Abstract
Background
Although many epidemiologic studies have investigated the CYP1A1 MspI gene polymorphisms and their associations with esophageal cancer (EC), definite conclusions cannot be drawn. To clarify the effects of CYP1A1 MspI polymorphisms on the risk of EC, a meta-analysis was performed in Chinese population.
Methods
Related studies were identified from PubMed, Springer Link, Ovid, Chinese Wanfang Data Knowledge Service Platform, Chinese National Knowledge Infrastructure (CNKI), and Chinese Biology Medicine (CBM) till October 2014. Pooled ORs and 95% CIs were used to assess the strength of the associations.
Results
A total of 13 studies including 1,519 EC cases and 1,962 controls were involved in this meta-analysis. Overall, significant association was found between CYP1A1 MspI polymorphism and EC risk when all studies in the Chinese population pooled into this meta-analysis (C vs. T: OR = 1.25, 95% CI = 1.04 to 1.51; CC + CT vs. TT: OR = 1.35, 95% CI = 1.06 to 1.72; CC vs. TT + CT: OR = 1.35, 95% CI = 1.03 to 1.76). When we performed stratified analyses by geographical locations, histopathology type, and source of control, significantly increased risks were found in North China (C vs. T: OR = 1.38, 95% CI = 1.12 to 1.70; CC vs. TT: OR = 1.72, 95% CI = 1.16 to 2.56; CC + CT vs. TT: OR = 1.52, 95% CI = 1.14 to 2.02; CC vs. TT + CT: OR = 1.55, 95% CI = 1.17 to 2.06), in the population-based studies (C vs. T: OR = 1.22, 95% CI = 1.05 to 1.42; CC vs. TT: OR = 1.38, 95% CI = 1.02 to 1.88; CC + CT vs. TT: OR = 1.36, 95% CI = 1.10 to 1.69; CC vs. TT + CT: OR = 1.43, 95% CI = 1.13 to 1.81) and ESCC (C vs. T: OR = 1.17, 95% CI = 1.04 to 1.32; CC + CT vs. TT: OR = 1.28, 95% CI = 1.08 to 1.52).
Conclusions
This meta-analysis provides the evidence that CYP1A1 MspI polymorphism may contribute to the EC development in the Chinese population.
Keywords: Meta-analysis, CYP1A1 MspI, Polymorphism, Esophageal cancer, Chinese
Review
Background
Esophageal cancer (EC) is the eighth most common cancer and sixth most deadly cancer worldwide, with an estimated 482,300 new esophageal cancer cases and 406,800 deaths in 2008 [1]. Its incidence rates vary internationally; China and southern and eastern Africa are the relatively high risk areas [1]. In China, more strikingly, esophageal cancer ranks the fifth most common diagnosed cancer and fourth leading cause of cancer-related mortality [2]. The mechanisms of esophageal carcinogenesis have not been fully illustrated. Tobacco use [3,4], alcohol consumption [3,5], low socioeconomic status, poor oral hygiene, and nutritional deficiencies [6-8] have been identified as risk factors for esophageal cancer. Yet, only a subset of individuals exposed to these risk factors eventually develop esophageal cancer, indicating a pivotal role of genetic factors in the esophageal carcinogenesis. In recent years, several common low-penetrance genes have been identified as potential esophageal cancer susceptibility genes. An important one is cytochrome P450 1A1 (CYP1A1), which plays an essential role in the metabolic activation of major classes of tobacco procarcinogen such as aromatic amines and polycyclic aromatic hydrocarbons (PAHs). So it may affect the metabolism of the environmental carcinogens and alter susceptibility to esophageal cancer.
CYP1A1 enzyme is a member of the CYP superfamily and prone to mutation [9]. Agundez [9] revealed an association between CYP1A1 enzyme activity and the risk of developing several types of cancers, including EC. CYP1A1 A2455G and T3801C are two most commonly studied polymorphism loci. CYP1A1 T3801C polymorphism (MspI, rs4646903), also known as the m1 allele, is a substitution of T with C in the non-coding 3′-flanking region which resulting in a highly inducible activity of the enzyme [10,11]. The first research of the association between CYP1A1 MspI polymorphism and EC was reported by Hori and co-workers in 1997 among the Japan population [12]. As a consequence, many studies analyzed the influence of CYP1A1 MspI polymorphism on EC risk; however, no clear consensus was reached. Meta-analyses of studies on the gene in other ethnic groups have been reported elsewhere and produced conflicting results [13-15]. In order to lessen the impact of different genetic background, we performed this meta-analysis to assess the relationship of CYP1A1 MspI polymorphism with risk of EC in Chinese population.
