Abstract
Biofilm formation in flea gut is important for flea-borne transmission of Yersinia pestis. There are enhancing factors (HmsHFRS, HmsCDE, and HmsT) and inhibiting one (HmsP) for Yersinia pestis biofilm formation. The RcsAB regulatory complex acts as a repressor of Yesinia biofilm formation, and adaptive pseudogenization of rcsA promotes Y. pestis to evolve the ability of biofilm formation in fleas. In this study, we constructed a set of isogenic strains of Y. pestis biovar Microtus, namely WT (RscB+ and RcsA-), c-rcsA (RscB+ and RcsA+), ΔrcsB (RscB- and RcsA-), and ΔrcsB/c-rcsA (RscB- and RcsA+). The phenotypic assays confirmed that RcsB alone (but not RcsA alone) had an inhibiting effect on biofilm/c-di-GMP production whereas assistance of RcsA to RcsB greatly enhanced this inhibiting effect. Further gene regulation experiments showed that RcsB in assistance of RcsA tightly bound to corresponding promoter-proximal regions to achieve transcriptional repression of hmsCDE, hmsT and hmsHFRS and, meanwhile, RcsAB positively regulated hmsP most likely in an indirect manner. Data presented here disclose that pseudogenization of rcsA leads to dramatic remodeling of RcsAB-dependent hms gene expression between Y. pestis and its progenitor Y. pseudotuberculosis, enabling potent production of Y. pestis biofilms in fleas.
Yersinia pestis is an extremely virulent pathogen causing severe invasive infections mainly manifested as bubonic plague in lymph nodes, septicemic plague in blood vessels, and pneumonic plague in lungs. Y. pestis is potent to synthesize biofilms, which are a population of bacterial colonies embedded in self-produced matrix1,2,3. Formation of attached Y. pestis biofilms in flea gut is important for flea-borne transmission of this pathogen1,2,3.
Y. pestis biofilm matrix is primarily composed of poly-B-1,6-N-acetylglucosamine exopolysaccharide1,2,3. The hmsHFRS operon is responsible for biosynthesis and translocation of biofilm exopolysaccharide through cell envelope4,5. HmsR and HmsS are located in inner membrane, whereas HmsH and HmsF are outer-membrane proteins5. HmsR has four transmembrane domains plus a cytoplasmic glycosyltransferase domain, while HmsS has two transmembrane domains; HmsR and HmsS form an enzymatic complex responsible for exopolysaccharide biosynthesis4,6,7. HmsH acts as a porin with β-barrel structure, and HmsF functions as a polysaccharide deacetylase; these two proteins form a complex for modification/export of partially deacetylated exopolysaccharide through outer membrane5,8.
The 3′,5′-cyclic diguanosine monophosphate (c-di-GMP), a small-molecule second messenger promoting exopolysaccharide biosynthesis, is produced from guanosine triphosphate by GGDEF-domain-containing diguanylate cyclases and degraded by EAL-domain-containing phosphodiesterases9. Y. pestis produces a total of two diguanylate cyclases HmsT and HmsD, and both of them are required for c-di-GMP biosynthesis and biofilm formation10,11. Although expression of both HmsT and HmsD is up-regulated in flea gut and upon temperature shift from 37°C (as in warm-blooded hosts) to 26°C (in flea gut), HmsD plays a major role in biofilm formation in fleas while the predominant effect of HmsT is on in vitro biofilm formation11. The hmsD gene is a member of the three-gene operon hmsCDE. HmsD is a trans-inner-membrane protein composed of three distinct domains, namely a periplasmic sensor domain, an HAMP signal converter domain and a cytoplasmic output GGDEF domain12,13. The periplasmic protein HmsC senses environmental signals and then interacts with HmsD periplasmic domain, which affects HmsD stability and thereby regulates cellular c-di-GMP levels12,13. In addition, Y. pestis expresses the sole c-di-GMP-specific phosphodiesterase HmsP, which is responsible for degradation of c-di-GMP and therefore has an inhibiting effect on biofilm formation7,14.
The hmsHFRS orthologs can be found in several bacterial species15, including the genetically close pgaABCD operon in Escherichia coli16. c-di-GMP binds to PgaC and PgaD (homologues of HmsR and HmsS, respectively), which stabilizes the PgaCD enzymatic complex and thereby activates its glycosyltransferase activity to produce exopolysaccharide17. Without c-di-GMP binding, PgaD fails to interact with PgaC and both of them are subject to proteolysis17. Y. pestis might employ the conserved c-di-GMP-HmsRS association mechanism to control exopolysaccharide production.
