Abstract
Non-coding variation within TCF7L2 remains the strongest genetic determinant of type 2 diabetes risk in humans. A considerable effort has been placed in understanding the functional roles of TCF7L2 in pancreatic beta cells, despite evidence of TCF7L2 expression in various peripheral tissues important in glucose homeostasis. Here, we use a humanized mouse model overexpressing Tcf7l2, resulting in glucose intolerance, to infer the contribution of Tcf7l2 overexpression in beta cells and in other tissues to the metabolic phenotypes displayed by these mice. Restoring Tcf7l2 expression specifically in beta cells to endogenous levels, in face of its overexpression elsewhere, results in impaired insulin secretion, reduced beta cell number and islet area, corroborating data obtained in humans showing similar phenotypes as a result of manipulations leading to Tcf7l2 loss of function. Interestingly, the persistent overexpression of Tcf7l2 in non-pancreatic tissues results in a significant worsening in glucose tolerance in vivo, indicating that Tcf7l2 overexpression in beta cells does not account for the glucose intolerance in the Tcf7l2 overexpression mouse model. Collectively, these data posit that Tcf7l2 plays key roles in glucose metabolism through actions beyond pancreatic beta cells, and further points to functionally opposing cell-type specific effects for Tcf7l2 on the maintenance of balanced glucose metabolism, thereby urging a careful examination of its role in non-pancreatic tissues as well as its composite metabolic effects across distinct tissues. Uncovering these roles may lead to new therapeutic targets for type 2 diabetes.
Introduction
Genetic variation within introns of TCF7L2 is strongly associated with increased risk of type 2 diabetes (T2D) (1–8). This non-coding region contains cis-regulatory elements that drive expression of TCF7L2 across a variety of tissues involved in glucose homeostasis, suggesting that the risk variants likely alter the expression of TCF7L2 (9–11). Notably, reports have shown that the most strongly T2D-associated single nucleotide polymorphism (SNP), rs7903146, resides within a region of open chromatin with demonstrated cis-regulatory activity in pancreatic beta cells, as well as other tissues (11–13). Furthermore, the T2D-risk (T) allele for this SNP drives stronger enhancer activity than the protective (C) allele (11–13). Together, these data suggest that the T2D-risk variant(s) leads to increased TCF7L2 expression, corroborating early observations that the T2D-risk allele of rs7903146 is associated with increased TCF7L2 mRNA expression in pancreas, even in non-diabetic individuals (14).
A link between increased expression of Tcf7l2 and glucose intolerance phenotypes has been further supported by mouse models. Mice homozygous for a Tcf7l2 null allele are born with dramatically low blood glucose levels and die perinatally partially due to hypoglycemia (9,15,16). Mice heterozygous for germline null Tcf7l2 alleles are viable and grow normally, mirroring the same pattern of reduced blood glucose and plasma insulin levels along with improved glucose tolerance and insulin sensitivity, even after a high fat diet regimen (9,15,16). Conversely, mice that overexpress Tcf7l2 display reciprocal phenotypes, including increased plasma insulin levels and glucose intolerance due to peripheral insulin resistance, indicating that overexpression of Tcf7l2 leads to a type 2 diabetic phenotype (9).
Despite these results, a significant body of evidence has accumulated over the past 7 years suggesting that genetic or molecular manipulations leading to Tcf7l2 loss of function in pancreatic beta cells lead to diabetogenic phenotypes. In vitro studies on human and rodent islets have found that silencing of TCF7L2 leads to decreased proinsulin production and processing, decreased insulin secretion, decreased islet number, decreased beta cell proliferation and increased apoptosis (17–21). Complementary studies suggest increased Tcf7l2 expression fosters beta cell regeneration (20,22). Collectively, these data suggest that reduced Tcf7l2 expression leads to beta cell depletion and malfunction, hyperglycemia and T2D-like phenotypes. Further work has recently demonstrated that Tcf7l2 is a master regulator of insulin production and processing in both rodent and human beta cells (21). This study from Zhou et al. identified direct targets of TCF7L2 including ISL1 and the subsequent downstream targets that direct proinsulin production and processing; these molecular targets are perturbed upon TCF7L2 silencing. Mouse in vivo experiments also seem to support a role for Tcf7l2 as a regulator of insulin production and secretion. Recent work from Guy Rutter's laboratory has shown that a beta cell-specific knock-out of Tcf7l2 leads to decreased beta cell volume, decreased insulin secretion, and subsequent glucose intolerance in mice fed high fat diet (23).
Together, these studies have painted a difficult pathophysiological picture to interpret. While human genetics and genomics data suggest that increased T2D risk is associated with increased TCF7L2 expression, a notion supported by various genetically engineered mouse models, other studies focused on the beta cell roles of TCF7L2 have pointed in the opposite direction, suggesting that a TCF7L2 loss of function in beta cells may represent the causal link between genetic variation in the TCF7L2 locus and T2D risk. To clarify these conflicting functional results, we sought to determine the relative contribution of Tcf7l2 in beta cells to the T2D phenotypes seen in mice overexpressing Tcf7l2. Toward that, we engineered mice with Tcf7l2 expression restored to endogenous baseline levels in beta cells while maintaining overexpression in peripheral metabolic tissues where Tcf7l2 is normally expressed, including brain, liver, gut and fat.
Our data show that reducing Tcf7l2 expression from high to normal levels in adult mouse beta cells results in blunted insulin secretion, reduced plasma insulin levels and selective loss of pancreatic beta cells, corroborating data from in vitro studies. Importantly, our results demonstrate that in face of this restored pancreatic expression of Tcf7l2, overexpression in other tissues still maintains the glucose intolerance phenotypes, including further amplifying these metabolic perturbations, strongly suggestive of important roles for Tcf7l2 in regulation of glucose metabolism beyond its well-established roles in beta cells.
