Abstract
In the present study we investigated the molecular mechanisms regulating the expression of RORγt, the central factor controlling IL-17 transcription and Th17 differentiation. In key studies we found that cells from mice with major deletions of the E-protein transcription factors, E2A and HEB, display greatly reduced RORγt/IL-17 expression and that E-protein-deficient mice exhibit greatly diminished IL-17-dependent inflammation in EAE models. In additional studies, we unexpectedly found that cells from mice with deletion of Id3, a protein that inhibits E-protein binding to DNA, display diminished RORγt/IL-17 expression and mice deficient in this protein exhibit decreased Th17-mediated inflammation in a cell-transfer colitis model. The explanation of these initially paradoxical findings came from studies showing that Id3 deficiency leads to increased IL-4-induced GATA-3 expression, the latter a negative regulator of RORγt transcription; thus, increased Id3 expression likely has a net positive effect on RORγt expression via its inhibition of IL-4 production. Finally, we found that both E-proteins and Id3 are up-regulated in tandem by the cytokines that induce Th17 differentiation, TGF-β and IL-6, implying that these transcription factors are critical regulators of Th17 induction.
Keywords: RORγt, E-proteins, Id3, GATA-3, IL-17, IL-4
Introduction
In recent year, there has been considerable progress in our understanding of the molecular factors regulating the Th17 response. However, most of this understanding centers around the regulation of IL-17 transcription by RORγt and relatively little around the regulation of RORγt itself. Thus, while it is known that Runx11 Stat32–3 and perhaps BATF4 exert a positive influence on RORγt transcription, it is not clear how these factors are related to the two cytokines known to up-regulate RORγt, namely TGF-β and IL-6. The same can be said for for HIF-1α, a factor with a positive effect on RORγt transcription that is induced by stress stimuli that do not necessarily include TGF-β and IL-65–7.
One class of transcription factors that has not been thoroughly explored as to their possible role in Th17 differentiation are the E-proteins, a family of proteins whose relation to Th17 is suggested by studies showing that members of this family regulate RORγt expression during thymocyte development8–9. E-proteins are homo- or heterodimeric transcriptional activators (or repressors) whose function is down-regulated by interaction with any of four Id proteins that inhibit their binding to DNA and thus their transcriptional activity10–12. In studies of thymic development it has been shown that interplay between E-proteins and Id proteins have important roles in the regulation of thymocyte differentiation at various thymocyte differentiation check-points and in shaping the fate of naïve cells13–15. In part, such regulation involves E-protein effects on RORγt expression since it has been shown that RORγt is decreased in E-protein deficient CD4+/CD8+ thymocytes9. In addition, pre-TCR signaling induces E-protein-dependent RORγt expression and this inhibits cell proliferation while promoting gene rearrangement8. With respect to peripheral effects of E-proteins, it has been shown that Id2-mediated suppression of E-proteins is necessary for lymphoid tissue inducing cell and mature NK cell development16; in addition, E-proteins may regulate the development of effector-memory CD8+ T cells since the latter are deficient in mice lacking Id317. However, there is no evidence presently available indicating that these negative peripheral effects of E-proteins involve RORγt. In the present study we show that E-proteins (E2A and HEB), acting in conjunction with Id3, positively regulate RORγt expression in peripheral T cells and that such regulation is a critical factor in the induction of Th17 differentiation.
Results
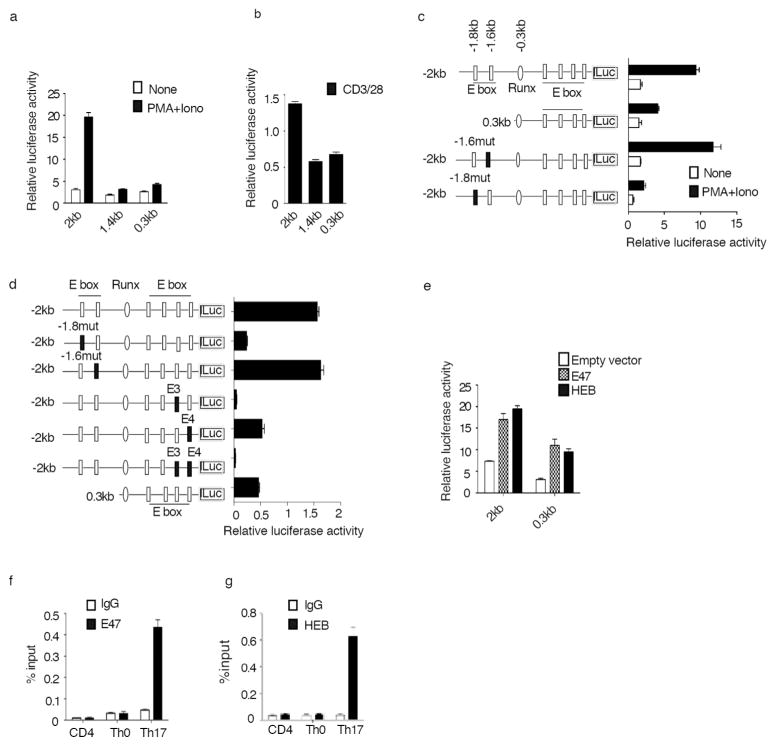
E-proteins are Required for RORγt Promoter-Reporter Activity
Prompted by the fact that RORγt expression is decreased in thymocytes of E2A null mice9, in initial studies we explored how E-proteins affect RORγt promoter activity in luciferase assays. To this end, we generated a PGL4 luciferase reporter plasmid containing RORγt promoter fragments of 2kb (−2kb--+109), 1.4kb(−1.4kb---+109) and 0.3kb(−0.3kb--+109) and then transfected the reporter constructs into EL-4 (LAF) cells or primary CD4+ T cells. We found that while the 0.3kb and 1.4kb fragments gave rise to minimal promoter-reporter activity, the 2kb fragment gave rise to high activity in both EL-4 cells (Fig. 1a) and primary CD4+ T cells (Fig. 1b).
Fig. 1. E-protein is required for RORγt promoter luciferase reporter activity.
(a, b), A 2kb mouse RORγt promoter fragment is required for maximal promoter activity. RORγt promoter fragments of various sizes inserted into luciferase reporter constructs were co-transfected with Renilla luciferase plasmid into EL4 (LAF) cells (a) or primary CD4+ T cells (b); following transfection cells were stimulated with PMA/ionomycin (P/I) (a) or anti-CD3/anti-CD28 (b) overnight after which luciferase activity was measured and normalized by Renilla luciferase activity.
(c) E-box binding sites are essential for RORγt promoter-reporter activity. A WT 2kb RORγt promoter constructs without and with mutated E- box binding sites at −1.8kb or −1.6kb were transfected into EL4 cells. Following transfection cells were stimulated overnight with P/I after which luciferase activity was measured and normalized as above.
(d) E-box binding sites E3 and E4 are also essential for RORγt promoter-reporter activity. A WT 2kb RORγt promoter construct without and with mutated E-box binding sites E3 and E4 were transfected into EL4 cells. Following transfection cells were stimulated overnight with P/I after which luciferase activity was measured and normalized as above.
