Abstract
X-chromosome inactivation (XCI) achieves dosage compensation between males and females through the silencing of the majority of genes on one of the female X chromosomes. Thus, the female X chromosomes provide a unique opportunity to study euchromatin and heterochromatin of allelic regions within the same nuclear environment. We examined the interplay of DNA methylation (DNAm) with CpG density, transcriptional activity and chromatin state at genes on the X chromosome using over 1800 female samples analysed with the Illumina Infinium Human Methylation450 BeadChip. DNAm was used to predict an inactivation status for 63 novel transcription start sites (TSSs) across 27 tissues. There was high concordance of inactivation status across tissues, with 62% of TSSs subject to XCI in all 27 tissues examined, whereas 9% escaped from XCI in all tissues, and the remainder showed variable escape from XCI between females in subsets of tissues. Inter-female and twin data supported a model of predominately cis-acting influences on inactivation status. The level of expression from the inactive X relative to the active X correlated with the amount of female promoter DNAm to a threshold of ∼30%, beyond which genes were consistently subject to inactivation. The inactive X showed lower DNAm than the active X at intragenic and intergenic regions for genes subject to XCI, but not at genes that escape from inactivation. Our categorization of genes that escape from X inactivation provides candidates for sex-specific differences in disease.
Introduction
X-chromosome inactivation (XCI) occurs early in female mammalian development to transcriptionally silence one of the pair of ∼156-Mb X chromosomes, thereby achieving dosage equivalency with males who have a single X chromosome and the sex-determining Y chromosome. XCI presents a unique opportunity to compare the epigenetic profile of the facultative heterochromatin of the inactive X chromosome (Xi) with the euchromatin of the active X chromosome (Xa) within the same cell. While the majority of genes on the X chromosome are subject to XCI, past surveys have suggested that ∼15% of genes escape from XCI and an additional ∼10% of genes continue to be expressed from the Xi to variable extents (1,2). As X/Y homologous genes in the pseudoautosomal region, and additional genes for which the ancestral Y homologue is retained, continue to be expressed from both the X and Y chromosomes, escape from XCI for these genes still results in dosage equivalency (3). Differences in X-linked gene expression between males and females may underlie some sexually dimorphic traits or disease predispositions (4), and such differences could result from differential changes in the expression or function of X/Y homologues or escape from XCI for genes that no longer have a functional Y homologue (5). Furthermore, a number of genes are variable in their XCI status (termed ‘variable escape genes’), escaping XCI in some females, but being subject to XCI in other females. Differences in the expression of a subset of these genes are thought to contribute to inter-female differences in disease or phenotype (6). Additional variability is observed between tissue types for the inactivation of some genes (7). XCI status for genes has previously been predicted through the analysis of the expression in somatic cell hybrids (1), imbalance in allelic expression patterns (2), changes in total expression between males and females (8) and DNA methylation (DNAm) of promoter sequences (7,9). Generation of a more complete catalogue of genes that escape from XCI would allow assessment of their contribution to disease, and the characterization of escape genes and their chromatin neighbourhood should also provide insights into factors underlying the spread of XCI and, by extension, the spread of heterochromatin across the genome.
Epigenome-wide studies have become widely used to identify potential mechanisms underlying disease-associated changes in gene expression, to identify biomarkers for disease and to identify regulatory regions affecting susceptibility to disease (10). Interest in the study of epigenetic marks has been heightened by their potential to be modified by environment, lifestyle or therapeutic interventions. DNAm has been the most commonly analysed epigenetic mark, as it is a relatively stable, well-studied mark that is readily interrogated in samples of limited quantity. DNAm in vertebrates is found predominantly at CpG dinucleotides and is generally observed to be biphasic, with limited DNAm at the CpGs found clustered in CpG islands and substantially more DNAm in non-island regions (11–13). Overall, there are estimated to be over 27 million CpGs in the human genome, ∼1.2 million of which are on the X chromosome. Over 15% of the genome's CpGs are found within CpG islands (11,14,15), with half of CpG islands found at transcription start sites (TSSs), generally of ubiquitously expressed ‘housekeeping’ genes, and DNAm of these promoter CpGs has been shown to be inversely correlated with expression (16). In contrast, within genes, the reverse trend has been reported with gene body DNAm correlated with increased expression, which has been hypothesized to reflect a suppression of transcriptional noise through co-recruitment with polymerase (17). However, other studies have not seen such a correlation, and the relationship of levels of gene body DNAm with expression is complex and sensitive to the quantification approach (18).