The Enterobacteriaceae Rcs phosphorelay system is an atypical two-component regulatory system composed of three proteins, RcsB, RcsC and RcsD18. RcsC and RcsD are membrane-bound proteins, while RcsB is a cytoplasmic one. RcsC acts as a sensor kinase catalyzing autophosphorylation of RcsD and RcsB, and the resulting phosphate group is then transferred to RcsD and finally to RcsB. Phosphorylated RcsB (RcsB-p) acts as a transcriptional regulator alone or upon binding with an auxiliary protein RcsA. The RcsAB complex recognizes a consensus box sequence TAAGAAT-ATTCTTA, which is a 7-7 invert repeat, within the promoter-proximal regions of its target genes mainly including those responsible for exopolysaccharide biosynthesis, flagellar mobility, and Rcs autoregulation (Table S1, and Fig S1).
The biofilm formation of Y. pestis and its genetically very closed progenitor Y. pseudotuberculosis is negatively regulated by the Rcs phosphorelay system19,21. The rcsA gene is inactivated in Y. pestis due to a 30 bp duplication insertion in its coding region, and replacing the rcsA pseudogene with functional rcsA allele of Y. pseudotuberculosis strongly represses Y. pestis biofilm formation and essentially abolished flea blockage19,21. The conversion of rcsA to a pseudogene during evolution from Y. pseudotuberculosis to Y. pestis is most likely a case of positive Darwinian selection19,21.
The present work discloses that the RcsAB complex acts as a major repressor of Y. pestis biofilm formation through directly repressing transcription of hmsCDE, hmsT and hmsHFRS meanwhile positively regulating hmsP in an indirect manner. The above results denote dramatic remodeling of biofilm-related hms gene expression between and Y. pestis and its progenitor Y. pseudotuberculosis due to adaptive pseudogenization of a regulatory gene rcsA.
Results
Bacterial strains and their biofilm phenotypes
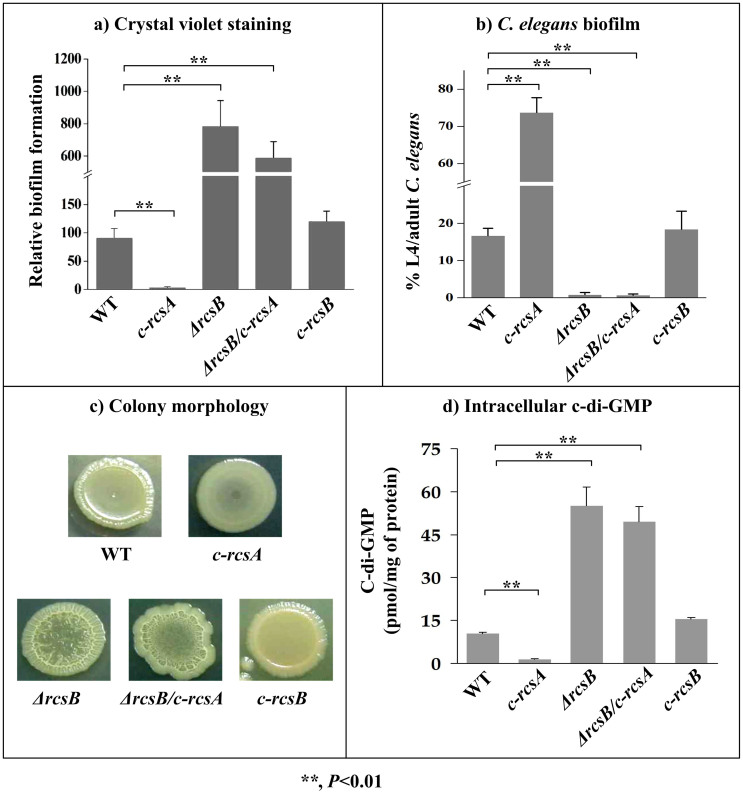
Transformation of pACYC184-rcsA into WT (wild-type, RscB+ and RcsA-) generated the rcsA-complemented strain c-rcsA (RscB+ and RcsA+), which led to a considerable decrease in c-di-GMP/biofilm production (Fig. 1).
Figure 1. Involvement of RcsAB in biofilm/c-di-GMP production.
Crystal violet staining of in vitro biofilm masses (a), C. elegans biofilms (b), bacterial colony morphology (c), and bacterial intracellular c-di-GMP concentration (d) were determined.
Deletion of rcsB from WT generated the rcsB-null strain ΔrcsB (RscB- and RcsA-), and further transformation of pACYC184-rcsA into ΔrcsB generated another rcsA-complemented strain ΔrcsB/c-rcsA (RscB- and RcsA+); compared to WT, both ΔrcsB and ΔrcsB/c-rcsA gave significantly enhanced c-di-GMP/biofilm production (Fig. 1).