(e) E-protein up-regulates RORγt promoter-reporter activity. The indicated RORγt promoter-reporter constructs were co-transfected with E47 or HEB expression plasmids or empty vector into EL4 cells and cultured overnight after which luciferase activity was measured and normalized as above.
Data shown are derived from mean ±SD of duplicate transfections in each experiment and are representative of at least three independent experiments.
(f,g) E-proteins bind to the RORγt promoter in primary Th17 cells. Chromatin from purified naïve CD4+ T cells, CD4+ T cells polarized under Th0 conditions or Th17 conditions were subjected to ChIP analysis for the detection of binding of E-proteins, E47 (g) and HEB (f) with anti-E47 and anti-HEB respectively. Results are representative of three independent experiments in each case. Data shown are mean±SD of duplicate PCRs.
DNA sequencing analysis of the RORγt promoter region revealed several consensus E-protein binding sites (E-boxes) that are conserved among mouse, rat and human RORγt promoters1. We found that mutation of the E-box at −1.6kb had no effect on promoter activity, whereas mutation of the E-box at −1.8kb reduced promoter activity to the level seen with the 0.3 kb promoter fragment (Fig. 1c). In addition, mutation of the E3 and/or E4 E-boxes 1 close to the transcription start site also led to reduced promoter activity of the 2kb fragment (Fig. 1d, Supplementary Fig. 1a), Thus, several E-boxes act in tandem to support optimal RORγt promoter-reporter activity.
In further studies we co-transfected reporter constructs with E-protein expression vectors. We found that co-transfection of the 2kb and 0.3kb RORγt promoter-reporter constructs along with either an E47 or HEB expression vector led to substantial increases in luciferase signal as compared to co-transfection of empty vector (Fig. 1e). In contrast, co-transfection of E-protein expression vectors with a 2kb promoter- reporter bearing various E-box binding site mutations, failed to reverse the complete or partial loss of the luciferase signal (Supplementary Fig. 1b). Collectively, the data derived from the above promoter-reporter studies suggested that E-proteins do have the potential to regulate RORγt transcription.
E2A and HEB Act as Transcription Factors that Regulate RORγt and IL-17 Expression
The results of the above luciferase reporter studies led us to investigate if E-protein act as RORγt transcription factors in developing Th17 cells. In initial studies we performed chromatin immunoprecipitation (ChIP) assays in CD4+ T cells and found that both E2A and HEB bind to the RORγt promoter in Th17 cells but not in naïve CD4+ cells or Th0 cells (Fig. 1f, 1g). Thus, E-proteins do in fact bind to the RORγt promoter in physiologic cells, but only in the context of Th17 differentiation.
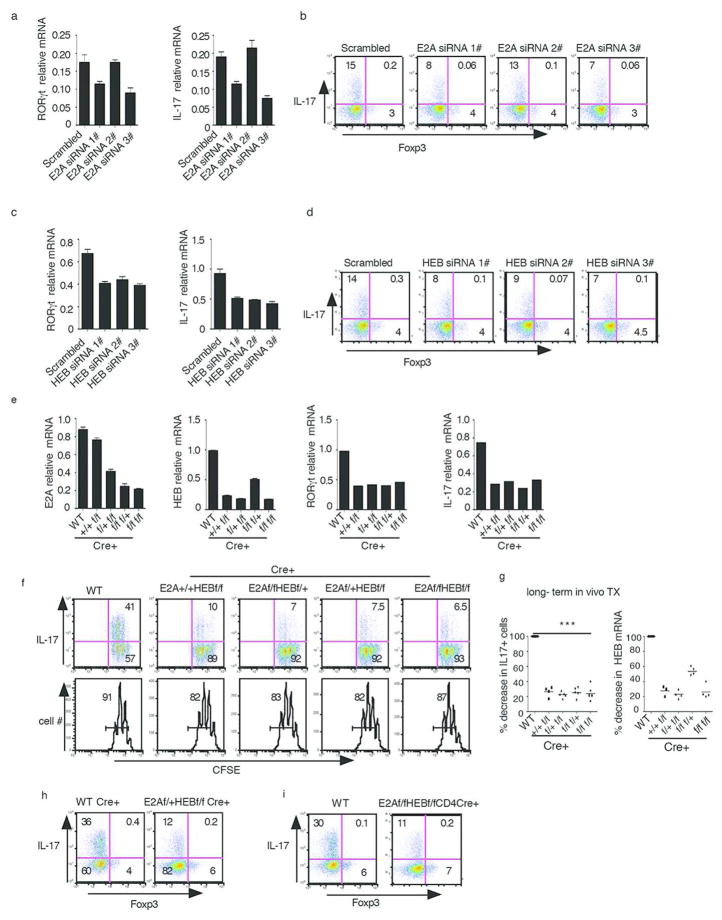
To determine if deletion of E-proteins affected RORγt transcription we next performed gene knock down studies using small interfering RNA (siRNA). Three different siRNAs for each gene were employed, of which at least two resulted in substantial reduction of mRNA expression (Supplementary Fig. 2a, 2b). We found that CD4+ T cells cultured under Th17 conditions transfected with E2A or HEB siRNA (but not scrambled siRNA) resulted in substantial attenuation of RORγt transcription and IL-17 expression at both the RNA (Fig. 2a, 2c) and protein level (Fig. 2b, 2d). These findings are in agreement with previous studies of the regulation of RORγt transcription and IL-17 expression by E2A siRNA18.
Fig. 2. E-proteins are required for Th17 cell differentiation.
(a,b) Purified naïve CD4+ T cells were transfected with E2A siRNA or control scrambled siRNA. Cells were then cultured under Th17 conditions after which RORγt (a) expression and IL-17 production was measured by RT-PCR and flow cytometry (a,b).
(c,d) Purified Naïve CD4+ T cells were transfected with HEB siRNA and control scrambled siRNA. Cells were cultured under Th17 conditions after which RORγt (c) and IL-17(c,d) were measured as indicated above.
Data above are representative of at least three independent experiments
(e) ER-cre-positive and WT mice were administered tamoxifen by gavage 6 times over a period of 30 days. Purified naïve CD4+ T cells were cultured under Th17 conditions after which they were analyzed for E2A, HEB, RORγt and IL-17 expression by real time RT-PCR.
Data are representative of three independent experiments. Error bars represent standard deviation of duplicated PCRs.
(f) ER-cre-positive and WT mice were administered tamoxifen by gavage 6 times over a one month period. Purified naïve CD4+ T cells obtained from these mice one week after the final dose of tamoxifen were labeled with CSFE and cultured under Th17 conditions for three days after which IL-17 expression was measured by flow cytometry. Data are representative of three independent experiments.
(g) Summary percentage of IL-17 decrease and percentage of E-protein decrease in Th17 cells from subjected to tamoxifen administration regimens. Data are derived from at least three independent experiments. in each case, the percentage of IL-17+ cells for WT mice were set as 100% and each dot represents data from one mouse. Bar represents mean value. ** P< 0.01, *** P< 0.001
(h) Purified CD4+ T cells from WT Cre+ or E2Af/+HEBf/f CD4 Cre+ mice treated with tamoxifen as in Fig. 2e were cultured under Th17 conditions; IL-17 expression was measured by flow cytometry. Data are representative of two independent experiments.