Extensive epigenome projects have documented DNAm, histone modifications and a wide variety of transcription factor binding sites in numerous cell lines and tissues. Cross-talk between histone marks and DNAm (reviewed in 19), as well as associations with expression levels, would be anticipated to result in correlations of DNAm with chromatin features. A recent study identified both positive and negative DNAm-expression correlations at all gene components and found that negative correlations were enriched in regions with marks of regulatory activity (13). The application of a high-throughput bioinformatics pipeline, which utilized hidden Markov modelling of data from 9 chromatin marks in 9 cell lines, resulted in the definition of 15 chromatin states, which included regulatory regions of promoters, enhancers and insulators, as well as transcribed regions of genes and repressed or inactive chromatin (20). Regulatory regions, as demarcated by DNAse hypersensitivity sites, have been shown to be enriched for epigenetically variable loci (13), providing support for the conjecture that epigenetic variability may underlie altered expression patterns and thus modulate disease. For example, there is growing evidence for the disruption of epigenetic regulators in cancer, in addition to the frequent alterations in DNAm that are seen in tumour cells. Thus, deciphering the normal epigenetic landscape of active and silenced regions of the genome is important for understanding the changes occurring with disease.
Previously, we have shown that DNAm can be used as a tool to identify genes that are subject to XCI (7,21), by identifying those TSSs with significantly more DNAm in females than males. A commonly used method to assess DNAm in humans is the Illumina bead technology, the latest platform of which is the Illumina Infinium HumanMethylation450 BeadChip or ‘450K array’ that assesses DNAm at >485 000 CpG sites across the genome, ∼40% of which are at CpG islands associated with TSSs. The CpGs on the 450K array are therefore biased towards TSSs and have been annotated by Illumina to identify not only CpG islands but also ‘shores’, ‘shelves’ and the rest of the genome—the ‘open sea’ (22). Shores were originally described as the 2-kb regions near CpG islands and have been shown to preferentially include differentially methylated regions between tissues (23), to show differential DNAm in cancers (23,24) as well as differential DNAm with respect to aging (25). Given the potential importance of these regions, an additional flanking 2 kb of DNA was defined by Illumina as the ‘shelf’. Recently, another annotation of the probes on the 450K array used a CpG density annotation based on a different classification system (26), which distinguished between high CpG density (HC) and intermediate CpG density (IC) CpG islands as well as low CpG density (LC) regions of the genome (22). A comparison of the two classification systems found that shores were typically of IC, and shelves were typically of LC (22). A recent study examined X-linked DNAm using the 450K array, with a focus on active X-specific DNAm (27). Probes that were hypermethylated on the Xa relative to the Xi were enriched for non-CpG islands relative to probes that were hypermethylated on the Xi relative to the Xa, or unmethylated on both Xs. Outside of promoter regions, the Xi has been reported to have lower levels of gene body DNAm compared with the Xa (12,28), whereas overall, Xa versus Xi DNAm levels have differed depending on the method of analysis (29–31).
Using 19 publically available 450 K datasets and one novel dataset for a total of 2928 samples from 27 different tissue types, we have explored the epigenetic landscape of the X chromosome and then categorized nearly 500 X-linked TSSs as subject to, escaping from, or showing variable escape from XCI in all 27 tissues using the differential DNAm between females (46, XX) and males (46, XY) as a surrogate for DNAm levels of the Xi. Overall the XCI status of a gene between different tissues was found to be relatively stable with 62% of genes found to be subject to XCI in all 27 tissues.
Results
Differences in DNAm on Xa and Xi reflect both CpG density and chromatin state
To investigate large-scale patterns of X-linked DNAm, we initially examined data from peripheral blood leukocytes (PB), as this was the tissue with the largest total number of samples (male n = 275, females n = 490) in the GEO 450K array data that matched our selection criteria (see Materials and Methods) (32). The initial analysis focussed on those 8527 X chromosome probes (hereafter called CpGs) from the 450K array that mapped to a unique location on the X chromosome and were not located in a repetitive element (22) (Supplementary Material, Table S1). Comparing the average level of male DNAm with the average female DNAm at each of these 8527 CpGs revealed two main clusters (Fig. 1A). A group of CpGs was highly methylated in both males (83–87% DNAm) and females (79–83% DNAm), and a second larger focal point was centred on CpGs with a female DNAm of 36–40% and a male DNAm of 7–11%. Previous examinations of X-linked DNAm (7,21) would suggest the latter CpGs reflect a pattern associated with the promoters of genes subject to XCI. The other cluster of CpGs that includes highly methylated CpGs in both males and females might reflect CpGs in non-island promoters or in promoters for the cancer-testis family of genes, known to be hypermethylated in males and females in the vast majority of tissues (33) or that fall outside of a promoter region.
Figure 1.