Results
Inducible excision of transgenic Tcf7l2 in beta cells
We utilized a mouse model of Tcf7l2 overexpression that we have previously described (9). These mice have been engineered to carry a human BAC corresponding to the TCF7L2 locus that further spans the 92 kb T2D-associated region. One copy of the mouse full-length cDNA was recombineered into the TCF7L2 translation start site within the BAC. Three copies of the BAC integrated randomly into the genome. Consequently, mice carrying these three BACs express transgenic mouse Tcf7l2 regulated by human cis-regulatory elements, in addition to the expression of the two endogenous copies of Tcf7l2 in the mouse genome. This results in a humanized model of overexpression, where Tcf7l2 is overexpressed at moderate levels in physiological tissues, determined by the endogenous regulatory landscape of the TCF7L2 human locus. This result in a 50–200% increase in the expression level of Tcf7l2 in transgenic mice compared with wild-type (9). Importantly, the Tcf7l2 cDNA recombineered in the human BAC is floxed, allowing for the conditional removal of the transgenic Tcf7l2 copies via Cre-recombination, and thereby restoring normal Tcf7l2 expression level in a tissue-specific manner. We favored this model of conditional gene excision over the routine conditional knock-out strategies because Tcf7l2 is a transcription factor with well-established developmental roles. Indeed, a conditional ablation of a transcription factor may lead to altered cell specification, proliferation, etc., which may confound the adult onset glucose metabolism phenotypes we are interested in.
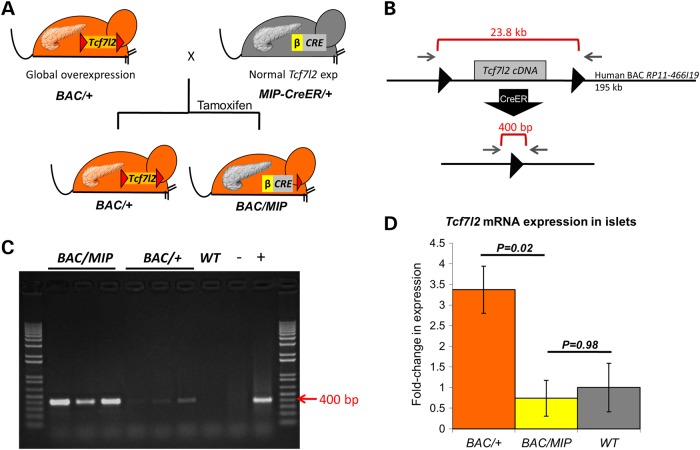
To selectively ablate Tcf7l2 overexpression in beta cells, we utilized an inducible beta cell-specific mouse insulin promoter-1 CreER (MIP-CreER) allele, previously described (24). Importantly, this CreER driver lacks hypothalamic Cre expression that other insulin promoter Cre have reported. We crossed the MIP-CreER mice to our Tcf7l2-BAC transgenic mice, creating F1 mice that are both BAC positive and MIP-CreER positive (BAC/MIP, Fig. 1A). Since the MIP-CreER is inducible, we injected BAC/MIP-CreER mice with tamoxifen to activate Cre, resulting in an adult-specific ablation of the Tcf7l2 transgenic copies in beta cells. Mice that do not inherit the MIP-CreER maintain global overexpression and are therefore referred to as BAC/+ henceforth (Fig. 1A).
Figure 1.
Beta cell-specific ablation of Tcf7l2 cDNA and restoration of Tcf7l2 expression. (A) Breeding scheme to create BAC/MIP mice. Hemizygous BAC/+ mice (orange) are shown carrying the Tcf7l2 cDNA in the BAC bordered by loxP sites (red triangles) and a pancreas with overexpression of Tcf7l2 (orange). Hemizygous MIP-Cre/+ mice (gray) are shown carrying the beta cell-specific MIP-Cre (gray) a pancreas with normal expression of Tcf7l2 (gray). Crossing these mice creates F1 offspring inheriting the BAC with no Cre (BAC/+) with continued overexpression of Tcf7l2, globally and including the pancreas (orange). This cross also produces mice that inherit both the BAC and the MIP-Cre, which upon activation by tamoxifen, cleaves at loxP sites to restore normal expression of Tcf7l2 in beta cells only (gray pancreas) while maintaining overexpression elsewhere (orange remainder of mouse). (B) Diagram of human BAC containing mouse Tcf7l2 cDNA floxed by loxP sites (black triangles). Gray arrows indicate PCR primers which can only amplify the 400 bp fragment after excision by Cre. (C) PCR amplification of 400 bp fragment in islets after excision by MIP-Cre. (D) Expression of Tcf7l2 in BAC/+, BAC/MIP and wild-type islets quantified by qRT-PCR.
To assay for the efficiency of CreER-mediated excision of the floxed 24 kb region of the BAC harboring the Tcf7l2 cDNA in BAC/MIP mice, we used primers outside the loxP sites to amplify the resulting 400 bp segment after CreER excision (Fig. 1B). As expected, PCR on islet DNA from BAC/MIP mice amplified the segment confirming an activated Cre and the subsequent removal of the floxed region containing the extra copies of Tcf7l2 cDNA (Fig. 1C).