Methods
Literature search
A comprehensive literature search was performed using the PubMed, Springer Link, Ovid, Chinese Wanfang Data Knowledge Service Platform, Chinese National Knowledge Infrastructure (CNKI), and Chinese Biology Medicine (CBM) for relevant articles published with the following Mesh terms: (‘Esophageal Neoplasms’ [MeSH] or ‘esophageal cancer’ or ‘esophageal tumor’ or ‘esophageal carcinoma’ or ‘esophageal squamous cell’ or ‘esophageal adenocarcinoma’) and (‘P4501A1’ or ‘CYP1A1’) and (China or Chinese or Taiwan). An upper date limit of 28 December 2014 was applied, and no lower date limit was used. The search was performed without any restrictions on language and focused on studies conducted in humans. Concurrently, the reference lists of reviews and retrieved articles were searched manually.
Inclusion/exclusion criteria
For inclusion, the studies must have met the following criteria: (1) case–control study or cohort study studying on association between the CYP1A1 MspI polymorphisms and EC susceptibility; (2) all patients with the diagnosis of EC confirmed by pathological or histological examination; (3) sufficient published data about sample size, odds ratio (OR), and their 95% confidence interval (CI); (4) all participants were Chinese; (5) containing complete information about all genotype frequencies. Studies were excluded when they were: (1) not case–control study or cohort study; (2) duplicate of previous publication; (3) based on incomplete data; (4) meta-analyses, letters, reviews, or editorial articles.
Data extraction
Information was extracted carefully from all eligible studies independently by two investigators according to the inclusion criteria listed above. Disagreements were resolved by discussion. The title and abstract of all potentially relevant articles were screened to determine their relevance. Full articles were also scrutinized if the title and abstract were ambiguous. The following data was collected from each study: first author’s surname, year of publication, geographical location, histopathology type, source of control, total numbers of cases and controls, and the numbers of cases and controls who harbored the MspI genotypes. If data from any category were not reported in the primary study, the items were designated ‘not stated’. We did not contact the author of the primary study to request the information.
Statistical analysis
Crude odds ratios (ORs) together with their corresponding 95% CIs were used to assess the strength of association between the CYP1A1 MspI polymorphism and the risk of EC. The pooled ORs were performed for allele model (C vs. T), dominant model (CT + CC vs. TT), recessive model (CC vs. CT + TT), heterozygous model (CC vs. CT), and homozygous model (CC vs. TT), respectively. Between-study heterogeneity was assessed by the Q-statistics with P values <0.1. Dependent on the results of heterogeneity test among individual studies, the fixed effect model (Mantel-Haenszel) or random effect model (DerSimonian and Laird) was selected to summarize the combined OR and their 95% CI. Hardy-Weinberg equilibrium (HWE) was calculated by using the goodness-of-fit test, and deviation was considered when P < 0.05. The significance of the pooled OR was determined by the z test. Sensitivity analysis was conducted to verify stability of the meta-analysis using both models (the fixed effect model and random effect model). Begg’s funnel plots and Egger’s linear regression test were used to assess publication bias. In addition to the comparison among all subjects, we also performed stratification analyses by geographical locations, histopathology type, and source of control. All the statistical analysis was conducted by using STATA statistical package (version 10, STATA, College Station, TX, USA).
Results
Eligible studies
Figure 1 graphically illustrates the trial flow chart. A total of 51 articles regarding CYP1A1 MspI polymorphism with respect to EC were identified. After screening the titles and abstracts, 24 articles were excluded because they were review articles, duplicates, not Chinese population, or irrelevant to the current study. In addition, of these published articles, 14 articles were excluded for full-text articles assessing due to case reports, reduplicate studies, or other polymorphisms of CYP1A1. Finally, 13 studies [16-28] including 1,519 EC cases and 1,962 controls were involved in this meta-analysis according to the inclusion criteria. The publication year of studies ranged from 2002 to 2014. Eleven of these studies were written in Chinese, two studies in English. The characteristics of the included studies are summarized in Table 1.
Figure 1.