DNAm landscape of the X chromosome by CpG density and chromatin state. (A) Average male versus average female DNAm at X-linked CpGs demonstrates two major clusters of DNAm. Each grey square represents a single CpG (n = 8527); thick black kernel density lines help visualize the number of CpGs. (B) Box and whisker plots of the average female (F = light grey) and male (M = dark grey) DNAm based on CpG density (number of CpGs, HC: n = 3725, ICshore: n = 849, IC: n = 1402, LC: n = 2551). Significance based on a Wilcox test comparison of means is as follow: *P-values 0.05–0.001, **P-values 0.001–2.2E−16 and ***P-values < 2.2E − 16. (C) Summary of the influence of CpG density and chromatin marks on different X-linked genomic regions. Xi≤≥Xa indicates that there is not a clear relationship explaining the interaction between DNAm, X inactivation density and chromatin marks.
To examine the impact of CpG density upon X-linked DNAm, CpGs were separated based on CpG density of the surrounding region (Fig. 1B). On average, CpGs in high-density CpG islands (HC) and flanking intermediate CpG density regions [IC shore regions, together abbreviated as high and intermediate CpG density (HIC) in figures] showed low male and moderate female DNAm. Isolated IC islands and non-island CpGs (LC) usually showed higher and more equal levels of male and female DNAm. To identify features that show distinct DNAm between the active euchromatin of the Xa and the inactive facultative heterochromatin of the Xi, we estimated the Xi DNAm by subtracting the male DNAm (representing the Xa), from the female DNAm (which contains an Xa and an Xi), and then plotted the distribution of DNAm subdivided by both CpG density (Supplementary Material, Fig. S1) and the chromatin states developed by Ernst et al. (20) (shown in Supplementary Material, Fig. S2 for different categorizations of CpG density). We choose to use the chromatin states established in the H1 cell line (20), a male embryonic cell line, as the H1 chromatin states most closely represented those present in a cell at the time before XCI and therefore when only an Xa was present. The 450K array is enriched for CpGs located in promoters, with 44% of X-linked CpGs on the array within promoters and 34% within the heterochromatic regions. Overall, the Xi showed less variability in DNAm (33–78%) than the Xa (9–83%), both across functional chromatin states and between CpG densities (Fig. 1C). Xi DNAm is higher than that of the Xa for promoters and enhancers, whereas Xa DNAm is higher than the Xi for transcribed regions and heterochromatin.
On the Xa, DNAm increased from strong promoters to weak promoters to poised promoters, and additionally, the average DNAm increased with decreasing CpG density. As such, high CpG density strong promoters had the lowest Xa DNAm (average 9%) and non-island poised promoters the highest Xa DNAm (average 47%) suggesting a synergy between CpG density and functional chromatin state of the Xa. At other regions, the interplay between CpG density and histone modifications was less clear-cut, with both having varying degrees of influence. These results demonstrated that in addition to island promoter CpGs, DNAm at active non-island promoters and island enhancers and insulators might also be useful for determining whether a gene would be subject to XCI. To maximize the CpGs that could be examined, we examined whether it was necessary to exclude probes that had been identified as potentially overlapping a non-unique location on the X chromosome (22) (Supplementary Material, Table S1). We compared the difference between female and male DNAm at each CpG density and chromatin state for these CpGs against those of the previously analysed unique CpGs and found only five significant (P-value < 0.05) differences in DNAm (Supplementary Material, Fig. S3), suggesting that for the majority of these CpGs, the potential ancestral repeat origin no longer impacted their DNAm. We therefore concluded that all 10 584 X-linked CpGs without autosomal homology on the 450K array (22) could be used to contribute to our knowledge of the XCI status of different TSSs and allow for the comparison of XCI status across tissues and females.
Xi promoter DNAm is correlated with relative expression level at escape genes
We created a subject and escape gene training set based on previous knowledge of XCI from expression studies (see Materials and Methods) and determined the male DNAm, female DNAm and difference in female and male DNAm (in PB) at all CpGs classes (combined CpG density and chromatin state) that had shown significant Xi hypermethylation (Supplementary Material, Fig. S4A). Comparing subject and escape TSSs, individual CpGs showed a significant difference (P-value < 2.2E − 16) in the average female DNAm but not in the average male DNAm. Using a moving average, we compared the difference between male and female DNAm in the subject and escape training sets based on distance of each CpG to the closest TSS (Supplementary Material, Fig. S4B) and found that within the −500- to 1000-bp range, the differences of the subject and escape training sets were distinct. Owing to the biased genic location of probes on the 450K array, in addition to states classified as promoters, the −500- to 1000-bp range contained over half of the X-linked CpGs classified as enhancers and insulators present on the array, despite the fact that only ∼8% of these states are within 2 kb of a TSS on average in the genome. A significant difference (P-value < 2.2E − 16) between the training sets was observed at female but not male DNAm when CpGs were averaged together by the closest annotated TSS instead of by individual CpGs (Supplementary Material, Fig. S4C) so from this point forward, all individual CpGs were averaged for the closest annotated TSS. The DNAm levels from the training sets were used to establish DNAm ranges that were then used to predict XCI status at all X-linked TSSs on the 450 K array (see Materials and Methods and Supplementary Material, Table S2).
