Abstract
Expression of the bacterial beta-galactosidase reporter gene (lacZ) in the vector used for the Knockout Mouse Project (KOMP) is driven by the endogenous promoter of the target gene. In tissues from KOMP mice, histochemical staining for LacZ enzyme activity can be used to determine gene expression patterns. With this technique, we have produced a comprehensive resource of gene expression using both whole mount (WM) and frozen section (FS) LacZ staining in 313 unique KOMP mutant mouse lines. Of these, ∼80% of mutants showed specific staining in one or more tissues, while ∼20% showed no specific staining, ∼13% had staining in only one tissue, and ∼25% had staining in >6 tissues. The highest frequency of specific staining occurred in the brain (∼50%), male gonads (42%), and kidney (39%). The WM method was useful for rapidly identifying whole organ and some substructure staining, while the FS method often revealed substructure and cellular staining specificity. Both staining methods had >90% repeatability in biological replicates. Nonspecific LacZ staining occurs in some tissues due to the presence of bacteria or endogenous enzyme activity. However, this can be effectively distinguished from reporter gene activity by the combination of the WM and FS methods. After careful annotation, LacZ staining patterns in a high percentage of mutants revealed a unique structure-function not previously reported for many of these genes. The validation of methods for LacZ staining, annotation, and expression analysis reported here provides unique insights into the function of genes for which little is currently known.
As part of a long-term effort to fully ascribe function to the entire genome, the International Knockout Mouse Consortium (IKMC; www.mousephenotype.org) (Bradley et al. 2012) was initiated in 2007 to create a resource of gene-specific knockout embryonic stem (ES) cells for all protein-coding genes in the mouse genome. To date, this global cooperative project has produced sets of targeted and/or gene-trapped cell lines for ∼17,000 unique genes, primarily using ES cells derived from the C57BL/6N inbred mouse strain. Mutant mouse phenotyping activities were initiated by the EUMODIC European Consortium for mouse phenotyping (Gates et al. 2011), which published the resulting data through Europhenome (www.europhenome.org) (Morgan et al. 2010), and by the Mouse Genetics Project at the Sanger Institute (www.sanger.ac.uk/mouseportal/). Collectively these efforts have produced mutant mice and generated physiological, behavioral, and morphological phenotyping data for more than 1000 genes. The next step, the International Mouse Phenotyping Consortium (IMPC; www.mousephenotype.org) (Brown and Moore 2012) was launched in 2011 as a coordinated, large-scale effort to convert ∼5000 knockout IKMC ES cells into adult homozygous (HOM) and heterozygous (HET) mutant mice to be functionally analyzed.
In order to accurately interpret the phenotypes of knockout mice for a specific gene, it is critical to identify the organs, substructures, and cells in which the gene is normally expressed. Traditionally, gene expression has been inferred using different methodologies revealing either transcript (e.g., in situ hybridization) or protein (e.g., immunofluorescence). A number of on-line accessible databases contain archived data on tissue surveys for mouse gene expression from in situ hybridization studies in embryo and adult mice (for review, see Geffers et al. 2012). However, these in situ atlases report gene expression restricted either to embryos or to only select adult organs (e.g., adult brain and kidney). In addition to in situ atlases, there are several sources for mouse gene expression surveys using transcriptome array methods with RNA extracted from homogenized tissue. Compendiums of adult tissue gene expression are available at GEO (www.ncbi.nlm.nih.gov/geo/), BioGPS (www.biogps.org/), the GXD Database (www.informatics.jax.org/expression.shtml), and Array Express (www.ebi.ac.uk/arrayexpress/). Although scientifically rich, hybridization array–based methods are limited since they provide no specificity with regard to the cell type or tissue substructure.
Alternatively, histochemical methods rely on chemical staining of endogenous promoter-driven “reporter” genes to provide anatomical specificity and spatial resolution of gene expression. A reporter gene utilizing a bacterial beta-galactosidase (lacZ) was first described in the early 1980s (Casadaban et al. 1980). Subsequently, the reporter gene approach detecting LacZ, another enzyme, or fluorescent proteins has been used in many studies to characterize gene expression patterns in animals. The lacZ reporter is particularly useful since it requires no special imaging equipment and requires only standard histochemical staining methods.
As part of the trans-NIH Mouse Initiative Knockout Mouse Project (KOMP; www.nih.gov/science/models/mouse/knockout/), we created 313 presumed null mutant mouse lines from targeted ES cells produced by the IKMC. The targeting vectors contain a bacterial beta-galactosidase reporter (lacZ), which, when inserted into the target gene, is expressed under the control of the target gene promoter (Skarnes et al. 2011). We completed LacZ staining analysis in 66 tissues using whole mount (WM) and frozen section (FS) histochemical staining in male and female adult mice for each of the mutant lines. The mouse lines are all available through the KOMP Repository (www.komp.org), and the LacZ-stained images and associated annotation are publicly available online (www.kompphenotype.org). Herein, we describe the general methods and results and also provide a more detailed analysis of the LacZ staining and gene expression patterns in selected cases. In many cases, the data generated suggest biological functions not previously known or associated with the gene.
Results
General pattern of LacZ staining in KOMP mutants
In the majority of the mutant mouse lines, WM and FS LacZ staining were each analyzed in at least one male and one female heterozygote (HET). Separate animals were evaluated for each staining modality and sex (∼n = 4 mice per mutation). WM and FS (Fig. 1) revealed that ∼80% of the mutant lines had specific LacZ staining in at least one tissue/organ. For 13% of the lines, only one tissue was stained, with testis accounting for more than half of these cases. In ∼25% of the mutants, six or more tissues were stained. Tissues with the highest frequency of staining were the CNS, testis, and kidney, with ∼40%–50% of mutants with reporter-gene staining in those tissues. Testis and ovary stained positively for LacZ in 42% and 23% of mutants, respectively. And ∼8% of the mutants had staining only in the testis, while staining exclusively in the ovaries was not observed. LacZ staining by WM and FS is tabulated by organs and tissues in Supplemental Figure 1.