Flow diagram of the literature search.
Table 1.
Characteristics of studies included in the meta-analysis
| Reference | Source of controls | Geographical locations | Histopathology type | Case number | Control number | Case | Control | HWE | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | CT | CC | TT | CT | CC | χ 2 | P | ||||||
| Wu 2002 [16] | HB | Taiwan | ESCC | 146 | 344 | 60 | 65 | 21 | 136 | 146 | 62 | 4.16 | 0.041 |
| Li 2002 [17] | HB | Shaxi | NS | 73 | 75 | 20 | 30 | 23 | 37 | 25 | 13 | 4.96 | 0.026 |
| Yin 2005 [18] | HB | Jiangsu | ESCC | 106 | 106 | 42 | 54 | 10 | 41 | 49 | 16 | 0.05 | 0.829 |
| Zhu 2005 [19] | HB | Shaxi | ESCC + EAC | 163 | 166 | 43 | 75 | 45 | 75 | 67 | 24 | 1.96 | 0.162 |
| Han 2005 [20] | HB | Shaxi | ESCC | 89 | 98 | 25 | 39 | 25 | 47 | 38 | 13 | 1.37 | 0.241 |
| Lu 2006 [21] | PB | Xinjiang | ESCC | 64 | 116 | 23 | 28 | 13 | 44 | 56 | 16 | 0.07 | 0.786 |
| Yin 2010 [22] | HB | Xinjiang | NS | 96 | 174 | 16 | 45 | 35 | 17 | 88 | 69 | 2.13 | 0.144 |
| Ji 2010 [23] | PB | Gansu | ESCC | 189 | 216 | 49 | 95 | 45 | 70 | 98 | 48 | 1.49 | 0.222 |
| Gao 2012 [24] | PB | Ningxia | ESCC | 40 | 80 | 15 | 17 | 8 | 28 | 41 | 11 | 0.43 | 0.511 |
| Huang 2012 [25] | PB | Guangxi | ESCC | 98 | 100 | 38 | 41 | 19 | 40 | 43 | 17 | 0.85 | 0.358 |
| Zhang 2013 [26] | PB | Shandong | ESCC | 138 | 170 | 34 | 78 | 26 | 59 | 83 | 28 | 0.02 | 0.896 |
| Yun 2014 [27] | PB | Henan | ESCC | 157 | 157 | 73 | 72 | 12 | 95 | 50 | 12 | 2.11 | 0.146 |
| Zhang 2011 [28] | PB | Henan | ESCC + EAC | 160 | 160 | 97 | 63 | 128 | 32 | - | |||
PB population-based, HB hospital-based, ESCC esophageal squamous cell carcinoma, EAC esophageal adenocarcinoma, NS not stated.
Meta-analysis results
Table 2 lists the primary results. Overall, a significantly elevated risk of EC was associated with two variants of CYP1A1 MspI (for CC and CT combined vs. TT: OR = 1.35, 95% CI = 1.06 to 1.72, P = 0.005 for heterogeneity; for CC vs. TT and CT: OR = 1.35, 95% CI = 1.03 to 1.76, P = 0.008 for heterogeneity). For the allele C vs. allele T, the pooled OR was 1.25 (95% CI = 1.04 to 1.51; P = 0.000 for heterogeneity) (Figure 2). However, there was significant heterogeneity between studies. Hence, we then performed subgroup analysis by geographical locations, histopathology type, and source of control. In the stratified analysis by geographical locations, significantly increased risks were found in the population from North China (C vs. T: OR = 1.38, 95% CI = 1.12 to 1.70; CC vs. TT: OR = 1.72, 95% CI = 1.16 to 2.56; CC + CT vs. TT: OR = 1.52, 95% CI = 1.14 to 2.02; CC vs. TT + CT: OR = 1.55, 95% CI = 1.17 to 2.06) but not found in the South. In the stratified analysis by source of controls, significantly increased risks were found in the population-based studies (C vs. T: OR = 1.22, 95% CI = 1.05 to 1.42; CC vs. TT: OR = 1.38, 95% CI = 1.02 to 1.88; CC + CT vs. TT: OR = 1.36, 95% CI = 1.10 to 1.69; CC vs. TT + CT: OR = 1.43, 95% CI = 1.13 to 1.81) but not found in the hospital-based studies. In the subgroup analysis by histopathology type, significantly increased association was found in esophageal squamous cell carcinoma (ESCC) (C vs. T: OR = 1.17, 95% CI = 1.04 to 1.32; CC + CT vs. TT: OR = 1.28, 95% CI = 1.08 to 1.52).