We predicted an XCI status at every TSS in each of the 486 individual female PB samples using the thresholds described earlier. We left an ‘uncallable’ XCI class between the subject and escape TSSs to account for variable mosaicism in females owing to the potential for skewing of XCI, and the heterogeneity that may exist between cells or within tissues owing to cell composition variability. There were 690 TSSs in PB for which there were DNAm data that we could use to predict XCI status. The majority of these TSSs (66%) were subject to XCI, 14% variably escaped from XCI, 11% were uncallable and 9% escaped from XCI (for further definition of each XCI status, see Materials and Methods), with 101 of these TSSs being ‘novel’ calls in that the TSS had not previously been assessed for XCI status. Ranking TSSs by average female DNAm revealed a continuum of DNAm, with subject TSSs ranging in average female DNAm from 31 to 57%, whereas escape TSSs ranged from 4 to 18% and uncallable TSSs between 21 and 32% (Fig. 2A). The variable escape TSSs were found within the ranges of all other XCI statuses, although they showed increased representation near the uncallable range of average DNAm. Over half (59%) of the TSSs we deemed uncallable or variable escape that had a previously determined XCI status (1,2) had been called as subject to XCI in those studies. The continuum of average TSS DNAm we observed was reminiscent of the continuum of relative expression levels from the Xi that we observed in previous studies using allelic imbalance (AI) in the expression to identify TSSs that escaped from XCI (2). TSSs that DNAm predicted were subject to XCI had an average AI of <0.05, which was well below the AI cut-off of 0.10 previously used to reflect 10% expression from the Xi in females with skewed XCI (2). We therefore wished to correlate DNAm with AI for TSSs, to determine whether female DNAm was predictive of partial expression from the Xi. There was a significant negative (P-value < 0.0001) correlation between the average level of female DNAm and AI for TSSs that escaped from XCI (green dots, Fig. 2B). In contrast, the average female DNAm of TSSs subject to XCI was not significantly correlated with AI, implying that once a certain level of DNAm was present, silencing was maintained but not enhanced by more DNAm (red squares, Fig. 2B).
Figure 2.
Female DNAm at TSSs shows a continuous range from subject to escape. (A) The average level of female DNAm in PB ranked from highest to lowest. XCI status of each TSS is denoted by colour (red: subject, purple: variable escape, grey: uncallable and green: escape) with error bars representing 1 SD. (B) Linear regression of AI and average female PB DNAm (red: subject, and green: escape). (C) Linear regression of average female DNAm on PB and BU. (D) Variable escape TSSs rarely disagreed in XCI status within a twin pair. A comparison of the DNAm within twin pairs and coloured according to WB twin XCI status revealed a high level of agreement in average twin TSS DNAm. Most TSSs had the same XCI status within a twin pair [subject in both: solid red circle, uncallable in both (solid grey circle) or escape in both (solid green circle)] or were uncallable in one twin but subject (open red circle) or escaped (open green circle) in the other twin. A limited number of variable escape TSSs were subject in one twin but escaped from XCI in the other (solid black circle). R2 and P-values for each linear regression are given on their respective graphs.
Variability in XCI status largely attributable to cis-acting factors with substantial genetic component
As the level of promoter DNAm was not correlated with the relative expression level from the Xi at subject TSSs, we questioned whether DNAm might be stochastic at these TSSs and therefore performed a linear regression of the average female DNAm in PB versus the next most abundant tissue (buccal; BU). This comparison of these unmatched samples revealed an extremely high level of correlation between female DNAm levels across tissues for hundreds of females (R2 = 0.9480, P-value <0.0001) (Fig. 2C) suggesting underlying genetic and epigenetic features may contribute to the extent of DNAm for individual TSSs. To help elucidate features that might be driving DNAm, TSSs that demonstrate variable escape from XCI within a single tissue were used to compare subject and escape at the same genomic location. Factors that underlie variable escape from XCI might be chromosome specific (i.e. cis-acting) or female specific, in which case they would likely be trans-acting. Our data suggested that cis-acting factors are of greater impact than trans-acting factors because: (i) we did not observe consistent bimodal patterns of DNAm for variably escaping TSSs and (ii) we did not see enrichment for the expression from the Xi of multiple TSSs that show variable escape from XCI in subsets of females, which is consistent with previous reports (1).