We next assayed the impact of the excision of the transgenic Tcf7l2 copies in beta cells at the transcription level. We isolated mRNA from both BAC/+ and BAC/MIP islets and performed quantitative reverse transcription PCR (qRT-PCR) to determine whether removal of Tcf7l2 cDNA from the BAC subsequently reduced Tcf7l2 expression in BAC/MIP islets. We observed overexpression of Tcf7l2 in BAC/+ islets matching the expression levels seen in the pancreas of the founder global overexpression mouse (9). Reflecting the excision of the transgenic copies of Tcf7l2 in BAC/MIP mice, we detected a significant decrease in Tcf7l2 expression in BAC/MIP islets, which now express Tcf7l2 at the same level as wild-type islets (Fig. 1D). These results illustrate our ability to generate a mouse model where we can ablate Tcf7l2 overexpression specifically in beta cells of adult mice, while preserving overexpression in peripheral tissues.
Glucose intolerance in Tcf7l2 overexpression mice
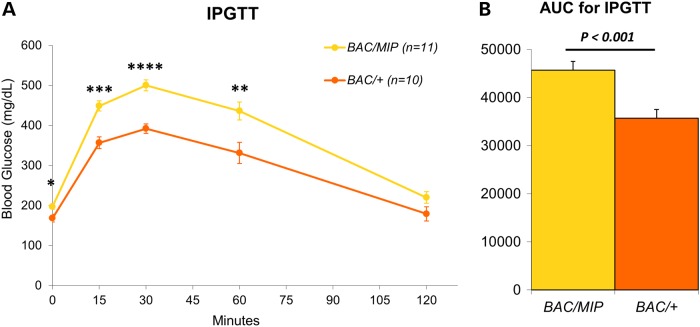
Tcf7l2 overexpression in BAC/+ mice results in increased insulin secretion and glucose intolerance after high fat diet (HFD) due to peripheral insulin resistance (9). Therefore, we assayed BAC/MIP mice for changes in glucose metabolism and insulin secretion to identify the specific contribution of Tcf7l2 overexpression in beta cell function.
We subjected both BAC/MIP and BAC/+ mice to HFD for 10 weeks. During this time, we tracked mouse weight gain and found no difference between BAC/MIP and BAC/+ mice. After HFD feeding, we performed an intraperitoneal glucose tolerance test (IPGTT). We observed that restoring wild-type levels of Tcf7l2 expression in the beta cells of BAC/MIP mice resulted in a significant worsening of glucose tolerance compared with BAC/+ littermates (Fig. 2A and B). We replicated this observation in a second cohort of mice, including BAC/+, BAC/MIP and wild-type littermates, where we injected half the amount of glucose during IPGTT. Wild-type mice displayed the strongest glucose tolerance among the three groups after 10 weeks of HFD. BAC/+ mice were glucose intolerant and BAC/MIP mice displayed the most severe glucose intolerance of the three groups, even with the attenuated glucose injection, confirming our earlier results (Supplementary Material, Fig. S1A and B). These results suggest that Tcf7l2 overexpression in beta cells of BAC/+ mice does not represent the main contributor to the glucose intolerance seen in these mice, stressing the role of extra-pancreatic tissues in the Tcf7l2-mediated glucose intolerance.
Figure 2.
Glucose tolerance of BAC/MIP mice. (A) IPGTT for BAC/+ (orange) and BAC/MIP (yellow) mice after 10 weeks high fat diet. Injected 2 g/kg dextrose. (B) AUC of the IPGTT plot from (A) with P values as shown. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
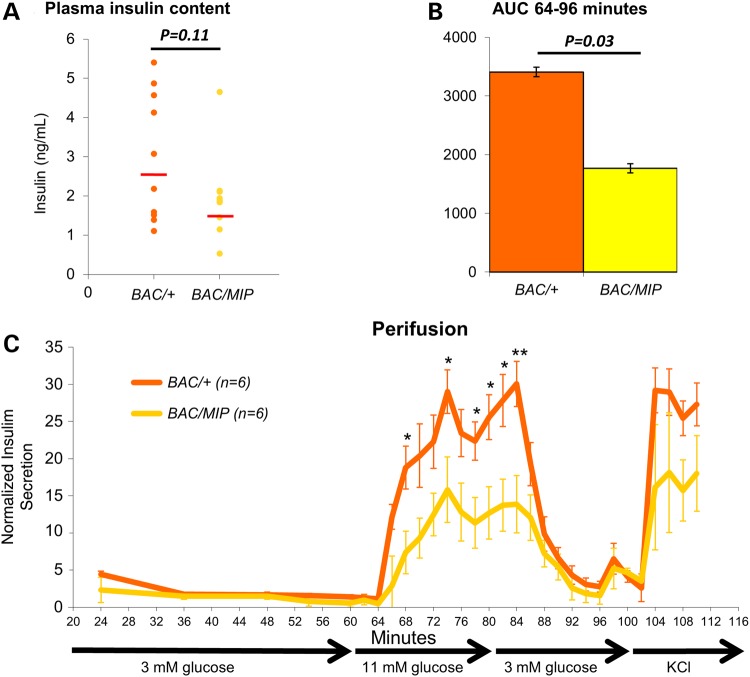
Next, we determined if the reduced Tcf7l2 expression in beta cells and the subsequent worsening of glucose intolerance are coupled with changes in plasma insulin content. Using an insulin enzyme-linked immunosorbent assay (ELISA), we observed BAC/MIP mice to have noticeably reduced circulating insulin compared with BAC/+ mice (Fig. 3A). This reduction of plasma insulin levels in BAC/MIP mice coincides with the worsening of glucose tolerance, suggesting this is the main mechanism by which glucose tolerance is worsened in BAC/MIP mice. This reduced plasma insulin content may further reflect impaired insulin secretion from islets. We subsequently assayed insulin secretion directly from BAC/+ and BAC/MIP islets in response to glucose stimulation using islet perifusion (25–27). When exposed to a concentrated glucose solution, BAC/MIP islets secrete significantly less insulin than BAC/+ islets (Fig. 3B and C), indicating impaired insulin secretion secondary to a reduction in Tcf7l2 expression in beta cells. Together, these results demonstrate a dose-dependent curve of insulin secretion related to Tcf7l2 expression levels, mimicking the blunted insulin secretion response described as a result of reduced Tcf7l2 expression in beta cells in humans and mice (17–21).