(i) Purified naïve CD4+ T cells from WT or E2Af/fHEBf/f CD4Cre mice were cultured under Th17 conditions for 3 days; IL-17 expression was measured by flow cytometry. Data are representative of two independent experiments
Then, in more definitive studies of the affect of E-protein deletion, we examined RORγt and IL-17 expression in ER-Cre-inducible E2A and HEB conditional knock out mice (E2Af/fHEBf/f ER-Cre-mice)13. Accordingly, we administered tamoxifen by gavage to ER-Cre-positive and WT mice every other day for ten days (five times) and then, one week after the final tamoxifen administration, obtained purified naïve CD4+ T cells from the mice and cultured them under Th17 conditions in the presence of 4-OH tamoxifen to evaluate the effects of deletion on Th17 differentiation. We found that cells from tamoxifen-treated ER-Cre+ mice exhibited about a 50% decrease in RORγt and IL-17 mRNA levels (Supplementary Fig. 2c, 2e) as well as similar reduction in the frequency of IL-17 producing cells (Supplementary Fig. 2d) as compared to tamoxifen-treated WT mice. Parenthetically, deletion of E2A or HEB had no effect on Foxp3 expression in the cells cultured under Th17 conditions (Supplementary Fig. 2d).
To achieve more profound E-protein deletion we next subjected E-protein conditional KO and WT mice to relatively long term tamoxifen administration. Accordingly, we administered tamoxifen 6 times over a period of one month and then, 6 days after the last injection, again cultured purified naïve CD4+ spleen T cells under Th17 conditions to evaluate effects of deletion on Th17 differentiation. Indeed, we found that this regimen led to an approximately 75%–80% decrease in the expression of E2A and HEB mRNA, accompanied by similar decreases in RORγt and IL-17 mRNA in cells from ER-Cre+ mice compared to WT mice (Fig. 2e). In addition, the frequency of IL-17 producing cells was also reduced by about 75%–80% in the conditional KO mice (Fig. 2f). Finally, it should be noted that E-protein mRNA decreases had a slightly greater effect on IL-17 expression than on RORγt expression; this could be due to the fact that IL-17 transcription is highly sensitive to decreases in RORγt.
Comparison of data derived from individual E-protein floxed mice (with partial or complete E2A or HEB deletions) in the long and short tamoxifen administration regimens described above (and shown in Fig. 2e and 2g as well as in Supplementary Fig. 2c and 2e) revealed that the percentage decrease in functional E-protein (a heterodimer of E2A and HEB that reflects both E2A and HEB mRNA levels) correlated with the percentage decrease in the number of IL-17-expressing cells. Thus, these the data strongly suggest that E-protein expression is in fact necessary for RORγt and subsequent IL-17 expression.
It has been reported that both E-proteins and RORγt are involved in regulating cell proliferation. To investigate this possibility, purified naïve CD4+ T cells from mice subjected to tamoxifen-treatment were labeled with CFSE and cultured under Th17 conditions. We found no significant changes in cell proliferation between WT and conditional KO cells (Fig. 2f lower panel) whereas IL-17 expression underwent a very substantial decrease (Fig. 2f upper panel). These results thus indicate that the effect of E-protein deletion on IL-17 expression is not due to an effect on cell proliferation. Finally, the above data on tamoxifen-mediated Cre-induced deletion of E-proteins was not an artifact of Cre toxicity since IL-17-producing cells were greatly reduced in E2Af/fHEBf/fCre+ Th17 cells compared to that of WT Cre+ Th17 cells (Fig. 2h).
Th17 Differentiation in E2Af/fHEBf/fCD4Cre-Mice
To verify the findings described above in mice with conditional E2A/HEB deletions we then determined Th17 differentiation in E2Af/fHEBf/fCD4Cre-mice. We found that purified naïve CD4+ T cells obtained from such mice and cultured under Th17 conditions give rise to greatly decreased numbers of IL-17-producing cells as compared with that of WT mice (Fig. 2i). Importantly, the deletion efficiency of E2A and HEB in CD4Cre-positive mice (75–80%) was similar to that of obtained with the one-month tamoxifen injection regimen (Supplementary Fig. 2f). These results again indicate that deletion of E2A and HEB leads to impairment of Th17 differentiation.
E-protein Control of RORγt Transcription is Regulated by Id Protein (Id3)
As mentioned above, Id proteins are a family of transcription co-factors that have been shown to bind to E-proteins and thereby inhibit the latter’s transcription activity by interfering with their DNA-binding activity11–12. Id2 and Id3 are the main Id family members involved in the regulation of E-protein function during lymphoid development19–20 and indeed we found that both of these Id proteins are expressed in Th17 cells (data not shown). We therefore conducted studies of Id protein effects on RORγt expression utilizing cells from mice with Id protein deletion, focusing mainly on mice with Id3 deletion (Id3−/− mice) since it has been shown that mice with Id2 deletion exhibit Id3 up-regulation and could thus yield equivocal results17.
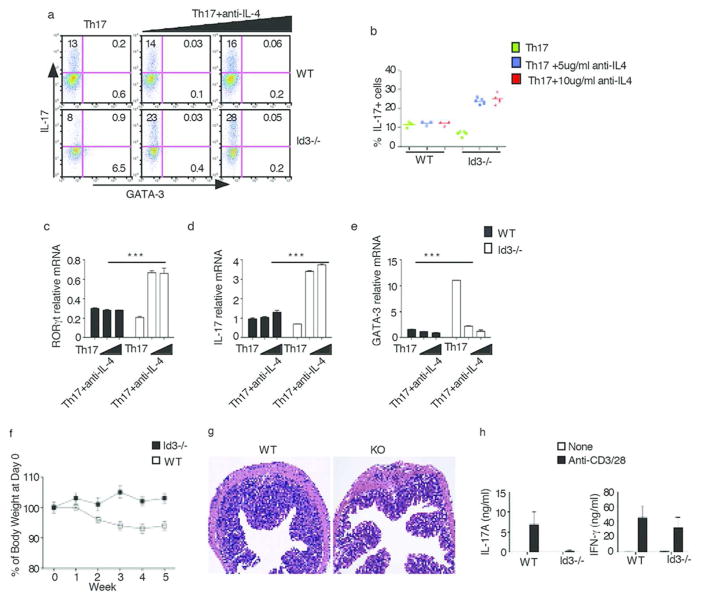
In initial studies along these lines, naive CD4+ T cells purified from Id3−/− and WT control mice were cultured under Th17 conditions for three days at which time IL-17 expression were measured by flow cytometry. Surprisingly, we observed that IL-17 expression in the Id3−/− Th17 cells was decreased rather than increased compared to WT Th17 cells (Fig. 3a left panels, 3b). We reasoned that this paradoxical result could have been due to the fact that the absence of Id3 led to the induction of an inhibitory factor normally suppressed by Id3 that has a countervailing negative effect on RORγt and IL-17 expression. One such possible inhibitory factor is IL-4 since production of the latter has been shown to be increased in Id3−/− mice13 and, in addition, has been shown to down-regulate IL-17 production21. Indeed, we found that cells from Id3−/− mice cultured under Th17 conditions in the presence of anti-IL-4 exhibited increased IL-17 expression (Fig. 3a, 3b, 3d). Furthermore, RORγt (Fig. 3c) and IL-17 mRNA (Fig. 3d) were also significantly increased in Id3−/− Th17 cells containing anti-IL-4, whereas GATA-3 levels were significantly decreased (Fig. 3a, right panels, Fig. 3e). These results thus suggested that IL-4 itself or GATA-3 induced by IL-4 does in fact inhibit RORγt expression in the absence of Id3.