Transformation of pACYC184-rcsB into ΔrcsB generated the rcsB-complemented strain c-rcsB (RscB+ and RcsA-), which had a biofilm/c-di-GMP production phenotype very similar to WT (Fig. 1).
Taken together, RcsB alone (but not RcsA alone) has an inhibiting effect on biofilm/c-di-GMP production, whereas assistance of RcsA to RcsB greatly enhances this inhibiting effect.
Regulation of hmsCDE, hmsT, hmsHFRS and hmsP by RcsAB
RcsAB box-like sequences could be found within the promoter-proximal regions of hmsCDE, hmsT and hmsHFRS, indicating that they might serve as direct RcsAB targets (Table S3), which promoted us to elucidate RcsAB-dependent expression of these candidate genes. hmsP was also included in the following gene regulation analyses.
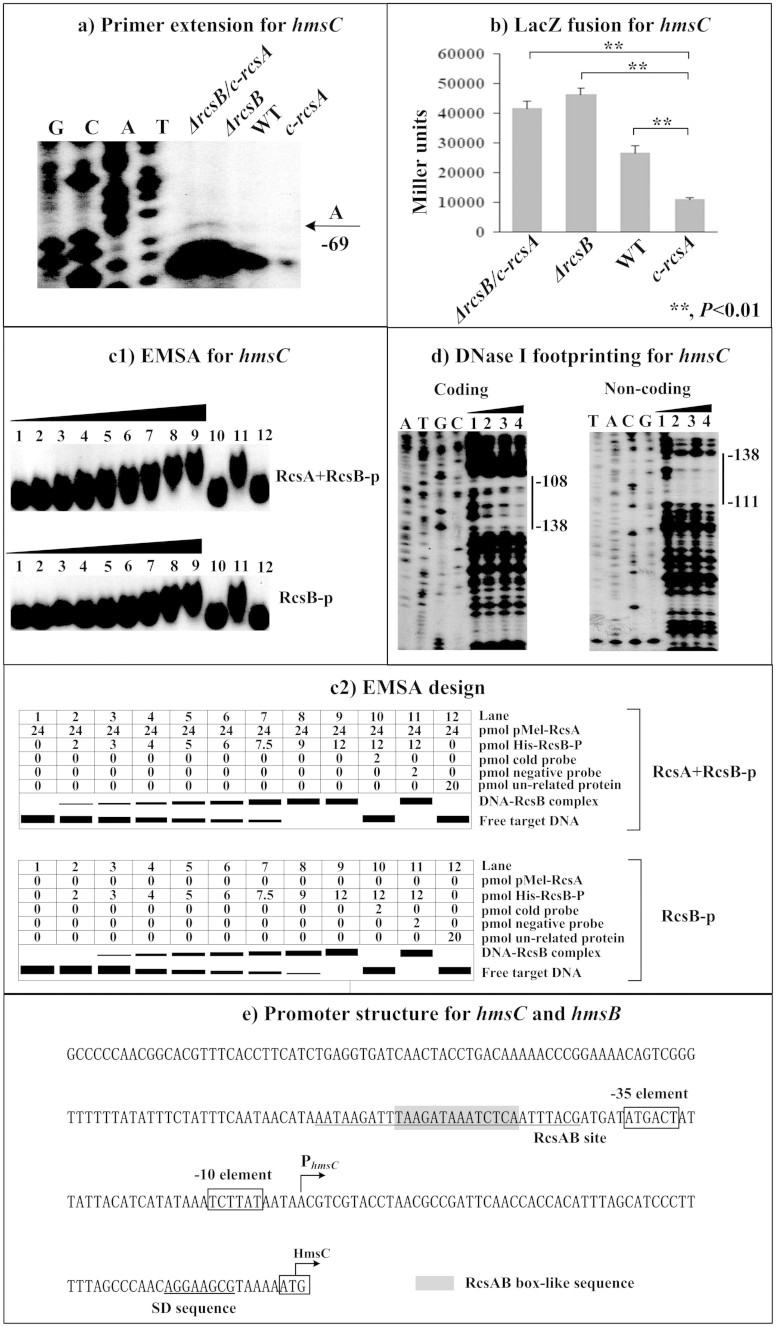
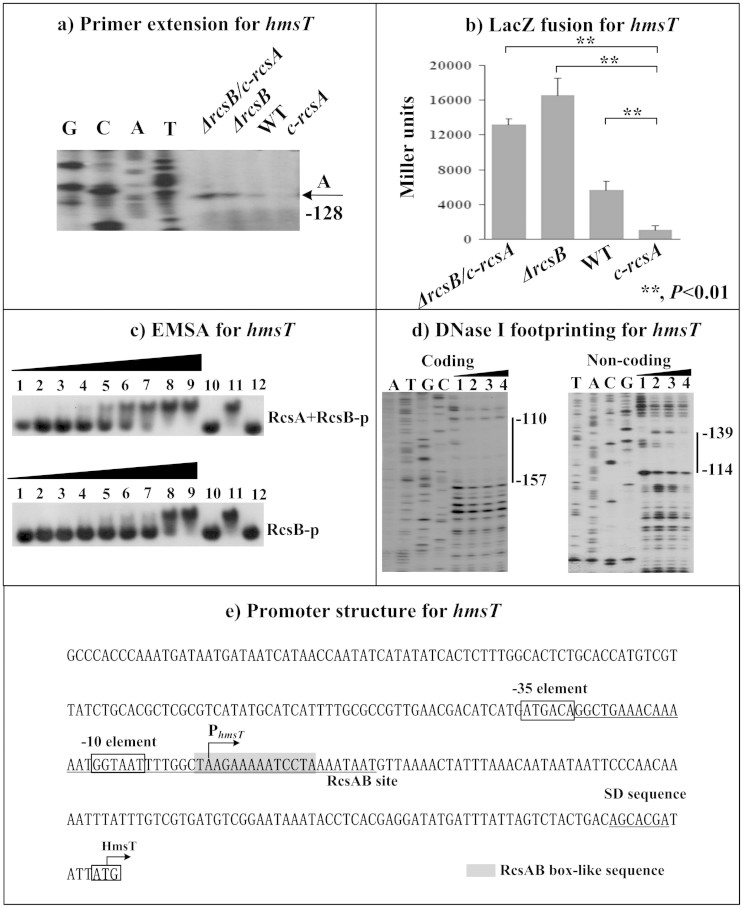
The primer extension assays indicated the relative mRNA levels of each of hmsC (Fig. 2a), hmsT (Fig. 3a) and hmsH (Fig. 4a) in the below four strains showed the following tendency: c-rcsA <WT <ΔrcsB ≈ ΔrcsB/c-rcsA. This observation was further confirmed by determination of the promoter activities of the above four genes by LacZ fusion (Fig. 2b, Fig. 3b, and Fig. 4b). As determined by electrophoretic mobility shift assay (EMSA), His-RcsB-p alone or mixed with excess MBP-RcsA could bind to the promoter-proximal region of each of hmsC (Fig. 2c), hmsT (Fig. 3c) and hmsH (Fig. 4c) in a dose-dependent manner; moreover, addition of excess RcsA could improve DNA-binding activity of RcsB-p. Further DNase I footprinting experiments showed that His-RcsB-p in presence of MBP-RcsA protected a single upstream region of each of hmsC (Fig. 2d), hmsT (Fig. 3d) and hmsH (Fig. 4d). The above observations indicated that RcsB-p in assistance of RcsA tightly bound to the corresponding promoter-proximal regions to achieve transcriptional repression of hmsCDE, hmsT and hmsHFRS.