Figure 1.

Patterns of LacZ staining in whole mount (WM, black) and frozen section (FS, gray) preparations. Male and female data are combined for this analysis. For calculating the percentage of organs/tissues stained, brain is counted as one organ whether one or six different brain regions stain. Overall, similar patterns and percentages of tissues stained using the two methods.
While the overall tissue distribution pattern of WM and FS staining was similar, some organs showed markedly different percentages of staining with these methods. There were three primary causes for these differences: (1) insufficient penetration of the staining solution into some organs in the WM preparations; (2) high levels of nonspecific staining that obscured specific staining in some tissues; and (3) some tissues were not prepared by both methods for comparative analysis. After excluding tissues with these explainable differences in staining, the overall correlation between WM and FS was 0.70 (R2) (Supplemental Fig. 2).
Reproducibility of LacZ staining
When excluding reproductive organs, biological replicates correlated within a staining modality (Supplemental Table 1). When including all tissues with and without specific staining, the concordance for WM staining replicates was 97%, while for FS staining the concordance was 91%. When examining the concordance only for tissues with specific reporter staining in one or both biological replicates, the concordance for WM and FS was 77% and 53%, respectively (Table 1). In order to further assess the repeatability of FS staining, we evaluated the FS staining in four mutant lines, each with four to five biological replicates (Supplemental Table 2). In this study using a higher number of biological replicates than the main survey, the concordance of annotation across all tissues for all four mutant lines was 92%, and it ranged from 88% to 98% among the lines.
Table 1.
Staining replication statistics

Background and nonspecific staining
Nonspecific staining in ∼90 wild-type (WT) C57BL/6N mice for both WM and FS was evaluated and summarized in Figure 2 (example images of nonspecific staining are presented in Supplemental Fig. 3A,B). In WM, there was WT staining in the gastrointestinal (GI) tract, presumably caused by the presence of luminal bacteria expressing beta-galactosidase. Specific GI staining could be distinguished from nonspecific, bacterial-associated GI staining by FS. Occasionally, nonspecific staining was observed in FS GI epithelial mucosa, presumably due to the presence of intracellular glycosides such as lactase. Nonspecific staining was observed in FS preparations of the tongue and esophagus, likely due to bacteria adherent on the surface epithelium. Nonspecific staining was also frequently observed in the testis (WM), vas deferens (WM and FS), prostate (FS), preputial gland (WM), coagulating gland (WM and FS) and epididymis (WM and FS) of the male reproductive tract, in the salivary gland of both sexes (WM) and less frequently found in kidney (WM), lymph nodes (FS), and thyroid (WM and FS) in both sexes.
Figure 2.

Frequency of nonspecific staining in wild-type (WT) control C57BL/6N male and female mice. Whole mount (WM), and frozen section (FS) nonspecific staining percentages are indicated in blue and red, respectively. Explanations of differences between WM and FS preparations for nonspecific staining for specific tissues are as follows: (1) bacteria on epithelium of tongue and esophagus in FS preparations; (2) luminal gut bacteria present in intact WM preparations; (3) prostate adjacent to preputial gland and inability to distinguish between preputial gland nonspecific staining and specific prostate gland staining in WM preparations; and (4) likely differences in staining conditions allowing the detection of endogenous galactoside activity in either WM or FS preparations, but not both. Nonspecific staining is particularly problematic in the GI tract and in the male sex organs.
Comparing LacZ staining between HOM and HET mutants
To determine whether reporter gene zygosity influenced the intensity and/or specificity of LacZ staining, HOM and HET mutants for the solute carrier family 7 (cationic amino acid transporter, y+ system), member 13 gene (Slc7a13), ninjurin 1 (Ninj1), and salt inducible kinase 1 (Sik1) were compared. These genes were chosen because of their known distinguishable gene-expression patterns. In general, LacZ staining in HOM mice was more intense and more structures/organs were stained compared with the patterns of staining in HET animals (Supplemental Fig. 4).
Examples of reporter staining for distributed structures: blood vessels, ducts, epithelium, muscle, and adipose, connective, and peripheral nervous system tissue
Blood vessels
WM preparations were particularly effective for identifying genes with extensive blood vessel staining in multiple tissues. A dramatic case study of blood vessel staining is presented for the WT1-interacting protein (Wtip) gene (Fig. 3A). We found extensive Wtip LacZ staining of blood vessels in many tissues, including brain, heart, adipose tissue, and muscle. FS revealed that all vessel types were positive for LacZ, including arteries, arterioles, veins, venules, and large caliber capillaries. A role for WTIP in vascular function has not been previously reported, although a role for the Wilms’ tumor protein (WT1) in vascular endothelial cells has been proposed (Scholz et al. 2009; Kirschner et al. 2010).
Figure 3.

(A) Wtip olfactory bulb, cerebral, and cerebellar blood vessels stain for LacZ in lateral brain whole mount (WM, left), and frozen section (FS, right) reveals staining of a branching cerebral blood vessel. In the Wtip brain, only blood vessels stain for LacZ. (B) Prr15l staining of lung in WM (left) and FS (right). In WM, the major airways clearly stain, but there is also a diffuse staining of the parenchymal tissue. In the FS image, the intense staining of the bronchiole epithelium (a), and the weaker punctate pattern of staining in the walls of the alveoli (b) are indicated. (C, left) Dram1 staining of mesenteric adipose tissue by WM showing diffuse uniform staining (a), while the ileum does not stain (b). (Right) FS of Dram1 mesenteric adipose tissue shows staining is restricted to the stromal, intercellular space (c). (D, left) Zfp12 WM staining of fundus (a), body (b), and antrum (c) of the stomach. WM staining of the pyloris and duodenum (d) and weak staining of the esophagus (e) are similar to that found in WT mice. (Right) FS indicates that the majority of Zfp12 LacZ staining is in the smooth muscle layer (f), although there is weak, scattered staining in the glandular epithelium (g). (E, left) Sec23a WM staining of cartilaginous ribs (a), intersternal plate cartilage (b), blood vessels (c), and a weak striated pattern of staining of the chest muscle (d). (Right) FS of striated Sec23a muscle indicates staining of connective tissue and/or blood vessels (e,f) between muscle fibers.