Table 2.
Main results in the total and subgroup analysis
| Analysis model | Study groups | n | Random-effect model | Fixed-effect model | Heterogeneity | |
|---|---|---|---|---|---|---|
| OR (95% CI) | OR (95% CI) | χ 2 | P | |||
| C vs. T | Total analysis | 12 | 1.25 (1.04 to 1.51) | 1.23 (1.11 to 1.37) | 33.46 | 0.000 |
| PB | 6 | 1.22 (1.05 to 1.42) | 1.22 (1.05 to 1.42) | 1.49 | 0.914 | |
| HB | 6 | 1.30 (0.90 to 1.90) | 1.25 (1.08 to 1.44) | 31.93 | 0.000 | |
| ESCC | 9 | 1.18 (1.00 to 1.39) | 1.17 (1.04 to 1.32) | 14.48 | 0.070 | |
| South China | 3 | 0.93 (0.76 to 1.14) | 0.93 (0.76 to 1.14) | 0.71 | 0.702 | |
| North China | 9 | 1.38 (1.12 to 1.70) | 1.37 (1.21 to 1.55) | 22.21 | 0.005 | |
| CC vs. TT | Total analysis | 12 | 1.42 (0.99 to 2.04) | 1.42 (1.15 to 1.75) | 29.24 | 0.002 |
| PB | 6 | 1.38 (1.02 to 1.88) | 1.38 (1.02 to 1.88) | 0.45 | 0.994 | |
| HB | 6 | 1.46 (0.70 to 3.04) | 1.45 (1.09 to 1.93) | 28.75 | 0.000 | |
| ESCC | 9 | 1.29 (0.94 to 1.77) | 1.27 (1.00 to 1.62) | 12.34 | 0.137 | |
| South China | 3 | 0.82 (0.54 to 1.25) | 0.82 (0.54 to 1.24) | 1.26 | 0.531 | |
| North China | 9 | 1.72 (1.16 to 2.56) | 1.73 (1.35 to 2.22) | 18.97 | 0.015 | |
| CC vs. CT | Total analysis | 12 | 1.09 (0.89 to 1.33) | 1.09 (0.89 to 1.32) | 10.90 | 0.452 |
| PB | 6 | 1.07 (0.80 to 1.43) | 1.06 (0.80 to 1.42) | 2.93 | 0.711 | |
| HB | 6 | 1.12 (0.78 to 1.59) | 1.11 (0.85 to 1.45) | 7.94 | 0.160 | |
| ESCC | 9 | 1.01 (0.80 to 1.28) | 1.01 (0.80 to 1.28) | 7.91 | 0.442 | |
| South China | 3 | 0.80 (0.53 to 1.21) | 0.80 (0.53 to 1.21) | 1.53 | 0.464 | |
| North China | 9 | 1.19 (0.95 to 1.50) | 1.19 (0.95 to 1.50) | 6.63 | 0.577 | |
| CC + CT vs. TT | Total analysis | 12 | 1.35 (1.06 to 1.72) | 1.37 (1.17 to 1.59) | 26.54 | 0.005 |
| PB | 6 | 1.36 (1.10 to 1.69) | 1.36 (1.10 to 1.69) | 4.10 | 0.536 | |
| HB | 6 | 1.39 (0.86 to 2.23) | 1.37 (1.11 to 1.70) | 22.44 | 0.000 | |
| ESCC | 9 | 1.28 (1.04 to 1.59) | 1.28 (1.08 to 1.52) | 11.73 | 0.164 | |
| South China | 3 | 0.97 (0.73 to 1.28) | 0.97 (0.73 to 1.28) | 0.11 | 0.947 | |
| North China | 9 | 1.52 (1.14 to 2.02) | 1.58 (1.32 to 1.89) | 18.30 | 0.019 | |
| CC vs. TT + CT | Total analysis | 13 | 1.35 (1.03 to 1.76) | 1.34 (1.13 to 1.59) | 26.94 | 0.008 |
| PB | 7 | 1.41 (1.06 to 1.89) | 1.43 (1.13 to 1.81) | 8.26 | 0.219 | |
| HB | 6 | 1.30 (0.79 to 2.14) | 1.25 (0.98 to 1.60) | 18.16 | 0.003 | |
| ESCC | 9 | 1.14 (0.88 to 1.47) | 1.12 (0.90 to 1.40) | 10.22 | 0.250 | |
| South China | 3 | 0.82 (0.56 to 1.20) | 0.81 (0.56 to 1.19) | 1.63 | 0.443 | |
| North China | 10 | 1.55 (1.17 to 2.06) | 1.53 (1.26 to 1.86) | 17.20 | 0.046 | |
PB population-based, HB hospital-based, ESCC esophageal squamous cell carcinoma, South China including Taiwan, Jiangsu, and Guangxi; North China including Shaxi, Xinjiang, Gansu, Ningxia, Shandong, and Henan.