Figure 2. RcsAB-dependent expression of hmsCDE.
(a) Primer extension. The relative mRNA levels of hmsC in indicated strains were determined by primer extension. The Sanger sequence ladders (lanes G, C, A, and T) and the primer extension products of hmsC were analyzed with an 8 M urea-6% acrylamide sequencing gel. The transcription start site of hmsC was indicated by arrow with nucleotide A, and the minus number under arrow indicated the nucleotide position upstream of hmsC start codon. (b) LacZ fusion. The hmsC:lacZ transcriptional fusion vector was transformed into in indicated strains, and then hmsC promoter activities (miller units of β-galactosidase activity) were determined in bacterial cellular extracts. (c) EMSA. The radioactively labeled DNA fragments were incubated with indicated purified proteins and then subjected to a native 4% polyacrylamide gel electrophoresis. (d) DNase I footprinting. Labeled coding or non-coding DNA probes were incubated with indicated purified proteins and then subjected to DNase I digestion. The digested DNA samples were analyzed in an 8 M urea-6% polyacrylamide gel. The footprint regions were indicated with vertical bars. Lanes C, T, A, and G represented Sanger sequencing reactions. The DNA-binding of His-RcsB-p in presence of MBP-RcsA (involved in EMSA and DNase I footprinting) and that of His-RcsB-p alone (in EMSA) were tested. (d) Promoter structure. Shown were with translation/transcription starts, core promoter −10 and −35 elements, SD sequences, RcsAB sites, and RcsAB box-like sequences.
Figure 3. RcsAB-dependent expression of hmsT.
Primer extension (a), LacZ fusion (b), EMSA (c), DNase I footprinting (d) experiments were performed as described in Fig 2.