Epithelium and ducts
Many mutant reporter genes had epithelial staining, including duct structures, skin, mucosa of the gut, and the airway. WM and FS staining both were very effective at identifying duct staining. One example of a reporter with LacZ staining exclusive to epithelia is the proline-rich 15-like gene (Prr15l). Prr15l (previously known as Atad4) is a gene of no known function, and there are no published papers describing any aspect of functional biology. We found the LacZ staining in Prr15l is almost entirely of epithelial cells in tissues, including gall bladder and bile ducts in the liver; mucosa of the entire GI tract from the stomach through the cecum; mucosal epithelia of the trachea; bronchiole epithelia (Fig. 3B); lymphatic ducts in the mesenteric adipose tissue; and ducts in the pancreas, submandibular, sublingual and parotid glands, and extensive staining of the kidney, primarily of tubules in both the renal cortex and the medulla.
Adipose tissue
WM staining of brown and white adipose tissue (BAT and WAT, respectively) demonstrated limited sensitivity for detecting the presence of LacZ due to dark pigmentation in BAT and difficulty in stain penetration through the lipid droplets in both BAT and WAT. WAT LacZ staining for the DNA-damage regulated autophagy modulator 1 (Dram1) gene provides an example of adipose tissue staining (Fig. 3C). WM staining revealed that all adipose depots were stained in Dram1, but no substructures were identifiable. FS revealed staining in both WAT and BAT tissues and that it was the intercellular stroma that was stained in adipose tissue; stroma also stained in several other glands and tissues. No published findings suggest a role for DRAM1 in stromal function of adipose tissue.
Skeletal and smooth muscle
WM and FS were equally sensitive for detecting LacZ staining in smooth and skeletal muscle, with WM able to distinguish blood vessel, nerve, and connective tissue staining within the muscle. In addition, FS identified many substructures within muscle, including blood vessels, muscle fibers, extracellular matrix, interfascial adipose, and possible myoblast and nerve cells. Generally, the reporters expressed in skeletal muscle often were coexpressed in connective tissue, smooth muscle, and heart muscle and were expressed in other organs/tissues such as the brain. The zinc finger protein 12 gene (Zfp12) codes for a transcription factor not yet associated with regulation of any specific set of genes. We found intense LacZ staining in the brain, but also staining in smooth muscles of blood vessels and the GI tract (Fig. 3D) and striated muscle.
Connective tissue and cartilage
WM was particularly sensitive for detecting LacZ staining in connective tissue in the trachea, the chest wall, and the joints. However, FS was often necessary to specifically identify if cartilage expressed the reporter gene. An example is SEC23A (Saccharomyces cerevisiae; Sec23a), known to be required for normal collagen production and secretion (Townley et al. 2008) and important for craniofacial development (Lang et al. 2006). Transillumination of the chest wall in the LacZ-stained Sec23a mutant revealed a pattern of muscle, vasculature, and cartilage staining (Fig. 3E). With this reporter gene we found extensive staining in multiple tissues, including connective tissue and cartilage throughout the body, brain, smooth and striated muscle, the kidney, liver, and blood vessels.
Case studies of organs and organ systems
Brain
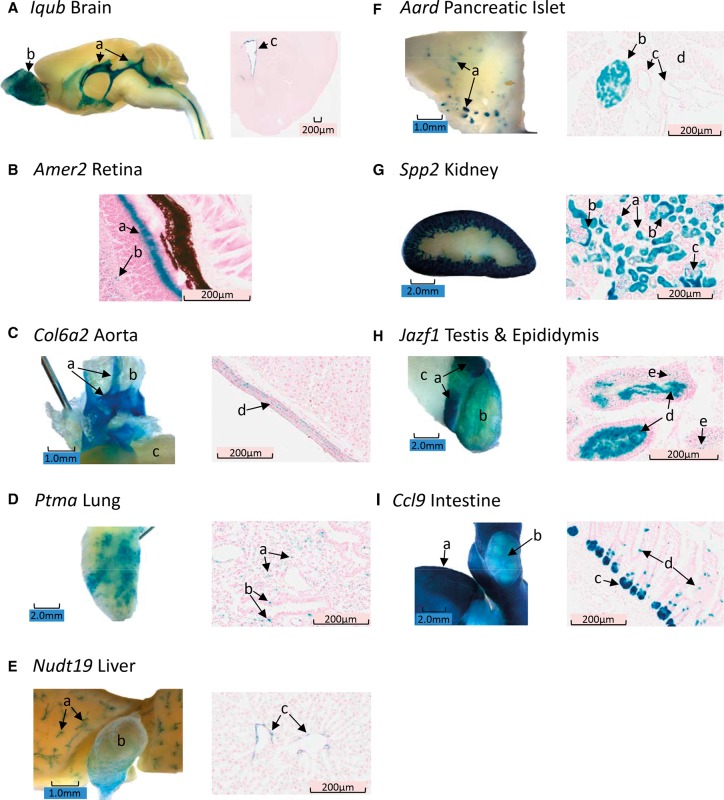
The IQ motif and ubiquitin domain containing gene (Iqub) is a putative ciliome protein gene and has recently been shown to be involved in cilia elongation using siRNA knockdown in cell culture (Lai et al. 2011). There are no other published reports describing a function for this gene. Available gene expression databases suggest this gene is expressed either at very low levels or with no expression in any mouse organ. LacZ staining in the CNS included the lateral, third, and fourth ventricle ependyma and the choroid plexus (Fig. 4A). The only nonventricular brain structure staining for LacZ was clusters of glomeruli in the olfactory bulb. LacZ identified Iqub reporter staining of epithelia in a number of other tissues, including the spinal canal, the trachea and lung bronchioles, the oviduct, and the ureter. These results reinforce the findings of Lai et al. (2011) that IQUB may be involved in cilia formation in a variety of structures and may be particularly important in cilia function in the ependyma of the CNS ventricular system.