Figure 3.
Beta cell analysis of insulin content and secretion. (A) Fasting plasma insulin levels in BAC/+ (orange) and BAC/MIP (yellow) mice after 10 weeks high fat diet (HFD). (B) AUC of insulin secretion measured by ELISA during insulin spike from 64 to 96 min of perifusion. (C) Normalized insulin levels secreted by isolated islets during glucose perifusion. Minutes indicate time since initial incubation of islets in glucose. Islets were incubated in varying glucose concentrations; 3 mm glucose from 0 to 60 min, 11 mm glucose from 60 to 80 min, 3 mm glucose from 80 to 100 min, and final incubation in KCl from 100 to 110 min. Insulin samples were collected every 12 min from 24 to 60 min and then every 2 min for the remainder. Insulin secretion from islets was quantified by insulin ELISA. *P < 0.05; **P < 0.01.
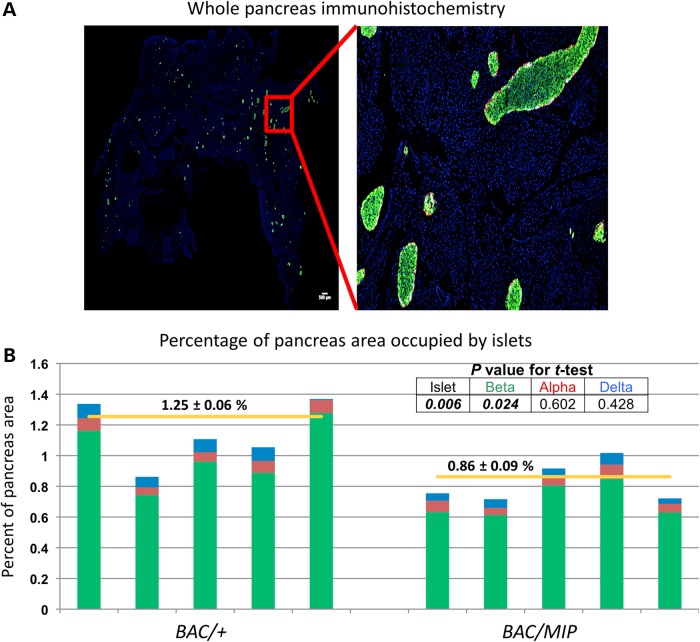
Reduced beta cell area in BAC/MIP mice
The blunted insulin secretion in BAC/MIP mice could be reflective of a decrease in beta cell number supported by recent data reporting a preferential loss of beta cells in patients with T2D (28). To assess this possibility, we utilized immunohistochemistry on whole pancreas from age- and weight-matched BAC/+ and BAC/MIP mice using antibodies against insulin, glucagon and somatostatin, as well as DAPI nuclear staining (Fig. 4A). Using a large-scale computer-assisted imaging method, we quantified endocrine cell mass and islet cellular composition. This study showed that a reduction of Tcf7l2 expression in BAC/MIP islets leads to a significant decrease in beta cell and total islet area (P < 0.006, two-sided Student's t-test) compared with BAC/+ overexpression mice (Fig. 4B). Importantly, we observed no decrease in alpha or delta cell area in BAC/MIP islets (P = 0.602 and 0.428, respectively, two-sided Student's t-test), indicating that our ablation of Tcf7l2 overexpression has a localized effect specifically in beta cells (Fig. 4B). These data further indicate that beta cells with less Tcf7l2 expression occupy less pancreatic area, supporting previous evidence that Tcf7l2 expression is important for beta cell survival and proliferation (17,18,20,22).
Figure 4.
Quantification of beta cell area. (A) Representative image of immunohistochemistry of whole-mouse pancreas with beta cells (green), alpha cells (red) and delta cells (white) and nuclei (blue). (B) Quantification of islet composition and area. The total endocrine cell area is normalized to the total pancreas area in each mouse; BAC/+ n = 5, BAC/MIP n = 5. Total islet area is then subdivided into its cellular components; beta cell area in green, alpha cell area in red, delta-cell area in blue. Yellow horizontal lines indicate the average total islet area for each group. BAC/+ and BAC/MIP mice are compared at total islet area as well as area of each cell type, with P values reported in table.
Discussion
Considerable efforts from various research groups, including our own, have highlighted a key role for Wnt-signaling in the regulation of energy metabolism, and these analyses have eluded to potential mechanisms of how misregulation of this conserved developmental pathway leads to increased susceptibility to T2D (29–31). However, most of these studies have largely concentrated on the roles of TCF7L2 in pancreatic beta cell. This limited focus was initially precipitated by the demonstration that reducing Tcf7l2 expression in rodent and human beta cells models can lead to impaired insulin secretion and production (17,18). Human genetics data have added further support by demonstrating that the T2D-risk allele is associated with impaired insulin secretion in humans, even in non-diabetics (32).
Nevertheless, the concerted effort to understand the link between beta cell TCF7L2 expression and T2D susceptibility has often met with contradictory results. At the heart of the contradiction is the direction of change in TCF7L2 expression leading to glucose intolerance and T2D susceptibility. Studies in human islets agree the T2D-risk allele is associated with increased TCF7L2 expression and decreased insulin secretion (12–14,33). However, in vitro analyses on isolated human and rodent islets suggest decreased TCF7L2 expression is associated with decreased insulin secretion (17–19,21,34). These studies also highlight other islet defects following TCF7L2 downregulation, including fewer GLP-1 and GIP receptors, decreased expression of genes involved in insulin vesicle fusion, and increased beta cell apoptosis (17–19,21). These results suggest decreased TCF7L2 expression drives beta cell malfunction, decreased insulin secretion and decreased glucose tolerance, but paradoxically the risk allele remains strongly associated with increased TCF7L2 expression.