Figure 2.
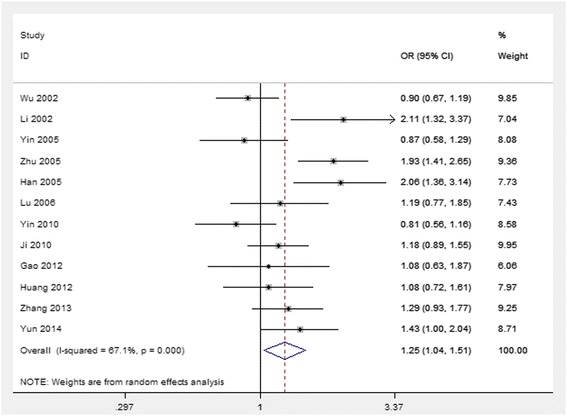
Forest plot (random-effect model) of lung cancer risk associated with CYP1A1 MspI polymorphism using the allele genetic model.
Sensitive analysis and bias diagnosis
In order to compare the difference and evaluate the sensitivity of the meta-analyses, we used both models (the fixed effect model and random effect model) to evaluate the stability of the meta-analysis. All the significant results were not materially altered (Table 2). Hence, results of the sensitivity analysis suggest that the data in this meta-analysis are relatively stable and credible. The Begg’s funnel plot and Egger’s test were performed to assess the publication bias of literatures. The shape of the funnel plots did not reveal obvious asymmetry (Figure 3). Then, the Egger’s test was used to provide statistical evidence of funnel plot symmetry. The Egger’s test indicated that there were no obvious publication bias under the allele model in overall analyses (t = −0.62, P = 0.550).
Figure 3.
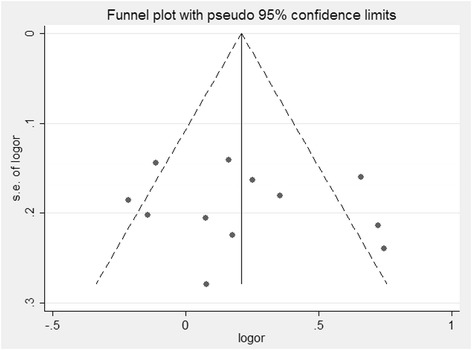
Begg’s funnel plot of CYP1A1 MspI polymorphism and lung cancer risk under the allele genetic model.
Discussion
CYP genes are large families of endoplasmic and cytosolic enzymes that catalyze the activation and detoxification, respectively, of reactive electrophilic compounds, including many environmental carcinogens (for example, benzo [a] pyrene). CYP1A1 is a phase I enzyme that regulates the metabolic activation of major classes of tobacco procarcinogens, such as aromatic amines and PAHs [29]. Thus, CYP1A1 may affect the metabolism of environmental carcinogens and alter the susceptibility to cancers, including EC. Although many studies analyzing the research results about the association between CYP1A1 MspI polymorphism and EC, definite conclusions cannot be drawn [13-15]. Therefore, we did this meta-analysis to estimate the relationship between CYP1A1 MspI polymorphism and susceptibility to EC among the Chinese population, in order to lessen the impact of different genetic background. This meta-analysis involved 13 articles with 1,519 EC cases and 1,962 controls. The results indicated a significant association between CYP1A1 MspI gene polymorphism and EC risk in the Chinese population. The sensitivity analysis confirmed the reliability and stability of the meta-analysis, and no publication bias was found among studies by Egger’s test. Therefore, the findings from our meta-analysis provide a strong evidence for the association between CYP1A1 MspI polymorphism and risk of EC in the Chinese population. Our results were inconsistent with a previously published meta-analysis in Chinese, which indicated that CYP1A1 Ile/Val genetic polymorphisms, but not CYP1A1 MspI polymorphisms, are associated with an increased digestive tract cancers risk in Chinese populations [30]. It was also inconsistent with the findings for Caucasians [13-15,31]. The exact mechanism for the ethnic discrepancy is uncertain but differences in underlying genetic backgrounds and social factors among different populations studied may be important. Ethnically diverse subjects may have unique cultures and lifestyles that can contribute to different genetic characteristics and susceptibility to specific cancers. Also, this inconsistency may be due to the smaller sample sizes for Chinese used in previous meta-analyses.