Figure 4.
(A) Iqub whole mount (WM, left) mid-sagittal section showing staining of ventricular ependyma throughout the brain including the lateral and fourth ventricles (a), as well as staining of the glomeruli in the olfactory bulb (b), and coronal frozen section (FS, right) staining of forebrain illustrating ependymal staining of the lateral ventricle (c). (B) Amer2 FS retina with intense staining of the photoreceptor cell layer (a) and diffuse scattered staining of the inner nuclear layer (b). (C, left) Col6a2 aorta staining in WM showing distinct staining of the aortic arch and the carotid arteries (a) with no staining of white adipose tissue (b) or the left ventricle (c). (Right) FS of the aorta shows smooth muscle only staining for LacZ (d). (D, left) Ptma WM lung staining showing a variegated staining pattern. (Right) This uneven, variegated pattern of staining is also found in FS with individual cells staining in the parenchyma (a), as well as nonciliated cells in the bronchial epithelium (b). (E, left) Nudt19 liver staining is shown for WM, indicating that the bile ducts (a) and gall bladder (b) stain for LacZ. (Right) FS staining shows that epithelial cells in the ducts are expressing the lacZ reporter. (F, left) Aard pancreas staining by the WM method shows a distributed punctate staining pattern suggestive of islets. (Right) Aard FS confirms the inference of islet staining from the WM studies and further shows that a subset of cells stains in the islet (b), while ducts (c) and exocrine tissue (d) do not stain in Aard. (G, left) Spp2 kidney staining by WM shows intense staining of the tubules in the cortex only, with no staining in the medulla or hilus. (Right) Spp2 FS shows renal cortical staining in tubules (a), Bowman's capsule (b), and some cells/membranes in the interior of Bowman's capsule (c). (H, left) Jazf1 WM testis staining shows the characteristic WT nonspecific pattern of staining in the epididymis (a) with a 2-mm-wide band of minimal staining separating caput LacZ and additional LacZ staining in the body of the epididymis. Although faint, there is above-background WM staining in the seminiferous tubules (b) within the intact tunica, while the associated adipose tissue (c) does not stain. (Right) FS staining of Jazf1 shows intense staining of seminiferous tubule lumen (d), suggesting that immature sperm express the lacZ reporter. Some, but not all spermatogonia also appear to stain in the FS (e). (I, left) Ccl9 intestinal staining by WM shows a uniform punctate pattern of staining in the wall of the small intestine (a). The vasculature is clearly demarked on the surface of the intestine since it does not stain. Peyer's patches (b) also do not stain, except for a small band of staining encircling each dome in the patch (b). (Right) FS staining of Ccl9 shows staining primarily in the Crypts of Lieberkuhn (c), but there are some cells in the microvilli also staining for LacZ (d).
We found brain staining exclusively or predominantly in the ventricular ependyma in a subset of our reporter mice, including alanine- and arginine-rich domain containing protein (Aard); coiled-coil domain containing 141 (Ccdc141); myosin XVIIIA (Myo18a); prolyl 4-hydroxylase, beta polypeptide (P4hb); solute carrier family 33 (acetyl-CoA transporter) member 1 (Slc33a1); pannexin 1 (Panx1); ubiquitin-conjugating enzyme E2E 2 (Ube2e2); and transmembrane protein 70 (Tmem70). Many of these same mutants also had staining of peripheral tissues, most notably in the kidney. In 10 mutants with brain ventricular ependymal and/or choroid plexus LacZ staining in our Atlas, eight also had marked and distinct LacZ staining in the kidney. This is not surprising since the choroid plexus and the ependyma of the brain share similar functions with the kidney, i.e., the filtering of blood to produce cerebrospinal fluid and urine, respectively (Lowery and Sive 2009; Wolburg and Paulus 2010).
Eye
Our WM eye preparation stained the intact eye and there was inadequate penetration of stain to internal eye structures. In contrast, FS revealed significant and detailed staining of the retina, and sometimes captured staining in associated eye structures such as the cornea, other internal eye structures, attached eye muscles, cartilage, adipose, vessels, and lacrimal glands. A typical example is provided by the LacZ staining for APC membrane recruitment 2 (Amer2; also known as Fam123a), as shown in Figure 4B. Very little is known about Amer2 gene family function, although it has been shown that AMER2 interacts with microtubular proteins (Siesser et al. 2012).
Heart ventricle, atrium, and aorta
WM preparations of the heart were useful for identifying those reporters with LacZ staining in the coronary arteries, aorta, and cardiac muscle. FS confirmed this localization and was very useful for distinguishing myocytes from other structures staining in large blood vessels. FS of the aorta could distinguish staining of the smooth muscle, intima, and supporting structures. For example, collagen, type VI, alpha 2 (Col6a2) is broadly expressed (BioGPS), and we also found extensive LacZ staining in the major blood vessels in the tunica media (Fig. 4C), but not in the ventricle or atrium, and staining in smooth muscle throughout the body. Allelic variants in Col6a have been implicated in aortic dissections in a Korean population (Suh et al. 2011), and overexpression may contribute to congenital heart defects (Grossman et al. 2011), confirming an important role for COL6A in maintaining the structural integrity of the major blood vessels.
Lung
WM of the lung was particularly useful for identifying LacZ staining in the bronchial tree when there was no staining of the alveoli and other lung cells/structures. For WM bronchial staining, subsequent FS studies routinely identified substructure staining within the airways (mucosa, epithelium, smooth muscle, cartilage, etc.) and also distinguished alveolar, parenchymal, and blood vessel staining. The prothymosin alpha gene (Ptma) codes for a precursor of thymosin, a hormone produced in the thymus and important for maintaining immune function, and is also expressed in many other tissues. LacZ staining was found in a specific punctate pattern in WM lung (Fig. 4D), which appeared to be initially due to incomplete stain penetration. However, FS also revealed a distributed punctate distribution pattern with staining of scattered cells in the interstitium of the alveoli. This general pattern of punctate, uneven distributed staining for Ptma was observed in brain, testis, smooth muscle of the GI tract, pancreatic acinar tissue, and several glands, suggesting a complicated spatial induction of gene expression, possibly related to its function, in many tissues.