Fig. 3. Id3 regulates Th17 cell differentiation.
(a) Naïve CD4+ T cells purified from Id3−/− and WT littermate control mice were cultured under Th17 conditions or Th17 conditions plus the addition of anti-IL-4 (5ug/ml, 10ug/ml) for three days. IL-17 was analyzed by flow cytometry. Data are representative of three independent experiments
(b) Naïve CD4+ T cells purified from individual Id3−/− and WT littermate control mice were cultured under Th17 conditions or Th17 conditions plus anti-IL-4 (5μg/ml, 10μg/ml) for three days; IL-17 was analyzed by flow cytometry after which the percentage of IL-17+ cells for each mouse was plotted. Each dot represents data from one mouse Data are pooled from two independent experiments.
(c,d,e) Naïve CD4+ T cells purified from Id3−/− and WT littermate control mice were cultured under Th17 conditions or Th17 conditions plus the addition of anti-IL-4 (5ug/ml, 10ug/ml) for three days. RORγt (c), IL-17 (d) and GATA-3 (e) expression were measured by real-time RT-PCR. *** P< 0.001. Data are representative of three independent experiments.
(f–i) Development of colitis in Rag2−/− mice transferred with naïve CD4+ T cells from WT or Id3−/− mice. (f) Change in body weight after cell transfer; (g) Hematoxylin and eosin staining of colon tissue sections from Rag2−/− mice 5 weeks after cell transfer; (i) IL-17 and IFN-γ production by mesenteric lymph node cells of Rag2−/− mice stimulated with anti-CD3/28; cytokines were measured in culture supernatants 48h after stimulation by ELISA.
In further studies to investigate the effect of IL-4 in Id3−/− mice on Th17 cell differentiation we found that naïve CD4+ T cells from Id3−/− mice produced increased amounts of IL-4 mRNA as compared to WT cells, but this increase was not accompanied by an increase in GATA-3 mRNA expression (Supplementary Fig. 3a). In contrast, CD4+ T cells from Id3−/− mice cultured (and activated) under both Th0 and Th17 conditions expressed increased amounts of IL-4 mRNA compared to WT cells (Supplementary Fig. 3b upper panels) and in this case GATA-3 mRNA was also expressed (Supplementary Fig. 3a, right panel and Fig. 3b, lower panels).
To examine the observations concerning Id3 in vivo we next examined IL-17 expression in Id3−/− mice with cell-transfer colitis. Accordingly, we monitored colitis development in RAG2−/− mice transferred purified CD4+/CD45Rbhi CD4+ T cells from Id3−/− or WT mice. We found that recipients of Id3−/− CD4+ T cells developed less severe colitis than recipients of WT T cells as indicated by both weight loss and colonic morphology (Fig. 3f, 3g). In addition, anti-CD3/CD28-stimulated T cells from the mesenteric nodes of mice transferred WT cells produced significantly greater amounts of IL-17 than T cells from mice transferred Id3−/− cells whereas, in contrast, these T cell populations produced equivalent amounts of IFN-γ (Fig. 3i). With the caveat that Id3 has multiple effects on lymphoid cell development in addition to those on Th17 differentiation, these results thus provided in vivo validation of our in vitro observation that cells from Id3−/− mice produce lower amounts of IL-17 than cells from WT mice.
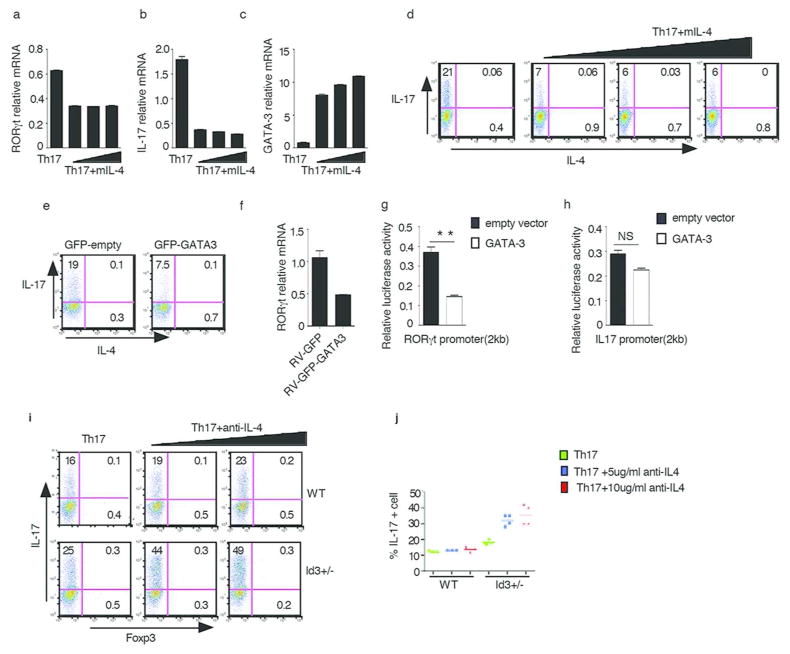
In related studies, we found first that WT CD4+ T cells cultured under Th17 conditions in the presence of exogenous IL-4 exhibited decreased RORγt and IL-17 mRNA expression and increased GATA-3 mRNA expression (Fig. 4a–c), as well as a decreased frequency of IL-17-producing cells (Fig. 4d). Second, developing Th17 cells transduced with a retrovirus expressing GATA-3 have a much lower frequency of IL-17-producing cells and RORγt expression as compared to cells transduced with empty vector (Fig. 4e, 4f). Third, RORγt promoter reporter activity but not IL-17 promoter reporter activity was inhibited by co-transfection of a GATA-3 expression plasmid (Fig. 4g, 4h). These studies thus provide further support for the idea that IL-4 and GATA-3 are negative regulators of E-protein/Id protein-mediated Th17 differentiation.
Fig. 4. GATA-3 Inhibits Th17 Differentiation.
(a–d) Purified CD4+ T cell from WT mice were cultured under Th17 conditions or Th17 conditions plus IL-4(10ng/ml, 20ng/ml and 40ng/ml) for three days. RORγt (a), IL-17(b), GATA-3(c) were measured by RT-PCR and IL-17 expression was analyzed by flow cytometry (d). Data are representative of three independent experiments
(e,f) CD4+ T cells were activated and transduced with GFP-empty (MIG) vector or GFP-GATA3 following which cells were cultured under Th17 conditions and analyzed 3 days after transduction. IL-17 was analyzed by flow cytometry (e), and RORγt expression was measured by RT-PCR (f). GFP+ cell were gated, numbers indicate percentage of IL-17 positive cells. Data are representative of two independent experiments.