Figure 4. RcsAB-dependent expression of hmsHFRS.
Primer extension (a), LacZ fusion (b), EMSA (c), and DNase I footprinting (d) experiments were performed as described in Fig 2.
By contrast, the relative mRNA levels (determined by primer extension, Fig. 5a) of hmsP showed the following tendency: c-rcsA> WT> ΔrcsB ≈ ΔrcsB/c-rcsA, which was further validated by quantitative RT-PCR (data not shown). However, LacZ fusion assay (Fig. 5b) indicated that RcsAB had no regulatory effect on promoter activity of hmsP. In addition, both EMSA (Fig. 5c) and DNase I footprinting (data not shown) indicated no association between RcsAB and hmsP upstream DNA. Therefore, RcsAB positively regulated hmsP most likely in an indirect manner.
Figure 5. RcsAB-dependent expression of hmsP.
Primer extension (a), LacZ fusion (b), and EMSA (c) experiments were performed as described in Fig 2.
Organization of RcsAB-dependent promoters
Transcription starts determined by primer extension were considered as transcribed promoters for indicated genes and, accordingly, core promoter −10 and −35 elements could be predicted. Each of hmsCD (Fig. 2e), hmsT (Fig. 3e), hmsP (Fig. 4e) and hmsHFRS (Fig. 5d) had a single transcribed promoter. It should be noted that all the above data were consistent with our previous report on regulation of hms genes by Y. pestis ferric uptake regulator Fur20.
The footprints determined by DNase I footprinting were considered as RcsAB sites for hmsCDE, hmsT, and hmsH; as expected, RcsAB box-like sequences (Table S3) could be found within all these RcsAB sites. The organization of RcsAB-dependent promoters of hmsCDE (Fig. 2e), hmsT (Fig. 3e), hmsHFRS (Fig. 4e), and hmsP (Fig. 5d) was constructed with translation/transcription starts, core promoter −10 and −35 elements, predicted Shine-Dalgarno (SD) sequences for ribosomal binding, RcsAB sites, and RcsAB box-like sequences.
Discussion
Transcriptional repression of genes for biofilm exopolysaccharide biosynthesis by RcsB with assistance of its auxiliary protein RcsA has been characterized in several bacterial species (Table S1). The present work confirms RcsAB-mediated tight inhibition of Y. pestis c-d-GMP/exopolysaccharide/biofilm production by using a set of isogenic strains of Y. pestis biovar Microtus, namely WT (RscB+ and RcsA-), c-rcsA (RscB+ and RcsA+), ΔrcsB (RscB- and RcsA-), and ΔrcsB/c-rcsA (RscB- and RcsA+). RcsAB acts as a major repressor of Y. pestis biofilm formation through directly repressing transcription of biofilm-enhancing genes hmsCDE, hmsT and hmsHFRS and meanwhile positively regulating biofilm-enhancing one hmsP in an indirect manner. RcsB in absence of RcsA does have residual regulatory effects on biofilm formation and hms gene expression and, moreover, RcsB-dependent regulation is greatly increased with assistance of RcsA, which was consistent with previous results19,21,22. The above regulatory circuit leads to different expression levels of each of hmsCDE, hmsT, hmsHFRS and hmsP in the above isogenic strains and thus distinct potencies of these strains to produce c-di-GMP/biofilm (summarized in Fig. 6).
Figure 6. RcsAB-dependent gene expression and phenotypes.
Shown were relative mRNA levels of each of hmsCDE, hmsT, hmsHFRS and hmsP in different isogenic Y. pestis strains, as well as relative potencies to produce c-di-GMP/biofilm of these strains.
Y. pseudotuberculosis (RscB+ and RcsA+, analogous to Y. pestis strain c-rcsA in this study] has a biofilm- phenotype in fleas19,21,22. In Y. pseudotuberculosis, biosynthesis of HmsCDE, HmsT, and HmsHFRS is tightly inhibited while HmsP is allowed to express. The pseudogenization of rcsA leads to inability of RcsAB complex in Y. pestis, which in turn alleviates RcsAB-mediated inhibition of expression of hmsCDE, hmsT, and hmsHFRS. As a prerequisite of potent Y. pestis biofilm formation, the adaptive pseudogenization of rcsA results in dramatic remodeling of hms gene expression patterns between Y. pseudotuberculosis and Y. pestis, finally enabling Y. pestis biofilm formation in fleas and thereby flea-borne transmission of this pathogen.