When we performed stratified analyses by geographical locations, histopathology type, and source of control, significant association with susceptibility for the development of EC was found in North China, in ESCC, and population-based studies; however, it was not found in South China and hospital-based studies. These can be influenced by some factors. First, the hospital-based studies usually have some biases because such controls may just represent a sample of ill-defined reference population and may not be representative of the general population. Second, genetic risk factors for EC in North and South China are different [32]. Last but not the least, for the relevant small sample size, only three studies from South China was included in the meta-analysis. With regard to heterogeneity, some of the factors extracted in this study were the main source of heterogeneity. But it might also make attributions for other unknown factors, such as dietary habits, dinking status, other environmental exposures (passive smoking and cooking oil fume), family history of cancer, other genetic-related respiratory diseases as well as other related genetic polymorphisms.
The pathways of carcinogen metabolism are complex, mediated by the activity of multiple enzymes. The effect of any single gene might have a limited impact on EC risk than have so far been anticipated. EC has some known major environmental determinants other than tobacco smoke, and large studies with detailed exposure information are needed to evaluate reliably any moderate genetic effects. Otherwise, some limitations should be acknowledged. Firstly, we did not perform subgroup analysis on smoking status because of the lack of sufficient data. Another potential limitation was that our results were based on unadjusted estimates. More precise analyses can be conducted if individual data were available, which would allow for the adjustment by other covariates including age, sex, race, and other factors. Finally, heterogeneity can interfere with the interpretation of the results of a meta-analysis. Although we minimized this likelihood by performing a careful search of published studies and subgroup analyses, significant inter-study heterogeneity nevertheless existed in the comparison of geographical locations. In spite of these limitations, our meta-analysis still had some advantages. We obeyed the inclusion and exclusion criteria strictly to reduce selection bias. A funnel plot and Egger’s linear regression test was used to assess publication bias. In addition, the impact of different genetic background was lessened by means of including the studies performed in the Chinese population only, and the test of the Hardy-Weinberg equilibrium for distribution of the genotypes in control groups suggested that there was almost no significantly different genetic background among the participants.
Conclusions
In conclusion, our meta-analysis supports that CYP1A1 MspI polymorphism might contribute to individual susceptibility to EC in the Chinese population. Concerning EC with multifactorial etiology, to further evaluate gene-gene and gene-environment interactions on CYP1A1 MspI polymorphism and EC, larger studies in selected populations with different environmental background or other risk factors are required. Such studies may eventually lead to have a better, comprehensive understanding of the association between the CYP1A1 MspI polymorphism and EC risk.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HZ and YZ participated in the design of this study; they both performed the data collection and statistical analysis. HZ drafted the manuscript. Both authors read and approved the final manuscript.
Contributor Information
Hui Zheng, Email: Huizheng2000@126.com.