(g) A 2kb RORγt promoter construct was co-transfected with empty vector or a GATA-3 expression vector into EL-4 cells after which the cells were stimulated overnight with PMA/Iono. RORγt promoter reporter activity was measured and normalized by Renilla luciferase activity. Data are representative of three independent experiments
(h) A 2kb IL-17 promoter construct was co-transfected with empty vector or a GATA-3 expression vector into Jurkat cells after which the cells were stimulated overnight with PMA/Iono. IL-17 promoter reporter activity was measured and normalized by Renilla luciferase activity. Data are representative of three independent experiments.
(i) Naïve CD4+ T cells purified from Id3+/− and WT littermate control mice were cultured under Th17 conditions or Th17 conditions plus anti-IL-4 (5μg/ml, 10μg/ml) for three days; IL-17 vs. Foxp3 expression was then analyzed by flow cytometry. Data are representative of two independent experiments.
(j) Naïve CD4+ T cells purified from individual Id3+/− and WT littermate control mice were cultured under Th17 conditions or Th17 conditions plus anti-IL-4 (5μg/ml, 10μg/ml) for three days; IL-17 was analyzed by flow cytometry after which the percentage of IL-17+ cells for each mouse was plotted. Each dot represents data from one mouse Data are pooled from two independent experiments.
Finally, we determined the effect of partial deletion of Id3 on Th17 differentiation with studies in which we cultured CD4+ T cells from Id3+/− mice under Th17 conditions. We found that partial deletion of Id3, in contrast to complete deletion, was associated with a modest increase in IL-17 expression compared to WT T cells that was further increased by addition of anti-IL-4 (Fig. 4i, 4j). To investigate this discrepancy further we compared the capacity of T cells from Id3+/− mice and Id3−/− mice to produce IL-4 and express GATA-3 and found that after TCR activation under Th0 or Th17 conditions, Id3+/− cells produce substantially less of these factors than Id3−/−cells (Supplementary Fig. 3c). These findings thus indicate that in mice with decreased but not absent amounts of Id3 protein (Id3+/− mice), there is more E-protein available to drive Th17 differentiation but not enough GATA-3 to block such differentiation; in contrast, in mice with total absence of Id3 protein (Id3−/− mice), high levels of IL-4 and GATA3 are produced which block the positive effect of E-protein on Th17 differentiation. Thus, RORγt expression requires calibrated up-regulation of Id3 to achieve an E-protein/Id3 protein ratio that balances the positive and negative effects of Id3.
In a separate series of studies we also determined the effect of Id2 deletion on RORγt and IL-17 mRNA expression despite the above mentioned concern about the possible effect of Id2 deletion of Id3 levels. We found that cells from Id2−/− mice stimulated under Th17 conditions for 3 days also expressed decreased RORγt and IL-17 mRNA as well as increased GATA-3 mRNA, but these effects were less than those observed in cells from Id3−/− mice (Supplementary Fig. 3d, 3e). Thus it is likely that Id2, under Th17 conditions has a similar but lesser effect on Th17 differentiation as Id3.
Synchronous Regulation of E-Proteins and Id Proteins by TGF-β and IL-6 Signaling
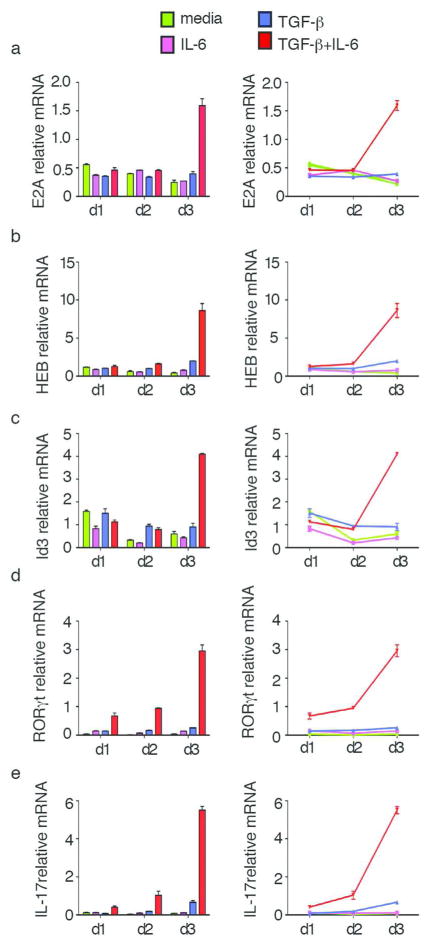
We next determined whether TGF-β and IL-6, the two major cytokines driving Th17 cell differentiation, act through E-proteins. To address this question, purified CD4+ T cells from WT mice were activated with anti-CD3/anti-CD28 and cultured with different combinations of cytokines. Cells were harvested on day 1, day 2 and day 3, at which time E-protein (E2A or HEB), Id3, RORγt and IL-17 expression were assessed by real-time RT-PCR (Fig. 5a–f). We found that on day 1 and day 2, there is no significant change of E-protein expression under all of the cytokine conditions. However, on day 3, cells cultured in the presence of TGF-β and IL-6 (i.e., Th17 conditions) exhibited a significant increase in both E2A and HEB expression as compared to cells cultured with no added cytokines (i.e., neutral conditions), or cells cultured with TGF-β or IL-6 alone (Fig. 5a, 5b). Interestingly, Id3 was also significantly increased on day3 under Th17 culture conditions but not under other conditions (Fig. 5c). In additional studies focusing on RORγt and IL-17 we found that while small increases of these proteins were seen on day 1 and day 2, large increases were seen only on day 3 accompanying changes in E-protein and Id3 expression (Fig. 5d, 5e). Thus, E-proteins and Id3 are synchronously up-regulated by cytokines that have been identified as the main inducers of the Th17 response and appear to be necessary for the induction of this response.
Fig. 5.
Kinetic studies of E-protein and Id protein expression under various cytokine conditions. Purified CD4+ T cells from WT mice were cultured with various combinations of cytokines involved in Th17 differentiation. Cells were harvested on d1, d2 and d3 respectively, after which expression levels of E2A (a), HEB (b), Id3(c), RORγt(d) IL-17(e) were analyzed by RT-PCR. Data are representative of at least three independent experiments.
In parallel studies we measured effect of IL1-β and IL-23 on HEB, Id3, RORγt and IL-17 levels. We again found that IL-6 and TGF-β (in combination) gave rise to increased levels of these factors on the day3 of culture. However, IL-1β and IL-23, alone or in combination (or both in combination with IL-6) was not associated with an increase in HEB and Id3 (Supplementary Fig. 4a–d).