RcsB, RcsC, and RcsD are still functional in Y. pesits and thus, there is residual RcsB-dependent repression of biofilm formation in this bacterium19. Preclusion of total inactivation of Rcs phosphorelay during Y. pestis evolution might be due to the following reasons: biofilm overproduction if rcsB is inactivated would has detrimental effects on flea as vectors as well as on bacterial growth and proliferation; Rcs phosphorelay plays roles during mammalian infections23.
As shown previously23, RcsAB binds to the promoter-proximal region of hmsT to repress hmsT transcription. As disclosed in this study, RcsAB inhibits transcription of hmsCDE, hmsT, and hmsHFRS through binding to the promoter-proximal regions of all these direct RcsAB targets. RcsAB sites overlap core promoter -10 elements and transcription start sites of hmsT and hmsHFRS. Association between RcsAB and the above target promoter regions will block entry of RNA polymerase to inhibit transcription of hmsT and hmsHFRS, which has been characterized for RcsAB-mediated transcriptional repression of an array of direct target genes in other Enterobacteriaceae organisms24,25,26. Notably, the RcsAB site is upstream of promoter −35 element of hmsCDE and, thus, inhibitory action of RcsAB on hmsCDE transcription appears to be highly unusual, which needs to be further elucidated.
Methods
Bacterial strains
The wild-type Y. pestis Microtus strain 201 (WT) is avirulent to humans but highly virulent to mice27. The partial coding region of each indicated gene was replaced by a kanamycin resistance cassette by using the one-step inactivation method based on the lambda phage recombination system28, to generate the corresponding mutant of Y. pestis (Table 1). For in trans complementation, a PCR-generated DNA fragment containing the coding region of each indicated gene together with its promoter-proximal region and transcriptional terminator-proximal region was cloned into the cloning vector pACYC184 (GenBank accession no. X06403), and the resulting recombinant vector was transformed into each indicated Y. pestis strain lack of the corresponding functional gene, generating the corresponding complemented mutant (Table 1). All the primers designed in this study are listed in Table S2.
Table 1. Y. pestis strains involved in gene deletion and complementation.
| Strain | Functional (+) or inactivated (-) | Feature | Reference | ||||
|---|---|---|---|---|---|---|---|
| rcsA | rcsB | hmsD | hmsT | hmsS | |||
| WT | − | + | + | + | + | The wild-type Y. pestis biovar Microtus strain 201. | [27] |
| c-rcsA | + | + | + | + | + | The vector pACYC184-rcsA# was introduced into WT. | This study |
| ΔrcsB | − | − | + | + | + | The base pairs 211 to 418 of rcsB gene was deleted from WT. | This study |
| c-rcsB | − | + | + | + | + | The vector pACYC184-rcsB was introduced into ΔrcsB. | This study |
| ΔrcsB/c-rcsA | + | − | + | + | + | The vector pACYC184-rcsA# was introduced into ΔrcsB. | This study |
| ΔhmsTΔhmsD | − | + | − | − | + | A reference c-di-GMP- strain. The base pairs -4 to 1179 of hmsT gene was deleted from WT, and then the base pairs 41 to 1238 of hmsD gene was deleted from ΔhmsT. | This study |
| ΔhmsS | − | + | + | + | − | A reference biofilm- strain. The base pairs 146 to 468 of hmsS was deleted from WT. | [20] |
#: functional Y. pseudotuberculosis rcsA.
Bacterial growth and RNA isolation
Overnight cell cultures in the Luria-Bertani (LB) broth with an optical density (OD620) of about 1.0 were diluted 1:50 into 18 ml of fresh LB broth for further cultivation at 26°C with shaking at 230 rpm to reach middle stationary phases (an OD620 of 0.8 to 1.2), followed by cell harvest for further gene regulation or phenotypic assays. Immediately before bacterial harvest for RNA isolation, double-volume of RNAprotect reagent (Qiagen) was mixed with one-volume of cell culture, and total RNA was extracted using TRIzol Reagent (Invitrogen). RNA quality was monitored by agarose gel electrophoresis, and RNA quantity was determined by spectrophotometry.
Primer extension assay
As described in our previous studies29,30, a 5′-32P-labeled oligonucleotide primer complementary to a portion of the RNA transcript of each indicated gene was employed to synthesize cDNAs from total RNA templates using Promega Primer Extension System. Sequence ladders were prepared with the same 5′-32P-labeled primers using AccuPower & Top DNA Sequencing Kit (Bioneer). Radioactive species were detected by autoradiography. If different Y. pestis strains were involved in a single experiment, equal amounts of the total RNA samples were used as the starting materials. The relative mRNA level was determined with the observed band intensity of the primer extension product of each target gene. The 5′-terminus of RNA transcript (i.e., transcription start) of each target gene was mapped according to the size of primer extension product.