Yun Zhao, Email: yunzhao1997@163.com.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, He Y, Zheng R, Zhang S, Zeng H, Zou X, et al. Esophageal cancer incidence and mortality in China, 2009. J Thorac Dis. 2013;5:19–26. doi: 10.1016/j.athoracsur.2012.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gammon MD, Schoenberg JB, Ahsan H, Risch HA, Vaughan TL, Chow WH, et al. Tobacco, alcohol, and socioeconomic status and adenocarcinomas of the esophagus and gastric cardia. J Natl Cancer Inst. 1997;89:1277–84. doi: 10.1093/jnci/89.17.1277. [DOI] [PubMed] [Google Scholar]
- 4.De Stefani E, Barrios E, Fierro L. Black (air-cured) and blond (flue-cured) tobacco and cancer risk. III: Oesophageal cancer Eur J Cancer. 1993;29A:763–6. doi: 10.1016/s0959-8049(05)80363-6. [DOI] [PubMed] [Google Scholar]
- 5.Lee CH, Wu DC, Lee JM, Wu IC, Goan YG, Kao EL, et al. Carcinogenetic impact of alcohol intake on squamous cell carcinoma risk of the oesophagus in relation to tobacco smoking. Eur J Cancer. 2007;43:1188–99. doi: 10.1016/j.ejca.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 6.Brown LM, Hoover R, Silverman D, Baris D, Hayes R, Swanson GM, et al. Excess incidence of squamous cell esophageal cancer among us black men: Role of social class and other risk factors. Am J Epidemiol. 2001;153:114–22. doi: 10.1093/aje/153.2.114. [DOI] [PubMed] [Google Scholar]
- 7.Taylor PR, Qiao YL, Abnet CC, Dawsey SM, Yang CS, Gunter EW, et al. Prospective study of serum vitamin e levels and esophageal and gastric cancers. J Natl Cancer Inst. 2003;95:1414–6. doi: 10.1093/jnci/djg044. [DOI] [PubMed] [Google Scholar]
- 8.Abnet CC, Lai B, Qiao YL, Vogt S, Luo XM, Taylor PR, et al. Zinc concentration in esophageal biopsy specimens measured by x-ray fluorescence and esophageal cancer risk. J Natl Cancer Inst. 2005;97:301–6. doi: 10.1093/jnci/dji042. [DOI] [PubMed] [Google Scholar]
- 9.Agundez JA. Cytochrome P450 gene polymorphism and cancer. Curr Drug Metab. 2004;5:211–24. doi: 10.2174/1389200043335621. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi S, Watanabe J, Nakachi K, Kawajiri K. Genetic linkage of lung cancer-associated MspI polymorphisms with amino acid replacement in the heme binding region of the human cytochrome P450IA1 gene. J Biochem. 1991;110:407–11. doi: 10.1093/oxfordjournals.jbchem.a123594. [DOI] [PubMed] [Google Scholar]
- 11.Kawajiri K, Nakachi K, Imai K, Yoshii A, Shinoda N, Watanabe J. Identification of genetically high risk individuals to lung cancer by DNA polymorphisms of the cytochrome P450IA1 gene. FEBS Lett. 1990;263:131–3. doi: 10.1016/0014-5793(90)80721-T. [DOI] [PubMed] [Google Scholar]
- 12.Hori H, Kawano T, Endo M, Yuasa Y. Genetic polymorphism of tobacco and alcohol-related metabolizing enzymes and human esophageal squamous cell carcinoma susceptibility. J Clin Gastroenterol. 1997;25:568–75. doi: 10.1097/00004836-199712000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Gong FF, Lu SS, Hu CY, Qian ZZ, Feng F, Wu YL, et al. Cytochrome P450 1A1 (CYP1A1) polymorphism and susceptibility to esophageal cancer: an updated meta-analysis of 27 studies. Tumour Biol. 2014;35:10351–61. doi: 10.1007/s13277-014-2341-y. [DOI] [PubMed] [Google Scholar]
- 14.Wu B, Liu K, Huang H, Yuan J, Yuan W, Wang S, et al. MspI and Ile462Val polymorphisms in CYP1A1 and overall cancer risk: a meta-analysis. PLoS One. 2013;8:e85166. doi: 10.1371/journal.pone.0085166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen FF, Zhou FY, Xue QS, Pan Y, Zheng L, Zhang H, et al. Association between CYP1A1 polymorphisms and esophageal cancer: a meta-analysis. Mol Biol Rep. 2013;40:6035–42. doi: 10.1007/s11033-013-2713-1. [DOI] [PubMed] [Google Scholar]
- 16.Wu MT, Lee JM, Wu DC, Ho CK, Wang YT, Lee YC, et al. Genetic polymorphisms of cytochrome P4501A1 and oesophageal squamous-cell carcinoma in Taiwan. Br J Cancer. 2002;87:529–32. doi: 10.1038/sj.bjc.6600499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li WY: A study of CYP1A1 gene polymorphism and the genetic susceptibility to esophageal carcinoma. Master’s Thesis of Shanxi Medical University 2002 (in Chinese).