A cis-acting effect could be genetic or owing to stochastic events at the individual gene level. Monozygotic twins provide a classic means of differentiating between heritable (genetic) and environmental influences as these twins share the same genotype. Two 450K DNAm studies in whole blood (WB) contained female monozygotic twins, so we compared the XCI status of all TSSs and identified 51 TSSs with variable escape from XCI across the 62 twin samples (31 pairs). DNAm at 24 of 31 twin pairs was most highly correlated with their respective twin compared with the other twins at these variable escape TSSs (Supplementary Material, Table S3). Overall, concordant XCI statuses between twins (solid red, green or grey dots in Fig. 3D) were significantly higher than expected by chance (χ2 analysis, P-value <0.0001), and only 3% of variable escape WB twin TSSs were subject to XCI in one twin and escaped from XCI in the other, suggesting a substantial contribution owing to genotype, but demonstrating that genotype does not explain all of the inter-individual variation observed in XCI status.
Figure 3.
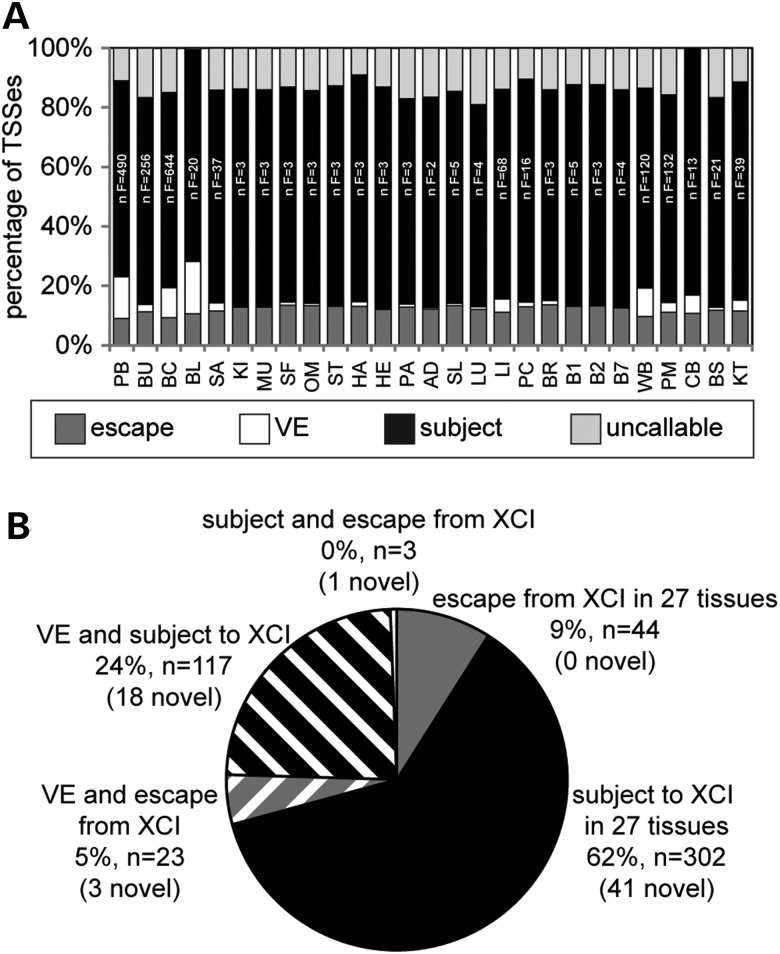
The majority of TSSs have the same XCI status in all examined tissues. (A) XCI status: escape (dark grey), variable escape (VE) (white), subject (black) and uncallable (light grey) in 27 tissues. Two letter tissue codes are found in Supplementary Material, Table S2 along with the complete tissue names. The number of female samples per tissue (n F) is given within each subject bar. (B) XCI status of the 489 TSSs that are informative in all 27 tissues, with the number of TSSs that had not been examined previously for XCI status listed in brackets (novel).
As an individual ages, an accumulation of environmental exposures acts in conjunction with the intrinsic process of aging itself, which has been suggested to result in increased changes in DNAm. We used a dataset of buffy coat samples to examine the impact of age on X-linked DNAm and XCI status, as this dataset was the largest in which age was documented in GEO (female: n = 88, male: n = 23). The XIST gene can be reactivated by loss of DNAm (34); however, we observed no significant correlation between age (in either males or females) and the average DNAm level of the XIST promoter (Supplementary Material, Fig. S5). We further found no correlation between age (47 to 92 years) and the average level of TSS DNAm or the overall female-specific level of escape from XCI in individual females, nor did we find any TSS that changed from an XCI status of subject or escape between the 10 youngest and the 10 oldest females. Previous studies (35–37) have found individual X-linked CpGs to show an age effect; however, a comparison of DNAm failed to identify a change in DNAm with age in either males or females for the 42 CpGs previously identified to show an age effect (Supplementary Material, Table S4). We therefore concluded that the effect of aging on X-linked DNAm and by extension, XCI status was negligible and unlikely the cause of variability in XCI statuses observed between females.