LacZ fusion and β-galactosidase assay
A promoter-proximal DNA region of each indicated gene was cloned into the low-copy-number transcriptional fusion vector pRW5031 that harbors a promoterless lacZ reporter gene. Y. pestis strains transformed with the recombinant plasmid or the empty pRW50 (negative control) were grown to measure β-galactosidase activity in cellular extract using β-Galactosidase Enzyme Assay System (Promega)29,30.
Protein expression and purification
The entire coding region of Y. pseudotuberculosis rcsA or Y. pestis rcsB was cloned into plasmid pMAL-c4X (Invitrogen)23 or pBADMyc-His A (New England Biolabs)23, respectively. The wild-type Y. pestis strain KIM6+ and the rcsB null mutant of KIM6+ were employed as host cells for expression of maltose-binding protein (MBP)-tagged RcsA (MBP-RcsA) and 6 × His-tagged RcsB (His-RcsB), respectively23. His-RcsB and MBP-RcsA were purified under native conditions using Ni-NTA Agarose Column (Qiagen) and Amylose Agarose Column (New England Biolabs), respectively23. Each purified protein was dialyzed and then concentrated to a concentration of about 0.1 mg/ml in phosphate buffered saline (pH 8.0) containing 20% glycerin.
EMSA
Each indicated 5′-32P-labeled target DNA fragment was incubated with increasing amounts of purified His-RcsB, or with increasing amounts of purified His-RcsB with addition of 24 pmol of purified MBP-RcsA, for 30 min at room temperature in a binding buffer29,30. To achieve RcsB phosphorylation, 25 mM fresh acetyl phosphate was incubated for 30 min with His-RcsB in the binding buffer, before labeled DNA probes were added. The resulting reactions were subjected to a native 4% (w/v) polyacrylamide gel electrophoresis. Each EMSA experiment included three controls, namely, cold probe as the specific DNA competitor (the same promoter-proximal DNA region unlabeled), negative probe as the nonspecific DNA competitor (the unlabeled coding region of the 16S rRNA gene), and nonspecific protein competitor (rabbit anti-F1-protein polyclonal antibodies)29,30. Detection of sequencing and radioactive species was as above.
DNase I footprinting
For DNase I footprinting29,30, the target DNA fragment with a single 32P-labeled end was incubated with increasing amounts of purified His-RcsB-p with addition of 24 pmol of purified MBP-RcsA, which was followed by partial digestion of RQ1 RNase-Free DNase I (Promega). The digested DNA samples were purified and analyzed in an 8 M urea-6% polyacrylamide gel. Detection of sequencing and radioactive species was as above. Footprints were identified by comparison with sequence ladders.
Biofilm and c-di-GMP assays
As described in our previous study32, three different methods were used to detect Y. pestis biofilms. First, in vitro biofilm masses, attached to well walls when bacteria were grown in polystyrene microtiter plates, were stained with crystal violet. Second, percentages of fourth-stage larvae and adults (L4/adult) of C. elegans after incubation of nematode eggs on Y. pestis lawns, negatively reflecting bacterial ability to produce biofilms, were determined. Third, rugose colony morphology of bacteria grown on LB agar plates, positively reflecting bacterial ability to synthesize exopolysaccharide, was observed. In addition, intracellular c-di-GMP levels were determined by a chromatography-coupled tandem mass spectrometry (HPLC-MS/MS) method as described in our previous study20.
Experimental replicates and statistical methods
For LacZ fusion, crystal violet staining of biofilms, and determination of L4/adult nematodes or c-di-GMP, experiments were performed with at least three independent bacterial cultures/lawns, and values were expressed as mean ± standard deviation. Paired Student's t-test was performed to determine statistically significant differences; P <0.01 was considered to indicate statistical significance. For primer extension and colony morphology observation, shown were representative data from at least two independent bacterial cultures.
Supplementary Material
Supplementary Information
Acknowledgments
This work is supported by Beijing Nova Program (Z121102002512049), National Natural Science Foundation of China (81201246, and 31430006), and National Basic Research Program of China (2013CB910800).
Footnotes
The authors declare no competing financial interests.
Author Contributions D.Z. and R.Y. designed experiments. N.F., H.Y., H.F., L.L, Y.Z., L.W., Y.H., D.Z. and R.Y. performed experiments. N.F., D.Z. and R.Y. analyzed data. N.F., H.Y., H.F., D.Z. and R.Y. contributed reagents, materials and analysis tools. D.Z. and R.Y. wrote this manuscript.