Liver
Generally, WM staining of liver revealed a diffuse pattern of hepatocyte staining, but also specific structures such as blood vessels and bile ducts were specifically stained, and intralobular organization of gene expression could be determined in some WM preparations. FS helped localize staining to hepatocytes or supporting cells or determined if the staining was in ductal or blood vessel cells. The nudix (nucleoside diphosphate linked moiety x)-type motif 19 gene (Nudt19) provides an example of liver staining. Nudt19 is a gene of unknown function that is highly expressed in the kidney and is also expressed in the liver and GI tract (BioGPS). In the pancreas and liver (Fig. 4E), Nudt19 LacZ staining was exclusively in the ducts. We also found very intense Nudt19 LacZ staining throughout the kidney, and in brain, the GI tract, and glands. This suggests that NUDT19 plays a role either in maintaining duct function or in regulating the composition of both bile and pancreatic secretions.
Pancreas
WM staining was particularly useful for identifying multifocal patterns of staining in the whole pancreas, and many of these WM multifocal patterns were confirmed to be islets by FS. The pattern of FS staining of islets often suggested that specific islet cell subtypes stained for LacZ, although we have not followed up these observations with islet cell-type–specific immunohistochemical staining. An excellent example of islet staining is for the alanine- and arginine-rich domain containing protein gene (Aard). Figure 4F shows the punctate, multifocal staining in pancreas in Aard WM preparations and the confirmation of islet LacZ staining by FS. There was interesting punctate staining in smooth muscle in multiple organs, staining in the cerebral ventricles and brain, and extensive LacZ staining in the testis for Aard. Published reports and expression surveys do not suggest a role for this gene in islet function.
Kidney
Both WM and FS revealed significant substructure in the cortex, medulla, and hilus of the kidney. For example, secreted phosphoprotein 2 (Spp2) is a lipid modulatory gene involved in signaling of S1p and is reportedly important in the developing kidney (Kirby et al. 2009). Although it is known to be expressed at high levels in the adult mouse kidney (BioGPS, GNF1M) and in the human heart and kidney (Ogawa et al. 2003), its function and anatomical localization in the kidney have not been previously reported. We show here (Fig. 4G) that LacZ staining for the Spp2 reporter is localized to the renal cortex and specifically to the convoluted proximal tubule and Bowman's capsule of the kidney. The functional significance of this localization remains to be explored.
Testis
Approximately 40% of the reporters expressed in the testis, and this was often staining localized to the seminiferous tubules. Testis nonspecific staining also occurred at a rate of ∼7% by WM, but this staining was diffuse and weak and could be distinguished from specific staining confirmed by FS. WM staining of the internal structures of the testis was limited prior to our routinely excising the tunica and extruding some of the seminiferous tubules in order to expose the internal testicular structures to the staining solution. With WM staining, we could conclude if the seminiferous tubules stained but no substructures were identifiable. FS revealed a wealth of structure, including interstitial cells, Leydig cells, spermatogonia, and spermatocytes. Testis was a unique structure in our LacZ screen since ∼8% of reporters only stained in the testis, and this was the highest percentage of unique staining for any organ except for the brain. These testis-specific genes were usually observed in spermatogonia, spermatocytes, or spermatids. For example, in the JAZF zinc finger 1 (Jazf1) mutant, LacZ staining was only observed in the testis (Fig. 4H), and staining was primarily in spermatogonia and spermatids. Jazf1 is reported to be expressed at only very low levels in the BioGPS databases, and a role in endometrial cancer has been suggested (Oliva et al. 2007), but a role in normal spermatogenesis or testis function has not been previously reported.
Gastrointestinal
WM staining of the GI tract was somewhat confounded by staining due to intraluminal bacteria galactosidase activity. However, this problem was greater in the colon and cecum and less problematic in the rest of the GI tract. For the tongue, WM staining was particularly useful for identifying squamous epithelial staining, and when the tongue was transected, it detected muscle, vascular, and glandular staining. The WM esophagus was used as an index of smooth muscle staining and correlated well with staining of smooth muscle in the rest of the GI tract identified by either the WM or FS method. WM stomach preparations could distinguish between staining of the antrum and pylorus, identify staining to subregions of the stomach (e.g., the transition zone between antrum and pylorus), and could also visualize major blood vessels. For the small intestine, a variety of substructures were identified by WM staining, including blood vessels, Peyer's patches (either staining when the rest of the intestine did not stain or vice versa), and punctate isolated staining. Overall, the sensitivity for WM staining to identify LacZ-positive structures in the GI tract was much less than that of FS. FS revealed a wealth of substructure staining, including mucosa and serosa, smooth muscle, crypts, glands, vasculature, Peyer's patches, and epithelial cells of the villi. An example of GI staining is presented for chemokine (C-C motif) ligand 9 (Ccl9). In our Ccl9 reporter, LacZ staining was exclusively in the GI tract, including glands in the tongue; in cells in the crypts of Lieberkuhn of the duodenum, jejunum, and ileum; and in the glandular epithelium of the colon (Fig. 4I). There was faint staining of the Ccl9 reporter in the dome of Peyer's patches. A role for CCL9 in GI function has not been described previously.
Discussion
Although reporter genes have been used in vivo for decades, there are no other published reports of a large survey comparing two different staining strategies, characterizing nonspecific staining, and analyzing the robustness and repeatability of the methods using hundreds of unique mutant mouse lines. This study reveals the utility of using two staining modalities to characterize substructure gene expression with LacZ staining. Furthermore, these results confirm the feasibility of using a lacZ reporter to reveal expression of a large number of poorly annotated genes in order to inform function in adult mice.