XCI status is consistent across 27 tissues for over 70% of TSSs
XCI status was determined across 27 tissues from a total of 1875 females (Fig. 3A and Supplementary Material, Table S5). Four hundred and eighty-nine TSSs were informative in all 27 tissues and, dramatically, for 71% of these TSSs, the same XCI status (subject: 62%, escape 9%) was observed in all 27 tissues (Fig. 3B). For the remaining TSSs that differed in XCI status across tissues (n = 143), most were subject to XCI (n = 117) or escaped from XCI (n = 23) in the majority of tissues but were either uncallable or showed variable escape from XCI in the remaining tissues. There were only three TSSs (CTPS2, WBP5 and LOC100129662) for which a call of subject was made in one or more tissues whereas a call of escape was made in one or more of the other tissues. For the CTPS2 gene, a designation of tissue-specific escape from XCI had also been made in a previous study of allelic expression (2); however, only one of three alternative TSSs was called subject to the inactivation in this study, and this call was made in only one of the multiple blood-derived tissue sets. The WBP5 gene had previously been called as a variable escape gene; however, the one tissue in which it was thought to be subject to XCI had only four female samples. Thus, we believe that it is more likely that WBP5 is a variable escape gene than a tissue-specific escape gene. The newly assessed LOC100129662 TSS was called escape only in blood (BL); in contrast to WBP5, there were 20 female samples in that set, and so such a call seems unlikely to have been by chance. In other tissues, the call was subject (15 tissues) or uncallable (11 tissues). Thus, overall, we see very limited evidence for complete tissue-specific escape from inactivation, rather variable escape that is frequently restricted to only a subset of the tissues examined.
The proportion of TSSs that escape from XCI was relatively consistent between tissues at 9–14%. The highest degree of escape from XCI was observed in brain (BR) whereas the lowest was in PB, and the five tissues with the least number of genes that escape from XCI were all derived from blood. These blood-derived tissues also showed the highest proportion of variable escape TSSs. The number of TSSs that were classed as variable escape from XCI differed considerably (from 0 to 18%), with 20 of the 27 tissues having 3% or lower TSSs with a call of variable escape from XCI. After cell composition analysis was performed on blood (BL), the degree of variable escape dropped from 18 to 12%, which is still substantially higher than the majority of other tissues (data not shown). This suggests that while individual differences in cell composition may in part contribute to the high level of variable escape from XCI in blood, heterogeneous cell composition cannot fully account for the much higher proportion of variable escape TSSs compared with other tissues. Consistent with previous studies, there were several continuous regions of genes that escape from XCI, particularly on the short arm of the X chromosome. However, single genes that escape from XCI were also observed, on both the short and long arm of the X chromosome. In total, 18 regions (containing from 1 to 14 TSSs) were called as predominantly escape from XCI across the 27 tissues (Supplementary Material, Table S5).
By increasing the number of TSSs, and by extension genes, for which we know an XCI status, we will be better able to study other aspects of XCI such as the spreading and maintenance of XCI and examining the role that genes that escape from XCI play in phenotypic variation between the sexes and between females. An XCI status was predicted for 63 TSSs where no previous expression data were available (labelled novel in Fig. 3B, the complete list of TSSs and their associated XCI statuses across tissues can be found in Supplementary Material, Table S5). The 63 TSSs with a novel XCI status were from 56 genes that were distributed along the length of the X chromosome.