E-proteins are Required for Th17-mediated Autoimmune inflammatory Disease
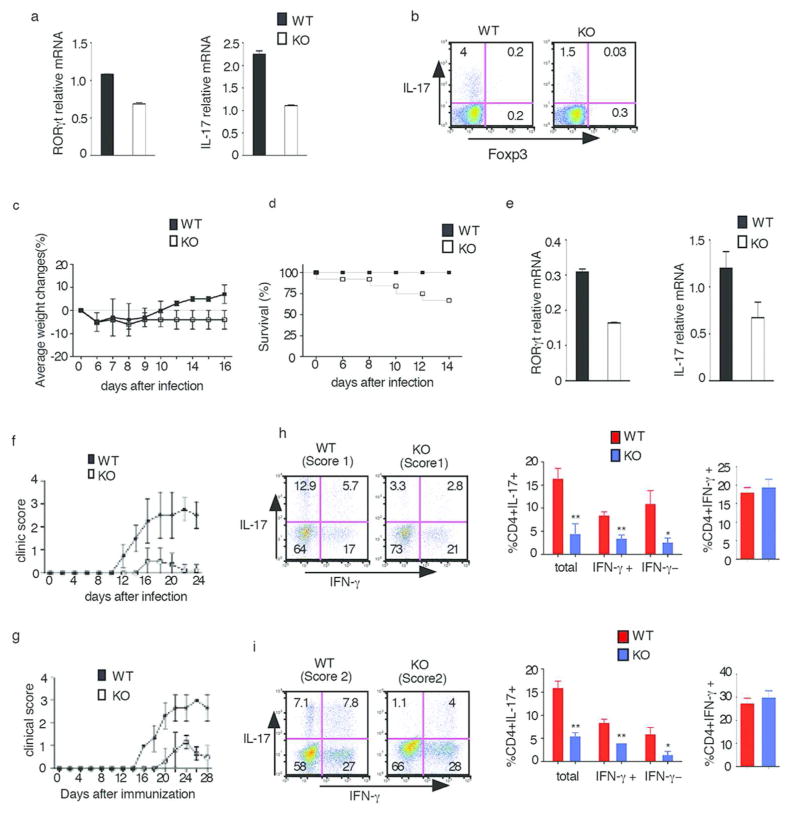
To determine whether E-protein is also essential for RORγt expression and Th17 differentiation in an in vivo setting, we first investigated the role of E-proteins in the generation of IL-17-producing T cells in the un-inflamed small intestine, a mucosal site previously shown to harbor such cells during commensal colonization22. Accordingly, ER-Cre-positive mice and WT mice were administered tamoxifen by gavage every other day over a period of 10 days and four weeks later the frequency of IL-17-producing cells in the small intestinal lamina propria (LP) was assessed by flow cytometry. We found that Er-Cre-positive mice exhibited a much lower level of RORγt and IL-17 mRNA (Fig. 7a) and lower frequency of IL-17 cells (Fig. 7b) than WT mice.
In further in vivo studies, we determined the effect of E-proteins on the Th17 response of mice subjected to small intestinal infection with Citrobacter rodentium, an infection in which an intact IL-23–IL-17 axis has been shown to be essential for host protection23. In these studies, ER-cre-positive and WT mice were administered tamoxifen by gavage as described above and one week later orally inoculated with C. rodentium. Whereas infected WT had a transient loss of weight followed by full recovery of weight by 10 days after inoculation, infected KO mice exhibited persistent loss weight over the 16 day observation period (Fig. 7c); furthermore, blinded studies of colons at day 14 after inoculation showed that C. rodentium-infected KO mice displayed greater mucosal inflammation, ulceration and fibrotic reaction than WT mice (data not shown) and, in pooled experiments, whereas approximately 20% of the cre-positive mice had died by day 14, none of the WT mice had died by this time point (Fig. 7d). These findings correlated with the fact that colonic tissues from KO mice exhibited decreased levels of RORγt and IL-17 compared to tissues from WT mice. These in vivo data, taken together, strongly suggest that E-protein deficiency leads to decreased IL-17 responses during both commensal colonization and pathogenic bacterial infection of the intestine.
We next investigated the role of E-protein in experimental allergic encephalitis (EAE), a model of inflammation previously shown to be associated with a robust Th17 response24. In an initial study, WT and E2Af/+HEBf/f Cre-positive (KO) mice were injected with tamoxifen 6 times as in Fig. 2e. EAE was induced in WT and KO mice by injecting MOG35-55 in complete Freund’s adjuvant and pertussis toxin (PTX) at day0 and day 2. WT mice exhibited initial evidence of disease on day 12 and evidence of severe disease (score 3) on day 18–20. In contrast, only about 50% of the E2Af/+HEBf/f Cre-positive mice exhibited disease and this consisted of mild disease (maximum score 1) on day 16–20, after which they recovered; the other 50% of mice in this group didn’t develop any disease. (Fig. 6c).
Fig. 6. E-proteins regulate Th17 responses in vivo.
(a,b) ER-cre-positive and WT mice were administered tamoxifen by gavage five times every other day. Four weeks after the final tamoxifen administration, lamina propria cells were purified from the small intestine; some of the cells obtained were directly analyzed by RT-PCR for RORγt or IL-17 (a), some of the cells were stimulated with PMA/Iono for 4hrs with Golgi stop in the last 3hrs and then analyzed for IL-17-expressing cells by flow cytometry gated on CD4+ cells (b). Data are representative of two independent experiments
(c,d) ER-cre-positive and WT were were orally administered with tamoxifen every other day for five times, One week after the final dose, mice were orally inoculated with C. rodentium and were weighed at the indicated time points (d) and followed for survival. Data are representative of two independent experiments.
(e) Colonic tissue obtained from WT or KO infected mice at 14 days after initiation of infection were subjected to real-time RT-PCR analysis of RORγt and IL-17 expression. Data are representative of two independent experiments.
(f) EAE disease course in WT and E2Af/+HEBf/f Cre+(KO) mice that had been treated with tamoxifen as above. Data are representative of two independent experiments. n=5(WT), n=6(KO).
(g) EAE disease course in Rag2-deficient mice reconstituted with 20×106 purified CD4+ T cells from WT and E2Af/+HEBf/f Cre+(KO) mice that had been treated with tamoxifen as above. n=6(WT), n=6(KO). Data are representative of two independent experiments.
(h) Cytokine production by cells isolated from the spinal cords of WT and E2Af/+HEBf/f Cre+(KO) mice on day 23 after EAE induction. The cells were stimulated for 4hr with PMA/Iono in the presence of Golgi stop and then analyzed for surface and intracellular cytokines by flow cytometry gated on CD4+ cells. Clinical score are shown in parentheses. Data are representative of two independent experiments. Tabulated Results from 2 independent experiments: Total: all IL-17+ cells; IFN-γ+: IL-17+IFN-γ+ cells; IFN-γ−:IL17+IFN-γ− cells. Error bars represent standard deviation; ** p<0.02, * p<0.05
(i) Cytokine production by cells isolated from the spinal cord of Rag2-deficient mice reconstituted with tamoxfen-treated WT or KO purified CD4+ T cells on day 20 after EAE induction. The cells were stimulated for 4hr with PMA/Iono in the presence of Golgi stop and then analyzed for surface and intracellular cytokines by flow cytometry gated on CD4+ cells. Clinical scores are shown in parentheses. Data shown are representative of two independent experiments. Tabulated results are from two independent experiments; Total: all IL-17+ cells; IFN-γ+: IL-17/IFN-γ+cells; IFN-γ−:IL17+IFN-γ− cells. Error bar represent standard deviation; ** p<0.01, *p<0.05
In complementary studies we employed an EAE model in which EAE is induced in RAG2-deficient mice (RAG2−/−) after the latter are re-constituted with purified CD4+ T cells from WT or E- protein conditional KO mice that has been treated with tamoxifen as described above. Transfer of 20×106 CD4+ T cells from WT mice to RAG2−/− mice, followed by induction of EAE 48h later, led to development of disease by 14 days which peaked at a score of 3. In contrast, transfer of the same number of CD4+ T cells from E-protein conditional KO mice resulted in delayed disease: most of the mice with disease displayed a disease score of 1, and only a few mice displayed a score of 2 (Fig. 6d).