References
- Hinnebusch B. J. & Erickson D. L. Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr. Top. Microbiol. Immunol. 322, 229–248 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby C. Uniquely insidious: Yersinia pestis biofilms. Trends Microbiol. 16, 158–64 (2008). [DOI] [PubMed] [Google Scholar]
- Zhou D. & Yang R. Formation and regulation of Yersinia biofilms. Protein Cell 2, 173–179 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov A. G., Kirillina O., Forman S., Mack D. & Perry R. D. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ. Microbiol. 10, 1419–1432 (2008). [DOI] [PubMed] [Google Scholar]
- Abu Khweek A., Fetherston J. D. & Perry R. D. Analysis of HmsH and its role in plague biofilm formation. Microbiology 156, 1424–1438 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman S. et al. Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology 152, 3399–3410 (2006). [DOI] [PubMed] [Google Scholar]
- Kirillina O., Fetherston J. D., Bobrov A. G., Abney J. & Perry R. D. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54, 75–88 (2004). [DOI] [PubMed] [Google Scholar]
- Perry R. D. et al. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J. Bacteriol. 186, 1638–1647 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondermann H., Shikuma N. J. & Yildiz F. H. You've come a long way: c-di-GMP signaling. Curr.Opin. Microbiol. 15, 140–146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov A. G. et al. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 79, 533–551 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C. et al. Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS One 6, e19267; 10.1371/journal.pone.0019267 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov A. G. et al. The Yersinia pestis HmsCDE regulatory system is essential for blockage of the oriental rat flea (Xenopsylla cheopis), a classic plague vector. Environ. Microbiol. 10.1111/1462-2920.12419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G. X., Yan H. Q., Zhu H., Guo X. P. & Sun Y. C. HmsC, a periplasmic protein, controls biofilm formation via repression of HmsD, a diguanylate cyclase in Yersinia pestis. Environ. Microbiol. 16, 1202–1216 (2014). [DOI] [PubMed] [Google Scholar]
- Bobrov A. G., Kirillina O. & Perry R. D. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247, 123–130 (2005). [DOI] [PubMed] [Google Scholar]
- Itoh Y., Wang X., Hinnebusch B. J., Preston J. F. 3rd & Romeo T. Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187, 382–387 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Preston J. F. 3rd & Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186, 2724–2734 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S., Lori C., Boehm A. & Jenal U. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 32, 354–368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N. & Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59, 379–405 (2005). [DOI] [PubMed] [Google Scholar]
- Sun Y. C., Hinnebusch B. J. & Darby C. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc. Natl. Acad. Sci. U S A 105, 8097–8101 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F. et al. Fur is a repressor of biofilm formation in Yersinia pestis. PLoS One 7, e52392; 10.1371/journal.pone.0052392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Positive selection, not negative selection, in the pseudogenization of rcsA in Yersinia pestis. Proc. Natl. Acad. Sci. U S A 105, E69; 10.1073/pnas.0806419105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang N. et al. Optimized methods for biofilm analysis in Yersinia pestis. Biomed. Environ. Sci. 26, 408–411 (2013). [DOI] [PubMed] [Google Scholar]
- Erickson D. L., Jarrett C. O., Wren B. W. & Hinnebusch B. J. Serotype differences and lack of biofilm formation characterize Yersinia pseudotuberculosis infection of the Xenopsylla cheopis flea vector of Yersinia pestis. J.Bacteriol. 188, 1113–1119 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Guo X. P., Hinnebusch B. J. & Darby C. The Yersinia pestis Rcs phosphorelay inhibits biofilm formation by repressing transcription of the diguanylate cyclase gene hmsT. J. Bacteriol. 194, 2020–2026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francez-Charlot A. et al. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49, 823–832 (2003). [DOI] [PubMed] [Google Scholar]
- Wehland M. & Bernhard F. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J. Biol. Chem. 275, 7013–7020 (2000). [DOI] [PubMed] [Google Scholar]
- Pescaretti Mde L., Lopez F. E., Morero R. D. & Delgado M. A. Transcriptional autoregulation of the RcsCDB phosphorelay system in Salmonella enterica serovar Typhimurium. Microbiology 156, 3513–3521 (2010). [DOI] [PubMed] [Google Scholar]
- Zhou D. et al. Genetics of metabolic variations between Yersinia pestis biovars and the proposal of a new biovar, microtus. J. Bacteriol. 186, 5147–5152 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A. & Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U S A 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Reciprocal regulation of pH 6 antigen gene loci by PhoP and RovA in Yersinia pestis biovar Microtus. Future Microbiol. 8, 271–280 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Autoregulation of PhoP/PhoQ and positive regulation of the cyclic AMP receptor protein-cyclic AMP complex by PhoP in Yersinia pestis. J. Bacteriol. 195, 1022–1030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge J., Fear J., Busby S., Gunasekaran P. & Kamini N. R. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 74, 271–276 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information