X-linked DNAm is associated with expression levels at CpG island promoters and inactive chromatin
Extensive gene body-specific DNAm has been reported for the Xa (28), consistent with some (e.g. 17), but not all (e.g. 18), studies of gene body DNAm at transcriptionally active or silent genes on the autosomes. To evaluate the possible effect of transcription on X-linked DNAm, we initially examined the correlation between genic expression levels and DNAm on the Xa by complementing the 450K PB DNAm data with expression data from a different set of GEO PB samples (GSE32280) and creating Xa expression quartiles using male expression levels. While the 450K probes are concentrated in promoters, we used the functional chromatin states (20) to separate CpGs into promoters, enhancers, transcribed regions and repressed and heterochromatic regions, with further subdivision by CpG density (Fig. 4 A, B, C and D, respectively). The promoters showed more Xa DNAm at the lowest expression quartile compared with the highest expression quartile (Fig. 4A); however, the generally low numbers of probes in enhancers, insulators and transcribed regions yielded no significant difference in Xa DNAm by expression quartile, whereas repressed and heterochromatic regions showed significantly more methylation at highly expressed genes at high CpG density but not at lower CpG densities (Fig. 4), yielding an inconclusive answer to the impact of transcription on gene body DNAm. We further examined the impact of transcription by comparing intragenic and intergenic regions across the X chromosome (Supplementary Material, Table S6). In general, intragenic CpGs showed higher DNAm than intergenic CpGs on the Xa, and no difference on the Xi; however, these comparisons were confounded by the fact that the distance of CpGs from the closest TSS differed greatly between intragenic and intergenic CpGs, with 81% of intergenic high-density CpGs within 1 kb of the closest TSS compared with only 23% of intragenic CpGs. Regardless of location or CpG density, CpGs within 1 kb of the closest TSS had lower DNAm (average DNAm of 22%) than CpGs farther from the TSS (average DNAm of 79%). Therefore, to compare the impact of gene transcription, we instead compared the intergenic transcription levels between CpGs within genes that escape from XCI and those subject to XCI (Fig. 4E). Gene bodies associated with TSSs that escape from XCI had significantly more Xi DNAm than gene bodies of genes subject to XCI (73 versus 67%, P-value < 0.0001). The regions of the X chromosome between genes demonstrated a difference between Xa and Xi DNAm similar to that seen at gene bodies of subject genes. Figure 4E summarizes the levels of DNAm seen at promoters for the Xa and Xi from the 10 584 CpGs on the 450K array across 2928 individuals.
Figure 4.
Transcription level has significant correlation with promoter DNAm as well as heterochromatin DNAm. Box and whisker plots comparing the average male (Xa) DNAm (PB) of the genes from the highest expression quartile and the lowest expression quartile by CpG density class (HIC, IC, LC; Ref. 22). (A) Promoters, (B) enhancers and insulators, (C) transcribed regions of genes and (D) repressed and heterochromatic regions were defined based on the chromatin states in Ref. 20. Significance based on a Wilcox test comparison of means is as follow: *P-values 0.05–0.001, **P-values 0.001–2.2E − 16 and ***P-values < 2.2E − 16). (E) Summary of X-linked DNAm patterns. The Xa and Xi DNAm patterns of females are summarized for genes that are subject to XCI and genes that escape from XCI. The average Xa and Xi DNAm values given are an average of all CpGs used in this analysis within the denoted category. The presence of individual CpGs (ovals) is meant only as a graphical representation and does not accurately denote CpG density. Chromatin states were combined with intergenic and intragenic data to separate CpGs within the gene body from CpGs between genes.
Discussion
A major goal of this study was to utilize DNAm to predict genes subject to XCI. Past expression-based studies (1,2) allowed the development of a training set of subject and escape genes to test whether the difference in DNAm between males and females would allow the calling of XCI status. Of the 489 TSSs that were informative across all 27 tissues examined, 63, or almost 13%, had not previously been assigned an inactivation status by transcriptional analyses, demonstrating that DNAm is an important complementary approach to determining XCI status for genes. Previous analyses (7,38) have shown tissue-specific differences in XCI in which a gene was subject to XCI in one tissue but escaped from XCI in another; however, this analysis of 27 tissues identified only three TSSs that showed this extreme level of tissue-specific XCI. Instead, differences in XCI status between tissues tended to involve a subset of tissues showing variable escape from XCI whereas the majority were either subject to XCI or escaped from XCI. The differences in the frequency of tissue-specific XCI observed between this study and our previous DNAm analysis using the Illumina Infinium HumanMethylation27 BeadChip (27K array) was likely due to more robust predictions with the 450K array, where there was an average of seven CpGs per TSS, whereas the 27K array only had an average of two CpGs per TSS. Furthermore, nearly one-third of genes previously (7) found to show tissue-specific XCI on the 27K array were within the uncallable range in this study. Therefore, we believe that the expanded number of CpGs used to determine XCI status in combination with a more stringent set of DNAm thresholds yields a better estimate of XCI status. While the 450K array provides robust predictions and a large public repository of data, it is also limited in the distribution of CpGs. Furthermore, recently identified methyl-cytosine derivatives such as hydroxyl-methyl-cytosine would not be distinguished from 5-methyl-cytosine, so our analysis was not sensitive to differential proportions of such variants, which have been reported to be enriched in brain (39).