Mouse models harboring both gain and loss of function in Tcf7l2 do not readily reconcile these differences. Mice overexpressing Tcf7l2 either in multiple tissues or specifically in the liver display impaired glucose tolerance (9,15). While germline null Tcf7l2 alleles have repeatedly been shown to result in improved glucose tolerance, beta cell-specific ablation of Tcf7l2 in mice often leads to the opposite phenotype, a worsening in glucose tolerance (9,16,23,35) or to no change in glucose tolerance at all (15). These data seem to be at odds with the observation that, in humans, the T2D-associated SNP within TCF7L2 is associated with impaired insulin secretion. Further studies will determine whether this is a fundamental biological difference between humans and mice.
In hopes of resolving these incongruencies, various explanations have been raised. Chief among those is the idea that perhaps mice do not represent a viable model of TCF7L2 biology in humans, a concept that has recently been demonstrated in connection to other genes involved in T2D susceptibility emerging from Genome-Wide Association Studies (36,37).
Our study helps to reconcile some of the conflicting data on the beta cell contribution to the Tcf7l2 glucose intolerance phenotype. We demonstrate that overexpression of Tcf7l2 in the multiple tissues where it is normally expressed leads to glucose intolerance, but reducing Tcf7l2 expression to wild-type levels in beta cells while maintaining overexpression elsewhere increases the severity of the glucose intolerance phenotype. Furthermore, this reduction of expression in beta cells is accompanied with decreases in insulin secretion and a reduction in beta cell area and numbers. These phenotypes are precisely those that have been systematically shown in human and mouse cell lines (17–19,21,34), suggesting that the biology of TCF7L2 as a central regulator of glucose metabolism is conserved between human and mouse. We posit that part of the seemingly contradictory data generated over the past several years is due to the evaluation of TCF7L2 impact on beta cells in an ex-vivo context. Specifically, we do see the same phenotypes associated with Tcf7l2 in beta cells, but within a whole-system mouse physiological context, Tcf7l2 is also expressed in multiple other tissues relevant to glucose homeostasis (38), and it seems that its roles in one or more of these peripheral tissues overrides the pancreatic actions of this factor, highlighting intricate higher-order multi-organ system interactions that can be faithfully observed within a complex multicellular experimental model.
Other studies support the conclusion that Tcf7l2 overexpression in the periphery leads to glucose intolerance. The T2D-associated genomic interval has been shown to be a complex regulatory region driving TCF7L2 expression in a variety of peripheral tissues, with the risk allele maintaining allele-specific effects (9–11). Recent data from Hans Clevers' laboratory indicate that TCF7L2 is an important regulator of liver metabolism and gluconeogenesis; liver-specific Tcf7l2 knock-out mice demonstrate decreased gluconeogenesis and improved glucose tolerance while transient overexpression of Tcf7l2 leads to increased gluconeogenesis (15).
Our data are also congruent with that from the Rutter laboratory, showing that reduced Tcf7l2 expression in beta cells in vivo leads to decreased beta cell volume, decreased glucose and Glp-1 stimulated insulin release and decreased glucose tolerance compared with wild-type (23,35). We observe similar phenotypes when we reduce Tcf7l2 expression from high to normal levels in our mice. In line with a worsening of glucose tolerance in our mouse models, we would predict that a beta cell-specific Tcf7l2 loss of function would lead to the same phenotypes described by Rutter and colleagues.
Our study highlights the complexities of whole-systems in vivo physiology, where multiple organs and tissues regulate the same biological function, often with opposing effects (39). Our data point to a classical example where manipulating a single gene, in this case Tcf7l2, may lead to a phenotype in one tissue, even when the overall in vivo phenotypic impact of that gene may be in direct opposition. Importantly, our work emphasizes the pressing need to better characterize the roles of TCF7L2 in other peripheral tissues. Even supposing the genetic association with T2D is demonstrated to arise from TCF7L2’s function in pancreatic beta cells, these data provide evidence that the role of TCF7L2 in regulating glucose metabolism from other tissues seems to be more robust. Harnessing the biology of TCF7L2 in these additional tissues may hold the key to developing novel and better therapeutic targets for T2D.
Materials and Methods
Animals
We used the CD1 Tcf7l2 overexpression mouse harboring the human BAC RP11–466I19 containing the full-length mouse Tcf7l2 cDNA (GenBank BC052022.1) as previously described (9). This BAC spans the entire T2D-associated LD block in humans, including all cis-regulatory elements within it. This results in a mouse that overexpresses Tcf7l2 only in tissues endogenously expressing this gene in humans. Hemizygous mice (BAC/+) contain three extra copies of floxed mouse Tcf7l2 cDNA leading to global Tcf7l2 overexpression (9). Hemizygous BAC mice were crossed to C57B/6 mice hemizygous for the inducible mouse insulin promoter Cre, MIP-CreER, as previously described (24). This cross of BAC/+ to MIP/+ generates BAC/+, MIP/+ wild-type (WT) and BAC/MIP-CreER F1 males. MIP/+ were excluded from further study. BAC/MIP-CreER males were injected with 200 µL of 10 mg/mL tamoxifen for 3 days at 5–7 weeks of age to activate the MIP-CreER. Active MIP-CreER in beta cells leads to excision of the floxed region of the BAC containing the extra copies of Tcf7l2 cDNA.