Although E-protein conditional KO mice and Rag2−/− mice that received CD4+ T cells from E-protein conditional KO mice developed very attenuated EAE, the number of infiltrating CD4+ T cells present in the spinal cords of these mice were similar to those in mice reconstitute with WT cells (data not shown). In both cases, IFN-γ producing cells were present at similar frequency among WT mice and E-protein conditional KO mice spinal cord, however, the frequency of the infiltrating T cells produced IL-17 was much lower in E-protein conditional KO mice (Fig. 6e, 6f) compared with WT mice. Together, these results indicated that E-proteins not only regulate IL-17 expression in vitro, they also regulate IL-17 in vivo.
Discussion
Previous studies of the molecular mechanisms governing Th17 differentiation have focused mainly on the factors regulating IL-17 transcription, particularly as the latter relates to the transcriptional activity of RORγt, the Th17 lineage-defining factor known to be under the control of Th17-inducing cytokines such as TGF-β and IL-625–26. In the present study, we address a perhaps more fundamental Th17 molecular mechanism, namely the mechanism by which RORγt itself is regulated. We show that E-proteins and Id3 are factors induced by TGF-β and IL-6 during Th17 differentiation that play distinct yet interactive roles in RORγt expression.
Our initial findings derived from luciferase reporter studies established that there are several E-protein binding sites in the RORγt promoter which when individually mutated, display greatly reduced promoter-reporter activity. Then, in more definitive studies utilizing ER-Cre conditional E2A and HEB double floxed mice we showed that a tamoxifen-induced 75%–80% reduction in E2A/HEB mRNA levels led to comparable reductions in RORγt and IL-17 expression levels. In addition, we showed that CD4+ T cells from E2Af/fHEBf/fCD4Cre mice with E2A/HEB deletion due to constitutive expression of Cre in CD4+ T cells exhibited a similar level of RORγt/IL-17 deletion. Finally, we demonstrated that E-proteins induction has a major impact on the capacity to mount Th17 responses in vivo under both non-inflammatory and inflammatory conditions. Taken together, these studies offer strong proof that RORγt is highly regulated by E-protein during in vitro Th17 differentiation. As such, they corroborate earlier preliminary studies showing that E-protein mRNA knock-down in vitro inhibits RORγt transcription and IL-17 expression18.
Id proteins are known to dimerize with E-proteins and thus block their ability to bind to DNA11–12. This suggested to us that the interaction between Id protein and E-proteins might be involved in RORγt transcription, and led us to conduct extensive studies of Th17 differentiation in Id3-deficient mice. Unexpectedly, we found that Id3−/− CD4+ T cells manifest decreased RORγt expression and IL-17 transcription rather than increased expression of these parameters. This initially paradoxical result was subsequently explained by the finding that Id3−/− cells cultured under Th17 conditions greatly up-regulate their production of a factor that has a strong negative effect on RORγt transcription, GATA-3, and such up-regulation, in turn, was explained by the finding that Id3−/− cells also increase their production of IL-4, a GATA-3 inducer previously shown to inhibit Th17 differentiation21. These in vitro results were subsequently confirmed in vivo in a cell-transfer colitis model in which Id3−/− naïve cells were found to induce less colitis and less IL-17 expression than WT naïve cells.
It should be noted that in a previous study it was found that Id3−/− cells exhibit increased rather than decreased Th17 differentiation as reported in the present study18. In extensive unpublished in vitro studies bearing on this issue we found that cells with partial deletion of Id3, i.e., Id3+/− cells, do in fact exhibit increase Th17 differentiation relative to WT cells because Id3+/− cells do not produce IL-4 at a level sufficient to induce inhibitory GATA-3 under Th17 conditions; thus, in this case increased E-protein RORγt transcriptional activity due to decreased Id3 is unopposed by increased GATA-3-mediated inhibition of RORγt transcription. Whether or not these data can explain previous findings awaits additional study; we can say with certainty, however, that the conclusions relative to Id3 effects reported here are based on reproduceable in vitro experiments that were subsequently corroborated by in vivo studies of Id3−/− mice.
Finally, the importance of both E-proteins and Id3 in the regulation of RORγt and IL-17 expression was highlighted by the fact that these proteins undergo up-regulation by TGF-β and IL-6 in CD4+ T cells during Th17 differentiation, but hardly at all in cultures containing either or these cytokines alone. This assertion follows from the fact that E-proteins and Id3 are the only factors so far shown to be involved in Th17 differentiation that are responsive to the key cytokines inducing such differentiation, other than RORγt itself whose transcription is being regulated by these proteins. The fact that E-proteins and Id3 up-regulation are synchronized also supports this assertion since, as we have seen, both factors are necessary to successfully execute the role of E-proteins in Th17 differentiation.
In conclusion, the up-regulation of E-proteins and Id proteins in nascent Th17 cells by TGF-β and IL-6 suggest that these transcription factors act in tandem to serve as a molecular “switch” that turns on Th17 differentiation. Given their up-regulation by factors specifically involved in the induction of the Th17 response, E-proteins and Id proteins appear to have a specific relation to this response.
Methods
Mice and Tamoxifen Administration
C57BL/6 WT mice were from the Jackson Laboratory. E2Af/fHEBf/fER-Cre mice and Id3−/− mice (all B6 background) were obtained from Dr. Yuan Zhuang, Duke University; all mice were bred in an NIAID animal facility under specific pathogen-free conditions. To induce Cre expression in E2Af/fHEBf/fER-Cre mice, the mice were administered tamoxifen (Sigma, Cat#H7904) dissolved in corn oil P.O. five times every other day (3mg/mice) or six times every 5 days. Cell culture or EAE studies were conducted one week after the final tamoxifen administration. All animal studies were performed under an animal use protocol approved by the Animal Use Committee of the NIAID.
Transient Transfection and Reporter Assays
The RORγt promoter was obtained by PCR using mouse genomic DNA as a template. The PCR product was cloned into the pGL4.10 basic luciferase vector (Promega) using the NheI and BglII entry sites. Promoter mutations were introduced using a Quickchange II site-directed mutagenesis kit (Stratagene). PCR primers for these mutations are listed in Supplementary Table 1 or E3, E4 mutation as previously published8. All constructs were verified by sequencing. EL4 cells were transfected using a Nucleofector II machine (Amaxa), then stimulated and subjected to luciferase assay as previously described1.