Overall, the reproducibility of WM and FS methods in a test-retest analysis was very good (>90% concordance), with WM having better reproducibility than FS. The reproducibility differences for specific staining by WM and FS are explained by a variety of factors, including the increased likelihood of missing staining due to the location of the captured section for FS and the greater technical difficulty of the FS staining process and therefore the potential for greater variability in staining results. In some cases, only one of four FS samples had specific staining in a tissue (see Supplemental Table 2), but this was often explained by weak staining of the reporter in that tissue and the possibility of biological variability in reporter gene expression. The data suggest that false-positive staining is relatively rare, but we certainly may miss staining in some tissues. Staining was highly reproducible in some tissues but not in others. For example, in the brain, reproducibility was very high for both WM and FS preparations because of the lack of any confounding pigmentation, and the depth of stain penetration by WM allowed us to detect a small number of stained cells and enhance sensitivity. Over the course of this study, we realized that some WM preparations were not adequately exposing specific structures to the stain, and WM scoring underestimated the frequency of staining in these structures. For example, there was poor penetration of the stain into intact eyes, lymph nodes that were surrounded by adipose tissue, adrenal glands, and some sex organs and glands in WM preparations (see Fig. 2). Modifications to our processes to better expose these structures to stain will improve results in future studies. For interpreting these results, one should compare and contrast the staining patterns in both the WM and FS preparations in the same mutant and also be mindful that this is a high-throughput screen to provoke hypothesis and it is not intended to be definitive.
Some large organs and structures gave remarkable anatomical detail and staining information with WM preparations not achievable by FS. For example, the mid-sagittal brain staining provided a gross overview of midline brain gene expression. Also, WM staining of the thoracic cage enabled an assessment of muscle, cartilage, bone, and vasculature in a single trans-illuminated sample not achievable by FS. Overall, combining both WM and FS analysis in the same mutants was very productive, with WM providing an excellent overview of organ and large substructure gene expression and FS localizing staining to substructures and specific cell types.
Cataloging of WT mouse nonspecific staining (see Fig. 2; Supplemental Fig. 3A,B) was important for distinguishing specific reporter-gene LacZ enzyme activity from background staining due to the presence of bacteria or endogenous galactosidase activity. The results indicate the need for FS, especially in the GI tract and male reproductive tract, to conclusively discriminate between specific and nonspecific expression. FS provides enough spatial resolution so that specific staining can be distinguished from nonspecific endogenous glycoside staining. Any studies utilizing LacZ staining should include WT controls since the degree of nonspecific staining will be highly dependent upon the methods used in each laboratory and the organs and tissues analyzed.
In the three cases we examined, HOM staining was more intense, and more unique structures were stained in HOM compared with HET animals. This could be explained by the additional copy of the reporter gene and by up-regulation of gene expression due to the complete absence of a protein product in the HOM mutants. In addition, the staining may not be linearly related to LacZ protein abundance, and there may be threshold levels for detection since the LacZ enzyme forms a tetramer and is more active in the tetrameric crystalline form (Juers et al. 2012) requiring complex kinetics for aggregation. However, using HOM reporter-gene mutants to characterize “normal” gene expression patterns could be problematic since aberrant gene regulation in a complete loss-of-function mutant might result in unusual gene expression patterns for the reporter, or the complete loss of reporter activity in specific cell types or tissues that develop poorly or do not develop as a result of the mutation.
Two tissues with particularly interesting LacZ staining patterns, testis and brain, deserve further discussion. The high percentage of reporter genes expressed in the testis likely is a consequence of several factors. Epigenetic reprogramming and extensive genome-wide demethylation occur in the primordial germ cells (Surani and Hajkova 2010) and may result in the expression of many genes as part of a proofreading routine during meiosis and spermatogenesis. In addition, the testis is a very complicated structure with duct cells, germ cells, juvenile sperm, and support cells for the spermatogonia and endocrine cells, and the high percentage of genes expressed in testis may partially reflect the complexity of the organ. Our finding of a high percentage of genes expressed in the testis is consistent with the report by Wu et al. (2004). The LacZ screen may contribute to identifying those genes involved in stem-cell maintenance and unique to the development of germ plasm since ∼8% of all genes surveyed were exclusively expressed in the testis. In contrast with the testis, there were no genes in our survey with LacZ staining unique to ovaries for either the WM or FS method (Fig. 1). Since the first stage of meiosis occurs in developing oocytes in the embryo, and only a few primary oocytes fully differentiate during each estrous cycle (Pinkerton et al. 1961), it is likely that we did not capture expression of these genes unique to oocyte development since many of the ovaries were not in estrus when sampled.
In the CNS, more mutants stained for LacZ than in any other tissue and only a small number of these genes were unique to the CNS. The data suggest that a high frequency of gene expression in the brain reflects pleiotropy. One possible example of pleiotropy is the set of genes coexpressed in the brain ventricular system and in the kidney presented in the results above, which likely have similar but also distinctly different functions in these two organs.
To our knowledge, there are only two other large reporter gene surveys in adult mouse using LacZ staining. Deltagen completed a large-scale, mouse-targeted mutation production and phenotyping effort in the late 1990s (Moore 2005). A lacZ reporter was incorporated into the targeting vector and staining was assessed by FS in a survey of adult tissues. Examples of their lacZ reporter gene survey are available at http://www.nih.gov/science/models/mouse/deltagenlexicon/. Also, at the Mouse Genetics Project at the Sanger Institute (http://www.sanger.ac.uk/mouseportal/), ∼500 mutant lines were produced from IKMC stem cells, and their comprehensive phenotyping data, including adult LacZ by WM staining in 336 mutants, are available (White et al. 2013). Cursory inspection of the LacZ staining data in these two surveys shows the results to be similar to that in the current study.