Verification of islet-specific conditional ablation of the transgenic Tcf7l2 copies
To verify active MIP-CreER and the subsequent excision of the floxed region in the BAC, we performed PCR on islet DNA using primers outside of the floxed region. Since the entire floxed region of the BAC is 23.8 kb, no excision of the region will prevent amplification. Proper excision of the floxed region results in a 405 bp fragment, easily amplified by PCR. Islets were isolated from 3 BAC/+, 3 WT and 3 BAC/MIP mice as previously described (40) and DNA was isolated using the Protein Precipitation Solution (Qiagen). Standard PCR conditions were used and the primer sequences are as follows: F—GTTTCTGGGTGAGCAAAAACA, R—CGTGAGACTACGATTCCATCAAT.
While this PCR technique validates CreER excision of the floxed region at the DNA level, we also verified removal of the overexpression at the RNA level. Islets isolated from 3 BAC/+, 3 WT and 3 BAC/MIP mice were used for RNA extraction using Tri-Reagent (Sigma) and then for cDNA generation using Superscript II reverse transcriptase (Invitrogen). Tcf7l2 exonic primers (F—ATCGTCACACCGACAGTCAA, R—TTGGAGTCCTGATGCTTTGAG) and Hprt housekeeping gene primers (F—TGTTGTTGGATATGCCCTTG, R – GCGCTCATCTTAGGCTTTGT) were used for qRT-PCR.
In vivo glucose tolerance experiments
Starting at 5–7 weeks of age, BAC/+ and BAC/MIP mice subsequent to tamoxifen injection were fed a high fat diet (55% fat, Harlan Teklad) for 10 weeks. After this 10-week period, BAC/+ and BAC/MIP mice were tested for glucose sensitivity by IPGTT. Prior to IPGTT, mice were fasted for 4 h and an initial blood glucose reading was taken. This fast was followed by intraperitoneal injection with 2 mg/kg dextrose, and subsequent blood glucose checks using an AccuChek Aviva glucometer (Roche). Blood glucose readings were taken at 15, 30, 60 and 120 min post dextrose injection. After IPGTT, mice resumed high fat diet. This experiment was later repeated including WT littermates.
Blood plasma collection and insulin quantification
After allowing the mice to restabilize for 3–7 days after IPGTT, we again fasted BAC/+ and BAC/MIP mice for 4 hours for blood collection from the tail vein. We collected 500 µl of blood in EDTA coated tubes followed by a 10-min centrifugation at 7000 × g at 4°C for plasma isolation. Plasma was diluted for insulin enzyme-linked immunosorbent assay (ELISA from Millipore) to determine plasma insulin content. BAC/+, BAC/MIP and WT mice were then sacrificed and whole pancreas was collected and collagenase digested to isolate the total pancreatic insulin content. Total pancreatic insulin content from 10 mice per genotype was quantified by insulin ELISA.
Dynamic islet perifusion
Islets were isolated from six high fat diet fed BAC/+and BAC/MIP mice for perifusion, which allows for dynamic assessment of islet function based on insulin secretion in response to changing concentrations of glucose (26–28). Islets were incubated in 3 mm glucose for 60 min with insulin samples collected every 12 min. At 60 min, glucose concentration was increased to 11 mm for 20 min and insulin samples were collected every 2 min. Then glucose concentration was returned to 3 mm for 20 min with continued sampling every 2 min. At the end of this 100 min time frame, islets were perifused with KCl for 10 min to release the intracellular insulin. All insulin samples were assessed using ELISA as before and were normalized to the lowest value of basal insulin release during first 60-min incubation in 3 mm glucose. Two insulin secretion peaks upon glucose stimulation are expected, representing current insulin stores and subsequent insulin production. Area under the curve (AUC) is measured from 64–96 min to capture the entirety of the glucose-stimulated insulin response curve.
Pancreatic histology and cellularity
Five BAC/+ and 5 BAC/MIP mice were sacrificed after 10 weeks high fat diet. Whole pancreas from each animal was dissected and fixed in 4% paraformaldehyde. Paraffin-embedded sections (5 µm) were stained with the following primary antibodies (all 1:500): polyclonal guinea pig anti-porcine insulin (DAKO, Carpinteria, CA), mouse monoclonal anti-human glucagon (Sigma-Aldrich, St. Louis, MO), polyclonal goat anti-somatostatin (Santa Cruz, Santa Cruz, CA) and DAPI (Invitrogen, Carlsbad, CA). The primary antibodies were detected using a combination of DyLight 488, 549 and 649-conjugated secondary antibodies (1:200, Jackson ImmunoResearch Laboratory, West Grove, PA).
Microscopic images were taken with an Olympus IX8 DSU spinning disk confocal microscope (Melville, NY) with imaging software StereoInvestigator (SI, MicroBrightField, Williston, VT). A modified method of ‘virtual slice capture’ was used (41,42). Briefly, the SI controls a XYZ-motorized stage and acquires consecutive images, which creates a high-resolution montage composed of images obtained from multiple microscopic fields of view. The entire tissue section was captured as ‘a virtual slice’ using a 10× objective. Each virtual slice taken at four fluorescent channels were further merged into one composite. Given this unique, unbiased, computer-assisted method of ‘virtual slice capture’ one section per pancreas is sufficient to provide representative pancreas composition. Quantification of cellular composition (i.e. each area of beta-, alpha- and delta-cell populations, and islet area by automated contouring of each islet) was carried out using a macro custom-written for Fiji/ImageJ (http://rsbweb.nih.gov/ij/). MATLAB (MathWorks, Natick, MA) was used for mathematical analyses.
Statistical analyses
Data are shown as the standard error of the mean (SEM). An unpaired two-sided Student's t-test was used to test for significance in all cases.