- 18.Yin L, Pu Y, Song Y, Hu X, Liu Y, Kai H. Polymorphism of susceptible genes for esophageal cancer risk in Huaian population in Jiangsu Province. Tumor. 2005;25:357–61. [Google Scholar]
- 19.Zhu SJ. Relationship of the polymorphism of CYP1A1 and SULT1A1 genes and susceptibility to esophageal carcinoma. Master’s Thesis of Shanxi Medical University 2005 (in Chinese).
- 20.Han Y, Feng X, Li P, Niu Z. Case–control study on relationship of CYP1A1 and GSTM1 polymorphism and susceptibility to esophageal squamous carcinoma. Chin J Public Health. 2005;21:3–6. [Google Scholar]
- 21.Lu H, Yang L, Li F. Genetic polymorphism of CYP1A1 MspI associated with susceptibility to esophageal cancer in the Kazakans. Nong Ken Med. 2006;28:81–3. [Google Scholar]
- 22.Yin D, Zhang G, Deng Y, Ma Y, Ju L, Chen Y. CYP1A1, GSTM1 genetic polymorphism and their combined role in susceptibility of esophageal cancer. Cancer Detect Prev. 2010;37:712–6. [Google Scholar]
- 23.Ji R, Wu J, Zhou Y, Zhang B, Zhang ZY, Yang Z. Relationship between CYP1A1, GSTM1 and GSTT1 genetic polymorphism and susceptibility of esophageal cancer in Wuwei, Gansu province. J Lanzhou Univ (Med Sci) 2010;36:29–34. [Google Scholar]
- 24.Gao P, Tian Y, Ye XF, Ge J, Zhang D, Xu WD. Study of CYP1A1, GSTT1, GSTM1 polymorphism and susceptibility on esophageal carcinoma in Ningxia Hui nationality. Ningxia Med J. 2012;34:196–9. [Google Scholar]
- 25.Huang X, Tan Z, Zhang Y. The relationship between CYP1A1, MspI genetic polymorphism and susceptibility of Guangxi Zhuang People esophageal carcinoma. J Guangxi Med Univ. 2012;29:224–6. [Google Scholar]
- 26.Zhang L, Ma W, Li Y, Jiang YZ, Ma GY. Genetic polymorphisms of cytochrome P450 and glutathione S-transferase in relation to human esophageal carcinoma. Chin J Public Health. 2013;29:1499–501. [Google Scholar]
- 27.Yun YX, Wang YP, Wang P, Cui LH, Wang KJ, Zhang JY, et al. CYP1A1 genetic polymorphism and risk for esophageal cancer: a case–control study in central china. Asian Pac J Cancer Prev. 2014;14:6507–12. doi: 10.7314/APJCP.2013.14.11.6507. [DOI] [PubMed] [Google Scholar]
- 28.Zhang CX, Guo LK, Guo XF. Correlation between drinking behavior and the polymorphisms of CYP1A1-MspI, aldehyde dehydrogenase-2 genes and esophageal carcinoma. Chin J Gerontol. 2011;31:1976–9. [Google Scholar]
- 29.Guengerich FP, Shimada T. Activation of procarcinogens by human cytochrome P450 enzymes. Mutat Res. 1998;400:201–13. doi: 10.1016/S0027-5107(98)00037-2. [DOI] [PubMed] [Google Scholar]
- 30.Liu C, Jiang Z, Deng QX, Zhao YN. Meta-analysis of association studies of CYP1A1 genetic polymorphisms with digestive tract cancer susceptibility in Chinese. Asian Pac J Cancer Prev. 2014;15:4689–95. doi: 10.7314/APJCP.2014.15.11.4689. [DOI] [PubMed] [Google Scholar]
- 31.Zhuo WL, Zhang YS, Wang Y, Zhuo XL, Zhu B, Cai L, et al. Association studies of CYP1A1 and GSTM1 polymorphisms with esophageal cancer risk: evidence-based meta-analyses. Arch Med Res. 2009;40:169–79. doi: 10.1016/j.arcmed.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Su M, Tian D, Liang S, Zhang J. Associations between CYP1A1 and CYP2E1 polymorphisms and susceptibility to esophageal cancer in Chaoshan and Taihang areas of China. Cancer Epidemiol. 2012;36:276–82. doi: 10.1016/j.canep.2011.10.008. [DOI] [PubMed] [Google Scholar]