Perhaps the most important result from this study is the anatomical localization of gene expression provided by combining the WM and FS methods. A selected subset is described in the results here, but many more examples are reported in the online atlas at www.kompphenotype.org. In many cases, LacZ staining revealed gene expression in substructures and tissues not reported in the BioGPS and GEO expression databases. This was often the case for genes expressed in distributed structures including blood vessels, peripheral nerves, and connective and epithelial tissues for which the gene expression databases often reported low ubiquitous staining.
Methods
Gene list
The list of 313 genes surveyed is presented in Supplemental Methods.
Mouse production
Mutants were created primarily with Knockout Mouse Project (KOMP) targeted ES cells derived from the C57BL/6N mouse strain. See Skarnes et al. (2011) for a description of the targeting strategy and cell lines produced, and visit the International Knockout Mouse Consortium (IKMC; www.mousephenotype.org) or the KOMP Repository (www.komp.org) and the project webpage (www.kompphenotype.org) for a description of the specific alleles for each of the mutants with LacZ staining in this survey. For all alleles, the lacZ reporter was designed to be under the control of the native promoter of the targeted gene.
Chimeras were backcrossed to C57BL/6NTac or C57BL/6NCrl inbred mouse strains and the resulting founding heterozygous mutants were expanded by additional backcrosses. Targeting was confirmed by long-range PCR (Skarnes et al. 2011) and zygosity for the targeted allele was confirmed by a qPCR assay specific for the lacZ sequence. Mutants and WT littermates were housed in an environmentally controlled animal facility on a 12:12-h light:dark cycle with lights on at 07:00 h. Mice were fed Harlan Teklad global rodent diet #2918 with a composition of 18% protein and 6% fat. Food and water were available ad libitum. Animal care and use procedures followed the guidelines of the Institute of Laboratory Animal Research Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the University of California, Davis.
LacZ staining and annotation
For each mutant line, our goal was to characterize two adult HET mice, one male and one female, by both WM and FS LacZ staining. WT littermate controls also were processed to identify patterns of nonspecific staining due to endogenous galactoside and resident bacterial enzyme activity. Approximately 50-day-old nonfasted animals were anesthetized with isoflurane and perfused using saline followed by ice-cold 4% paraformaldehyde (Invitrogen). Tissues and organs were dissected and post-fixed in 4% paraformaldehyde for 30 min. For details of the FS and WM staining, see Supplemental Methods.
Staining and annotation procedures were adapted from Adams and Gale (2006) and modified to improve sensitivity of staining. Briefly, some of the WM tissues were completely bisected for better penetration of stain (e.g., liver, spleen, adrenal gland), and the testicular tunica was incised to better expose the seminiferous tubules to the staining solution. The brain was removed from skull and cut mid-sagittally into two halves using a brain mold. One brain half was saved for mid-sagittal WM staining, and the other half was cut into 2-mm-thick coronal sections prior to WM staining. For preparing adipose tissue and brain for frozen sectioning, Neg50 OCT was used for adipose and tissue freezing medium for brain. Whole blood was smeared onto glass slides, briefly fixed in 95% ethanol, and then processed with the other frozen sections. The pH of the PBS was adjusted to 7.7 and used for both washes and to make the LacZ staining solution. Nuclear fast red solution was used to counter-stain FS after LacZ staining. As positive controls for FS staining, we kept a bank of frozen OCT reference blocks containing HET mutant tissues with staining of known intensity. These reference blocks were sectioned and stained in parallel with new mutants in order to validate that the staining process remained uniform.
LacZ staining was annotated in 66 different tissues and organs (see Supplemental Methods). Anatomical terms and substructure nomenclature generally conform to the Anatomical Dictionary for the Adult Mouse (http://www.informatics.jax.org/searches/AMA_form.shtml). Tissues were scored as nonspecific if the pattern of staining was judged to be the same as that in WT controls (see Results) due to either resident bacteria or endogenous glycoside activity.
To compare the pattern and intensity of LacZ staining between HOM and HET mice, two HOM and two HET mice were processed for Slc7a13, Ninj1, and Sik1 mutants. The tissues were collected as the mice became available and were stained with the same methods used for the HET mutants.
Statistical analysis
To evaluate the repeatability of staining, we compared the tissues scored as positive for LacZ between one male and one female mouse, within each strain and for each staining method (303 and 298 mutant lines had replicates for WM and FS staining, respectively). We also compared the consensus staining for each mutant between the WM and FS staining methods. If there were more than one animal of each sex characterized for a specific gene, then a representative animal for each sex was chosen. Correlations between staining modalities were calculated using a Pearson product-moment correlation.
Comparison of cases with other expression databases
We routinely compared the LacZ staining tissue distribution patterns against two databases in BioGPS (www.biogps.org) (Wu et al. 2009). These two databases were the GNF1M and the MOE430 (Lattin et al. 2008) arrays for mouse tissues. In addition, we routinely checked the GEO database, specifically the GSE1986 mouse tissue survey results. In the text, our conclusions regarding the expression of specific genes in specific tissues is based upon our interpretation of the preponderance of the evidence from these expression surveys, since different expression surveys did not always agree.
Data access
LacZ staining data for 313 mutant lines are available on the project webpage (https://www.kompphenotype.org). These data are presented as a heatmap organized alphabetically by gene ID, and the main webpage indicates if there was positive reporter gene staining for each mutant for either the FS or WM staining modality. LacZ results for all of the mutants can be viewed by organ system and by specific organs. The LacZ summary for each mutant indicates the presence of staining for each tissue and delivers representative WM and FS images with positive, specific LacZ staining for each mutant. Metadata include targeted clone used for mouse production, age at LacZ staining, genotype, sex, and animal identification number.
Supplementary Material
Acknowledgments
We thank Nick Gale and Niels Adams for assistance in setting up the LacZ staining and annotation protocols in our laboratory and the Mouse Biology Program staff for the production and husbandry of the mutant mice. This work was supported by NIH grants U42OD011175, U54HG006364, and U01HG004080.