RNA Interference
CD4+ T cells were obtained using a naïve CD4 T cell purification kit (Miltenyi), and then transfected by nucleofection with Stealth E2A- or HEB-specific siRNA or control (scrambled) siRNA (Invitrogen) (Supplementary Table 1) as previously described1.
In vitro Th17 cell Differentiation and Flow Cytometric Analysis
CD4+ T cells were purified by magnetic sorting using mouse anti-CD4 beads (Miltenyi) or by naïve CD4 T cell purification kit (Miltenyi). Purified cells were cultured under Th17 condition as previous described1. Some of cells were cultured in the presence of 1μM 4-OH-tamoxifen (4-OHT, Sigma cat #T5648). Flow cytometric analysis of intracellular cytokine was performed as previously described1. The following antibodies were used for flow cytometry: anti-CD4-PE or -Percp, anti-IL-17-PE or -APC (BD Bioscience), anti-Foxp3-FITC or -APC, anti-GATA3-PE (eBioscience).
mRNA isolation and Quantitative Real-Time PCR
mRNA Isolation and Quantitative Real-Time PCR were performed as previously described1. List of primers and probes from Applied Biosystems is provided in Supplementary Table I.
ChIP Assay
Chromatin was obtained by using a ChIP-IT™ Express Enzymatic kit (Active motif cat# 53009). The chromatin was incubated with anti-E47 (Santa Cruz cat#’sc-763), anti-HEB (Santa Cruz cat#sc-357) or normal Rabbit IgG antibody as previously described1. The DNA obtained was subjected to real-time PCR using primer pairs and probes listed in Supplemental Table 1.
Retroviral Transduction
Empty vector and GATA-3 retroviral transduction were performed as previously described1. The cells were cultured under Th17 condition after transduction and analysis as previously described1.
Transfer colitis
CD4+CD45RBhi naïve T cell obtained from WT or Id3−/− mice were purified by FACS sorting following which 4X105 purified cells were transferred into Rag2−/− mice. The mice were then monitored for weight loss and tissue collected for analysis at the time indicated.
C. rodentium-induced colitis
ER-cre-positive mice and ER-Cre-negative mice were administered tamoxifen every other day for 10 days (5 times) as described above. C. rodentium (ATCC51459) was prepared by incubation with shaking at 37° C for 6 hours in LB broth. The relative concentration of bacteria was assessed by measuring absorbance at OD600. Mice were inoculated with 1×109 cfu bacteria by oral administration one week after final the tamoxifen treatment. Tissue was collected post-inoculation for histology and/or cytokine analysis at the times indicated.
Isolation of lamina propria lymphocytes
Purified small intestinal or large intestinal lymphocytes cells were obtained as previously described27.
Experimental Allergic Encephalitis (EAE) Induction
EAE was induced by subcutaneous immunization on day 0 with MOG35-55 peptide (0.2mg/mouse, Biosynthesis) emulsified in CFA (CFA supplemented with 8mg/ml Mycobacterium tuberculosis, Gibco) and I.P. injection on day 0 and 2 with pertussis toxin (200ng/mouse, Calbiochem). Scoring system criteria: 0-no disease, 1-flaccid tail, 2-weak/partially paralyzed hind legs, 3-completely paralyzed hind legs, 4-completely paralyzed hind legs and partially paralyzed front legs.
CD4+ T cell Transfers
CD4+ T cells were purified by anti-CD4 magnetic microbeads (purity >95%) from WT Cre+ and E2A/HEB/Cre mice that had been treated with Tamoxfen as described above, and were transferred into RAG2−/− mice (20×106 cells per mouse) intravenously. 48h after adoptive transfer, the mice were subjected to EAE induction.
Isolation of Lymphocytes from Spinal Cords
Spinal cords were extracted from dissected spinal columns by flushing the latter with PBS using a 19G needle. The columns were then cut into small pieces and were subjected to lymphocyte extraction using a Neutral Tissue Dissociation Kit (Miltenyi, Cat #130-092-628). The cell suspension obtained was then washed with PBS, re-suspended in 10ml of ice cold HBSS containing 0.9M sucrose and centrifuged at 850g for 10 min; finally, the pelleted cells were washed with media, re-stimulated with PMA/Iono, and analyzed for intracellular cytokines by flow cytometry as above.
Supplementary Material
Acknowledgments
We thank Dr. Yuan Zhuang (Duke University) for E2Af/fHEBf/f Er-cre mice and Id3−/− mice, Dr. William Paul (NIAID/NIH) for a GATA3 construct and Dr. Kersh Gilbert (Emory University) for E47, HEB and Id3 constructs. We also thank Dr. Qian Chen (NIH/NIAID) for help with the EAE model.
Footnotes
Authorship
F.P. planned and performed studies and wrote/edited the manuscript; I.F. performed studies and managed animal breeding; Z.Y. performed studies; W.S. planned studies and edited the manuscript. The authors have no competing financial interests.
References
- 1.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei L, Laurence A, Elias KM, O’Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007;48:34605–10. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–74. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 4.Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, Hatton RD, Stormo GD, Weaver CT, Russell JH, Murphy TL, Murphy KM. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–9. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet VR, Johnson RS, Haddad GG, Karin M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–11. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, Pan F. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–84. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–76. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–26. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 9.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–70. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–86. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 11.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–84. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 12.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 13.Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braunstein M, Anderson MK. HEB-deficient T-cell precursors loss T-cell potential and adopt and alternate pathway of differentiation. Mol Cell Biol. 2011;31:971–82. doi: 10.1128/MCB.01034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones-Mason ME, Zhao X, Kappes D, Lasorella A, Iavorone A, Zhuang Y. E protein transcription factors are required for the development of CD4(+) lineage T cells. Immunity. 2012;36:348–361. doi: 10.1016/j.immuni.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boos MD, Yokota Y, Eberl G, Kees BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–30. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D’Cruz LM, Watowich SS, Murre C, Goldrath AW. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–9. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maruyama T, Li J, Vaque JP, Konkel JE, Wang W, Zhang B, Zhang P, Zamarron BF, Yu D, Wu Y, Zhuang Y, Gutkind JS, Chen W. Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nat Immunol. 2011;12:86–95. doi: 10.1038/ni.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–6. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 20.Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- 21.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–32. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 22.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Symonds EL, Riedel CU, O’Mahoney DO, Lapthorne S, O’Mahoney L, Shanahan F. Involvement of T helper type 17 and regulatory T cell activity in Citrobacter rodentium invasion and inflammatory damage. Clin Exp Immunol. 2009;157:148–154. doi: 10.1111/j.1365-2249.2009.03934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zepp J, Wu L, Li X. IL-17 Receptor Signaling and T Helper 17-Mediated Autoimmune Demyelinating Disease. Trends Immunol. 2011;32:232–239. doi: 10.1016/j.it.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–4. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 26.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;19:179–89. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, Fuss IJ, Watanabe T, Asano N, Davey MP, Rosenbaum JT, Strober W, Kitani A. NOD2 transgenic mice exhibit enhanced MDP-mediated down-regulation of TLR2 responses and resistance to colitis induction. Gastroenterology. 2007;133:1510–21. doi: 10.1053/j.gastro.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.