Supplementary material
Funding
This work was supported by the National Institute of Health (Grants DK093972, HL123857, HL118758 to M.A.N; DK020595 to G.I.B) and a gift from the Kovler Family Foundation (MH). Funding to pay the Open Access publication charges for this article was provided by Discretionary funds provided to M.A.N. by the University of Chicago.
Supplementary Material
Acknowledgements
We would like to thank Guy Rutter for his critical reading of the manuscript; members of the Witkowski laboratory for assisting with the perifusion studies; and Honggang Ye for his guidance with glucose tolerance testing.
Conflict of Interest statement. None declared.
References
- 1.Grant S.F., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 2.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 3.Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., et al. A genome-wide association study of type 2 diabetes in Finns detect multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 5.Zeggini E., Weedon M.N., Lindgren C.M., Frayling T.M., Elliott K.S., Lango H., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cauchi S., Meyre D., Dina C., Choquet H., Samson C., Gallina S., Balkau B., Charpentier G., Pattou F., Stetsyuk V., et al. Transcription factor TCF7L2 genetic study in the French population: expression in human beta cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55:2903–2908. doi: 10.2337/db06-0474. [DOI] [PubMed] [Google Scholar]
- 8.Saxena R., Elbers C.C., Guo Y., Peter I., Gaunt T.R., Mega J.L., Lanktree M.B., Tare A., Castillo B.A., Li Y.R., et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am. J. Hum. Genet. 2012;90:410–425. doi: 10.1016/j.ajhg.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savic D., Ye H., Aneas I., Park S.Y., Bell G.I., Nobrega M.A. Alterations in TCF7L2 expression define its role as a key regulator of glucose metabolism. Genome Res. 2011;21:1417–1425. doi: 10.1101/gr.123745.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savic D., Bell G.I., Nobrega M.A. An in vivo cis-regulatory screen at the type 2 diabetes associated TCF7L2 locus identifies multiple tissue-specific enhancers. PLoS One. 2012;7:e36501. doi: 10.1371/journal.pone.0036501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savic D., Park S.Y., Bailey K.A., Bell G.I., Nobrega M.A. In vitro scan for enhancers at the TCF7L2 locus. Diabetologia. 2013;56:121–125. doi: 10.1007/s00125-012-2730-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaulton K.J., Nammo T., Pasquali L., Simon J.M., Giresi P.G., Fogarty M.P., Panhuis T.M., Mieczkowski P., Secchi A., Bosco D., et al. A map of open chromatin in human pancreatic islets. Nat. Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stitzel M.L., Sethupathy P., Pearson D.S., Chines P.S., Song L., Erdos M.R., Welch R., Parker S.C., Boyle A.P., Scott L.J., et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyssenko V., Lupi R., Marchetti P., Del Guerra S., Orho-Melander M., Almgren P., Sjögren M., Ling C., Eriksson K.F., Lethagen A.L., et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J. Clin. Invest. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boj S.F., van Es J.H., Huch M., Li V.S., Jose A., Hatzis P., Mokry M., Haegebarth A., van den Born M., Chambon P., et al. Diabetes risk gene and Wnt effector Tcf7l2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell. 2012;151:1595–1607. doi: 10.1016/j.cell.2012.10.053. [DOI] [PubMed] [Google Scholar]
- 16.Yang H., Li Q., Lee J.H., Shu Y. Reduction in Tcf7l2 expression decreases diabetic susceptibility in mice. Int. J. Biol. Sci. 2012;8:791–801. doi: 10.7150/ijbs.4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shu L., Sauter N.S., Schulthess F.T., Matveyenko A.V., Oberholzer J., Maedler K. Transcription factor 7-like 2 regulates beta cell survival and function in human pancreatic islets. Diabetes. 2008;57:645–653. doi: 10.2337/db07-0847. [DOI] [PubMed] [Google Scholar]
- 18.Shu L., Matveyenko A.V., Kerr-Conte J., Cho J.H., McIntosh C.H., Maedler K. Decreased TCF7L2 protein levels in type 2 diabetes mellitus correlate with downregulation of GIP- and GLP-1 receptors and impaired beta-cell function. Hum. Mol. Genet. 2009;18:2388–2399. doi: 10.1093/hmg/ddp178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Silva Xavier G., Loder M.K., McDonald A., Tarasov A.I., Carzanige R., Kronenberger K., Barg S., Rutter G.A. TCF7L2 regulates late events in insulin secretion from pancreatic islet beta-cells. Diabetes. 2009;58:894–905. doi: 10.2337/db08-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rulifson I.C., Karnik S.K., Heiser P.W., ten Berge D., Chen H., Gu X., Taketo M.M., Nusse R., Hebrok M., Kim S.K. Wint signaling regulates pancreatic beta cell proliferation. Proc. Natl. Acad. Sci. USA. 2007;104:6247–6252. doi: 10.1073/pnas.0701509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y., Park S.Y., Su J., Bailey K.A., Ottosson-Laakso E., Shcherbina L., Oskolkov N., Zhang E., Thevenin T., Fadista J., et al. TCF7L2 is a master regulator of insulin production and processing. Hum. Mol. Genet. 2014 doi: 10.1093/hmg/ddu359. 10.1093/hmg/ddu359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu L., Zien K., Gutjahr G., Oberholzer J., Pattou F., Kerr-Conte J., Maedler K. TCF7L2 promotes beta cell regeneration in human and mouse pancreas. Diabetologia. 2012;55:3296–3307. doi: 10.1007/s00125-012-2693-z. [DOI] [PubMed] [Google Scholar]
