5793(99)00471-8. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Zheng CX, Lin MH, Huang ZX, Chen RH, Li QG, Li Q, Chen P. Distinct quasispecies characteristics and positive selection within precore/core gene in hepatitis B virus HBV associated acute-on-chronic liver failure. J Gastroenterol Hepatol. 2013;28:1040–1046. doi: 10.1111/jgh.12109. [DOI] [PubMed] [Google Scholar]
- 42.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 43.Astrovskaya I, Tork B, Mangul S, Westbrooks K, Măndoiu I, Balfe P, Zelikovsky A. Inferring viral quasispecies spectra from 454 pyrosequencing reads. BMC Bioinformatics. 2011;12 Suppl 6:S1. doi: 10.1186/1471-2105-12-S6-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin KT, Shann YJ, Chau GY, Hsu CN, Huang CY. Identification of latent biomarkers in hepatocellular carcinoma by ultra-deep whole-transcriptome sequencing. Oncogene. 2014;33:4786–4794. doi: 10.1038/onc.2013.424. [DOI] [PubMed] [Google Scholar]
- 45.Gong L, Han Y, Chen L, Liu F, Hao P, Sheng J, Li XH, Yu DM, Gong QM, Tian F, et al. Comparison of next-generation sequencing and clone-based sequencing in analysis of hepatitis B virus reverse transcriptase quasispecies heterogeneity. J Clin Microbiol. 2013;51:4087–4094. doi: 10.1128/JCM.01723-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt S, Glebe D, Alving K, Tolle TK, Linder M, Geyer H, Linder D, Peter-Katalinic J, Gerlich WH, Geyer R. Analysis of the pre-S2 N- and O-linked glycans of the M surface protein from human hepatitis B virus. J Biol Chem. 1999;274:11945–11957. doi: 10.1074/jbc.274.17.11945. [DOI] [PubMed] [Google Scholar]
- 47.Ni Y, Sonnabend J, Seitz S, Urban S. The pre-s2 domain of the hepatitis B virus is dispensable for infectivity but serves a spacer function for L-protein-connected virus assembly. J Virol. 2010;84:3879–3888. doi: 10.1128/JVI.02528-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sayiner AA, Ozcan A, Sengonul A. Naturally occurring MHR variants in Turkish patients infected with hepatitis B virus. J Med Virol. 2008;80:405–410. doi: 10.1002/jmv.21104. [DOI] [PubMed] [Google Scholar]
- 49.Carman WF, Mimms LT. Pre-S/S gene variants of hepatitis B virus. Rizetto M, Purcell RH, Gerin JL, Verne G, editors. Viral hepatitis and liver disease. Turin: Edizioni Minerva Medica; 1997. pp. 108–115. [Google Scholar]
- 50.Melegari M, Bruno S, Wands JR. Properties of hepatitis B virus pre-S1 deletion mutants. Virology. 1994;199:292–300. doi: 10.1006/viro.1994.1127. [DOI] [PubMed] [Google Scholar]
- 51.Pourkarim MR, Sharifi Z, Soleimani A, Amini-Bavil-Olyaee S, Elsadek Fakhr A, Sijmons S, Vercauteren J, Karimi G, Lemey P, Maes P, et al. Evolutionary analysis of HBV “S” antigen genetic diversity in Iranian blood donors: a nationwide study. J Med Virol. 2014;86:144–155. doi: 10.1002/jmv.23798. [DOI] [PubMed] [Google Scholar]
- 52.Suwannakarn K, Tangkijvanich P, Thawornsuk N, Theamboonlers A, Tharmaphornpilas P, Yoocharoen P, Chongsrisawat V, Poovorawan Y. Molecular epidemiological study of hepatitis B virus in Thailand based on the analysis of pre-S and S genes. Hepatol Res. 2008;38:244–251. doi: 10.1111/j.1872-034X.2007.00254.x. [DOI] [PubMed] [Google Scholar]
- 53.Pollicino T, Amaddeo G, Restuccia A, Raffa G, Alibrandi A, Cutroneo G, Favaloro A, Maimone S, Squadrito G, Raimondo G. Impact of hepatitis B virus (HBV) preS/S genomic variability on HBV surface antigen and HBV DNA serum levels. Hepatology. 2012;56:434–443. doi: 10.1002/hep.25592. [DOI] [PubMed] [Google Scholar]
- 54.Huang FY, Wong DK, Seto WK, Zhang AY, Lee CK, Lin CK, Fung J, Lai CL, Yuen MF. Sequence variations of full-length hepatitis B virus genomes in Chinese patients with HBsAg-negative hepatitis B infection. PLoS One. 2014;9:e99028. doi: 10.1371/journal.pone.0099028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hadziyannis S, Gerber MA, Vissoulis C, Popper H. Cytoplasmic hepatitis B antigen in “ground-glass” hepatocytes of carriers. Arch Pathol. 1973;96:327–330. [PubMed] [Google Scholar]
- 56.Wang HC, Wu HC, Chen CF, Fausto N, Lei HY, Su IJ. Different types of ground glass hepatocytes in chronic hepatitis B virus infection contain specific pre-S mutants that may induce endoplasmic reticulum stress. Am J Pathol. 2003;163:2441–2449. doi: 10.1016/S0002-9440(10)63599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsieh YH, Su IJ, Wang HC, Chang WW, Lei HY, Lai MD, Chang WT, Huang W. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis. 2004;25:2023–2032. doi: 10.1093/carcin/bgh207. [DOI] [PubMed] [Google Scholar]
- 58.Pollicino T, Cacciola I, Saffioti F, Raimondo G. Hepatitis B virus PreS/S gene variants: pathobiology and clinical implications. J Hepatol. 2014;61:408–417. doi: 10.1016/j.jhep.2014.04.041. [DOI] [PubMed] [Google Scholar]