Author contributions: D.B.W. and R.K.P. analyzed the data and wrote the manuscript. B.B., E.D., and A.T. performed LacZ staining and annotation and provided valuable insights and innovations in LacZ processing. R.K.P. completed detailed brain annotation. S.M.G. provided histology review and annotation training. E.K.E., J.R., and B.L. programmed the database management system and constructed the webpage. K.C.K.L., P.J.D.J., and D.B.W. conceived the research plan, supervised the work, and edited the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.184184.114.
References
- Adams AC, Gale NW. 2006. High resolution gene expression analysis in mice using genetically inserted reporter genes. In Mammalian and avian transgenesis: new approaches (principles and practice) (ed. Pease S, Lois C), pp. 131–161 Springer-Verlag, Berlin. [Google Scholar]
- Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, Bottomley J, Brown SD, Bürger A, Bult CJ, et al. 2012. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome 23: 580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Moore MW. 2012. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm Genome 23: 632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban MJ, Chou J, Cohen SN. 1980. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol 143: 971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates H, Mallon AM, Brown SD; EUMODIC Consortium. 2011. High-throughput mouse phenotyping. Methods 53: 394–404. [DOI] [PubMed] [Google Scholar]
- Geffers L, Herrmann B, Eichele G. 2012. Web-based digital gene expression atlases for the mouse. Mamm Genome 23: 525–538. [DOI] [PubMed] [Google Scholar]
- Grossman TR, Gamliel A, Wessells RJ, Taghli-Lamallem O, Jepsen K, Ocorr K, Korenberg JR, Peterson KL, Rosenfeld MG, Bodmer R, et al. 2011. Over-expression of DSCAM and COL6A2 cooperatively generates congenital heart defects. PLoS Genet 7: e1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juers DH, Matthews BW, Huber RE. 2012. LacZ β-galactosidase: structure and function of an enzyme of historical and molecular biological importance. Protein Sci 21: 1792–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby RJ, Jin Y, Fu J, Cubillos J, Swertfeger D, Arend LJ. 2009. Dynamic regulation of sphingosine-1-phosphate homeostasis during development of mouse metanephric kidney. Am J Physiol Renal Physiol 296: F634–F641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner KM, Sciesielski LK, Scholz H. 2010. Wilms’ tumour protein Wt1 stimulates transcription of the gene encoding vascular endothelial cadherin. Pflugers Arch 460: 1051–1061. [DOI] [PubMed] [Google Scholar]
- Lai CK, Gupta N, Wen X, Rangell L, Chih B, Peterson AS, Bazan JF, Li L, Scales SJ. 2011. Functional characterization of putative cilia genes by high-content analysis. Mol Biol Cell 22: 1104–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang MR, Lapierre LA, Frotscher M, Goldenring JR, Knapik EW. 2006. Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nat Genet 38: 1198–1203. [DOI] [PubMed] [Google Scholar]
- Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, et al. 2008. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Sive H. 2009. Totally tubular: the mystery behind function and origin of the brain ventricular system. Bioessays 31: 446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MW. 2005. High-throughput gene knockouts and phenotyping in mice. Ernst Schering Res Found Workshop 50: 27–44. [DOI] [PubMed] [Google Scholar]
- Morgan H, Beck T, Blake A, Gates H, Adams N, Debouzy G, Leblanc S, Lengger C, Maier H, Melvin D, et al. 2010. EuroPhenome: a repository for high-throughput mouse phenotyping data. Nucleic Acids Res 38 (Database issue):D577–D585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa C, Kihara A, Gokoh M, Igarashi Y. 2003. Identification and characterization of a novel human sphingosine-1-phosphate phosphohydrolase, hSPP2. J Biol Chem 278: 1268–1272. [DOI] [PubMed] [Google Scholar]
- Oliva E, de Leval L, Soslow RA, Herens C. 2007. High frequency of JAZF1-JJAZ1 gene fusion in endometrial stromal tumors with smooth muscle differentiation by interphase FISH detection. Am J Surg Pathol 31: 1277–1284. [DOI] [PubMed] [Google Scholar]
- Pinkerton JHM, McKay DG, Adams EC, Hertig AT. 1961. Development of the human ovary: a study using histochemical techniques. Obstet Gynecol 18: 152–181. [PubMed] [Google Scholar]
- Scholz H, Wagner KD, Wagner N. 2009. Role of the Wilms’ tumour transcription factor, Wt1, in blood vessel formation. Pflugers Arch 458: 315–323. [DOI] [PubMed] [Google Scholar]
- Siesser PF, Motolese M, Walker MP, Goldfarb D, Gewain K, Yan F, Kulikauskas RM, Chien AJ, Wordeman L, Major MB. 2012. FAM123A binds to microtubules and inhibits the guanine nucleotide exchange factor ARHGEF2 to decrease actomyosin contractility. Sci Signal 5: ra64 10.1126/scisignal.2002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh JH, Yoon JS, Kwon JB, Kim HW, Wang YP. 2011. Identification of genomic aberrations by array comparative genomic hybridization in patients with aortic dissections. Korean J Thorac Cardiovasc Surg 44: 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Hajkova P. 2010. Epigenetic reprogramming of mouse germ cells toward totipotency. Cold Spring Harb Symp Quant Biol 75: 211–218. [DOI] [PubMed] [Google Scholar]
- Townley AK, Feng Y, Schmidt K, Carter DA, Porter R, Verkade P, Stephens DJ. 2008. Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J Cell Sci 121(Pt 18):3025–3034. [DOI] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, et al. 2013. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154: 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolburg H, Paulus W. 2010. Choroid plexus: biology and pathology. Acta Neuropathol 119: 75–88. [DOI] [PubMed] [Google Scholar]
- Wu SM, Baxendale V, Chen Y, Pang AL, Stitely T, Munson PJ, Leung MY, Ravindranath N, Dym M, Rennert OM, et al. 2004. Analysis of mouse germ-cell transcriptome at different stages of spermatogenesis by SAGE: biological significance. Genomics 84: 971–981. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge SL, Haase J, Huss JW, Su AI. 2009. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.