- 23.da Silva Xavier G., Mondragon A., Sun G., Chen L., McGinty J.A., French P.M., Rutter G.A. Abnormal glucose tolerance and insulin secretion in pancreas-specific Tcf7l2-null mice. Diabetologia. 2012;55:2667–2676. doi: 10.1007/s00125-012-2600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wicksteed B., Brissove M., Yan W., Opland D.M., Plank J.L., Reinert R.B., Dickson L.M., Tamarina N.A., Philipson L.H., Shostak A., et al. Conditional gene targeting in mouse pancreatic β-cells: analysis of ectopic Cre transgene expression in the brain. Diabetes. 2010;59:3090–3098. doi: 10.2337/db10-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabrera O., Jacques-Silva M.C., Berman D.M., Fachado A., Echeverri F., Poo R., Khan A., Kenyon N.S., Ricordi C., Berggren P.O., et al. Automated, high-throughput assays for evaluation of human pancreatic islet function. Cell Transplant. 2008;16:1039–1048. [PMC free article] [PubMed] [Google Scholar]
- 26.Marek N., Krzystyniak A., Ergenc I., Cochet O., Misawa R., Wang L.J., Goła˛b K., Wang X., Kilimnik G., Hara M., et al. Coating human pancreatic islets with CD4(+)CD25(high)CD127(-) regulatory T cells as a novel approach for the local immunoprotection. Ann. Surg. 2011;3:512–519. doi: 10.1097/SLA.0b013e31822c9ca7. [DOI] [PubMed] [Google Scholar]
- 27.Kizilel S., Scavone A., Liu X., Nothias J.M., Ostrega D., Witkowski P., Millis M. Encapsulation of pancreatic islets within nano-thin functional polyethylene glycol coatings for enhanced insulin secretion. Tissue Eng. Pt. A. 2010;7:2217–2228. doi: 10.1089/ten.TEA.2009.0640. [DOI] [PubMed] [Google Scholar]
- 28.Kilimnik G., Zhao B., Jo J., Periwal V., Witkowski P., Misawa R., Hara M. Altered islet composition and disproportionate loss of large islets in patients with type 2 diabetes. PLoS One. 2011;6:e27445. doi: 10.1371/journal.pone.0027445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basile K.J., Johnson M.E., Xia Q., Grant S.F. Genetic susceptibility to type 2 diabetes and obesity: follow-up findings from genome-wide association studies. Int. J. Endocrinol. 2014;2014:769671. doi: 10.1155/2014/769671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.da Silva Xavier G., Bellomo E.A., McGinty J.A., French P.M., Rutter G.A. Animal models of GWAS-identified type 2 diabetes genes. J. Diabetes Res. 2013;2013:906590. doi: 10.1155/2013/906590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nobrega M.A. TCF7L2 and glucose metabolism: time to look beyond the pancreas. Diabetes. 2013;62:706–708. doi: 10.2337/db12-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosengren A.H., Braun M., Mahdi T., Andersson S.A., Travers M.E., Shigeto M., Zhang E., Almgren P., Ladenvall C., Axelsson A.S., et al. Reduced insulin exocytosis in human pancreatic β-cells with gene variants linked to type 2 diabetes. Diabetes. 2012;61:1726–1733. doi: 10.2337/db11-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Bacquer O., Kerr-Conte J., Gargani S., Delalleau N., Huyvaert M., Gmyr V., Froguel P., Neve B., Pattou F. TCF7L2 rs7903146 impairs islet function and morphology in non-diabetic individuals. Diabetologia. 2012;55:2677–2681. doi: 10.1007/s00125-012-2660-8. [DOI] [PubMed] [Google Scholar]
- 34.Loder M.K., da Silva Xavier G., McDonald A., Rutter G.A. TCF7L2 controls insulin gene expression and insulin secretion in mature pancreatic β-cells. Biochem. Soc. Trans. 2008;36:357–359. doi: 10.1042/BST0360357. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell R.K., Mondragon A., Chen L., McGinty J.A., French P.M., Ferrer J., Thorens B., Hodson D.J., Rutter G.A., Xavier G.D. Selective disruption of Tcf7l2 in the pancreatic β cell impairs secretory function and lovers β cell mass. Hum. Mol. Genet. 2014 doi: 10.1093/hmg/ddu553. 10.1093/hmg/ddu553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flannick J., Thorleifsson G., Beer N.L., Jacobs S.B., Grarup N., Burtt N.P., Mahajan A., Fuchsberger C., Atzmon G., Benediktsson R., et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat. Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson E. Zinc transport and diabetes risk. Nat. Genet. 2014;46:323–324. doi: 10.1038/ng.2934. [DOI] [PubMed] [Google Scholar]
- 38.Sakabe N.J., Nobrega M.A. Genome-wide maps of transcription regulatory elements. WIREs Syst. Biol. Med. 2010;4:422–437. doi: 10.1002/wsbm.70. [DOI] [PubMed] [Google Scholar]
- 39.Sakabe N.J., Savic D., Nobrega M.A. Transcriptional enhancers in development and disease. Genome Biol. 2012;13:238. doi: 10.1186/gb-2012-13-1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park S.Y., Wang X., Chen Z., Powers A.C., Magnuson M.A., Head W.S., Piston D.W., Bell G.I. Optical imaging of pancreatic beta cells in living mice expressing a mouse insulin I promoter-firefly luciferase transgene. Genesis. 2005;43:80–86. doi: 10.1002/gene.20157. [DOI] [PubMed] [Google Scholar]
- 41.Kilimnik G., Jo J., Periwal V., Zielinski M.C., Hara M. Quantification of islet size and architecture. Islets. 2012;4:167–172. doi: 10.4161/isl.19256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim A., Kilimnik G., Guo C., Sung J., Jo J., Periwal V., Witkowski P., Dilorio P., Hara M. Computer-assisted large-scale visualization and quantification of pancreatic islet mass, size distribution and architecture. J. Vis. Exp. 2011;49:e2471. doi: 10.3791/2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.