- 59.Zhang X, Gao L, Liang X, Guo M, Wang R, Pan Y, Liu P, Zhang F, Guo C, Zhu F, et al. HBV preS2 transactivates FOXP3 expression in malignant hepatocytes. Liver Int. 2015;35:1087–1094. doi: 10.1111/liv.12642. [DOI] [PubMed] [Google Scholar]
- 60.Qu LS, Liu JX, Liu TT, Shen XZ, Chen TY, Ni ZP, Lu CH. Association of hepatitis B virus pre-S deletions with the development of hepatocellular carcinoma in Qidong, China. PLoS One. 2014;9:e98257. doi: 10.1371/journal.pone.0098257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liaw YF, Leung NW, Chang TT, Guan R, Tai DI, Ng KY, Chien RN, Dent J, Roman L, Edmundson S, et al. Effects of extended lamivudine therapy in Asian patients with chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. Gastroenterology. 2000;119:172–180. doi: 10.1053/gast.2000.8559. [DOI] [PubMed] [Google Scholar]
- 62.Bartholomeusz A, Locarnini SA. Antiviral drug resistance: clinical consequences and molecular aspects. Semin Liver Dis. 2006;26:162–170. doi: 10.1055/s-2006-939758. [DOI] [PubMed] [Google Scholar]
- 63.Borroto-Esoda K, Miller MD, Arterburn S. Pooled analysis of amino acid changes in the HBV polymerase in patients from four major adefovir dipivoxil clinical trials. J Hepatol. 2007;47:492–498. doi: 10.1016/j.jhep.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 64.Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–1608.e1-2. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]
- 65.Torresi J, Earnest-Silveira L, Civitico G, Walters TE, Lewin SR, Fyfe J, Locarnini SA, Manns M, Trautwein C, Bock TC. Restoration of replication phenotype of lamivudine-resistant hepatitis B virus mutants by compensatory changes in the “fingers” subdomain of the viral polymerase selected as a consequence of mutations in the overlapping S gene. Virology. 2002;299:88–99. doi: 10.1006/viro.2002.1448. [DOI] [PubMed] [Google Scholar]
- 66.Villet S, Pichoud C, Villeneuve JP, Trépo C, Zoulim F. Selection of a multiple drug-resistant hepatitis B virus strain in a liver-transplanted patient. Gastroenterology. 2006;131:1253–1261. doi: 10.1053/j.gastro.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 67.Ahn SH, Park YK, Park ES, Kim JH, Kim DH, Lim KH, Jang MS, Choe WH, Ko SY, Sung IK, et al. The impact of the hepatitis B virus polymerase rtA181T mutation on replication and drug resistance is potentially affected by overlapping changes in surface gene. J Virol. 2014;88:6805–6818. doi: 10.1128/JVI.00635-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pang R, Tse E, Poon RT. Molecular pathways in hepatocellular carcinoma. Cancer Lett. 2006;240:157–169. doi: 10.1016/j.canlet.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 69.Tang H, Oishi N, Kaneko S, Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977–983. doi: 10.1111/j.1349-7006.2006.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benn J, Schneider RJ. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao Y, Hu X, Chen J, Cai B, Tang J, Ying B, Wang H, Wang L. Precore mutation of hepatitis B virus may contribute to hepatocellular carcinoma risk: evidence from an updated meta-analysis. PLoS One. 2012;7:e38394. doi: 10.1371/journal.pone.0038394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, Cao G. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066–1082. doi: 10.1093/jnci/djp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madden CR, Finegold MJ, Slagle BL. Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J Virol. 2001;75:3851–3858. doi: 10.1128/JVI.75.8.3851-3858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sirma H, Giannini C, Poussin K, Paterlini P, Kremsdorf D, Bréchot C. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene. 1999;18:4848–4859. doi: 10.1038/sj.onc.1202867. [DOI] [PubMed] [Google Scholar]
- 75.Yang CY, Kuo TH, Ting LP. Human hepatitis B viral e antigen interacts with cellular interleukin-1 receptor accessory protein and triggers interleukin-1 response. J Biol Chem. 2006;281:34525–34536. doi: 10.1074/jbc.M510981200. [DOI] [PubMed] [Google Scholar]
- 76.Jammeh S, Tavner F, Watson R, Thomas HC, Karayiannis P. Effect of basal core promoter and pre-core mutations on hepatitis B virus replication. J Gen Virol. 2008;89:901–909. doi: 10.1099/vir.0.83468-0. [DOI] [PubMed] [Google Scholar]
- 77.Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Makris A, Thomas HC. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;2:588–591. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 78.Omata M, Ehata T, Yokosuka O, Hosoda K, Ohto M. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N Engl J Med. 1991;324:1699–1704. doi: 10.1056/NEJM199106133242404. [DOI] [PubMed] [Google Scholar]
- 79.Li J, Buckwold VE, Hon MW, Ou JH. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J Virol. 1999;73:1239–1244. doi: 10.1128/jvi.73.2.1239-1244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