By comparing the level of DNAm for TSSs against previous expression analyses (2), we were able to demonstrate that there was an average Xi DNAm level of ∼50% above which TSSs appeared to always be subject to XCI. Having more DNAm did not make a TSS more silent, or less likely to show variable escape from XCI. Below ∼15% Xi DNAm, TSSs were predicted to escape from XCI, and we observed a relationship between promoter DNAm and the predicted expression level from the Xi relative to the Xa (2). We described TSSs as ‘escaping’ from XCI; however, it is unclear whether these genes truly avoid XCI or are initially subject to XCI and then undergo reactivation, as the limited mice studies would suggest (40). Additional studies in mice have suggested that DNAm of the promoters of genes subject to XCI occurs several days after XCI is established (41). Therefore, if escape from XCI is truly an escape rather than reactivation, it is likely that the ongoing expression of these genes contributes to their avoidance of DNAm. Alternatively, if escape from XCI is in fact caused by reactivation, then an initial lack of stable DNAm may contribute to their unstable silencing. Further experiments examining the early stages of XCI will be necessary to evaluate the complex relationship between a lack of DNAm and escape from XCI.
We also continue to observe a class of variable escape TSSs that were subject to XCI in some females and escaped in others. Variable escape TSSs were found to have average levels of DNAm that ranged from subject to escape (Fig. 2A), although they were enriched near the thresholds of DNAm of subject and escape TSSs. While we cannot exclude the influence of trans-acting factors such as polymorphisms in the DNAm machinery proteins DNMT3B, DNMT1o and SMCHD1 that are involved in the establishment of X-linked DNAm (42–44), we favour a predominantly cis-effect for two reasons. First, female DNAm of variable escape TSSs was normally distributed and not bimodally distributed, which would have been the expected pattern of DNAm with a trans-effect. Second, the DNAm of twins for variable escape TSSs supported the existence of a genetic component to XCI status.
In addition to using DNAm to predict XCI, it may also be possible to identify a DNAm pattern supporting the presence of human X-linked imprinted genes. The study of X chromosome monosomy (45,46) has not identified such genes in humans, although mouse has been shown to have X-linked imprinted genes (47). We hypothesized that the DNAm of imprinted genes would differ depending on the parental imprinting status. If the maternal X was hypomethylated, then males, who have only a maternal X, would be unmethylated. Females would have intermediate DNAm in situations where the paternal X was always the Xi or would be fully methylated if the maternal X was always the Xi. The former pattern would resemble the normal pattern for subject genes; however, the latter pattern should be distinctive. Thus, we compared the DNAm patterns of highly skewed female lymphoblast samples against females with random XCI and found no evidence for a bimodal distribution or shift towards hypermethylation (data not shown). Alternately, if the maternal X was hypermethylated for an imprinted gene, then males would show high DNAm and females intermediate levels, similar to the pattern of DNAm found at the XIST intermediate CpG density promoter. We therefore searched for additional X-linked CpGs that showed a pattern of DNAm similar to XIST and found only three additional genes (GPM6B, ARX and BCOR) (27), which had at least three CpGs that demonstrated an ‘XIST-like’ pattern of DNAm; however, again none of these genes showed a bimodal DNAm pattern in the highly skewed female lymphoblast samples. In the absence of imprinting, it is unknown why these genes show male hypermethylation. While XIST was initially described as being unique in its expression solely from the Xi, recent reports have found three [XIST, LOC286467/FIRRE (48) and LOC550643 (49)] female-specific X-linked DNase Hypersensitive sites across the X chromosome. Further examination of the DNAm at the promoters of FIRRE and LOC550643 found that FIRRE had two CpGs with ‘XIST-like’ DNAm whereas LOC550643 was hypomethylated in both males and females.
An additional goal of this study was to compare the DNAm landscape across the euchromatic Xa and the heterochromatic Xi. CpG density is known to be an important factor in determining DNAm, and CpG island shores have been shown to contain many of the CpGs that show differential DNAm between tissues or cancers (23,24). We therefore integrated CpG density {using both [HC, IC and LC classification of (22)] and the Illumina/UCSC definition of a CpG island [island, shore, shelf and sea]}, with functional chromatin states (20), revealing a set of CpGs, including non-island promoters as well as island enhancers, which show female hypermethylation and are useful for assessing XCI status of a gene using DNAm. We found only limited evidence supporting that active transcription on the X chromosome resulted in increased DNAm in gene bodies (Fig. 4E); however, overall female DNAm was ∼4% lower than that of males for the heavily methylated CpGs, suggesting that the Xi is hypermethylated relative to the Xa. Interestingly, we see a similar difference between the Xa and Xi in intergenic regions as we do at gene bodies of genes subject to XCI, but not those that escape from XCI. Perhaps retention of the differential DNAm outside of gene bodies reflects more widespread transcription of the genome (e.g. 50), although it may also reflect a general increased accessibility for the DNAm machinery of the euchromatic Xa relative to the heterochromatic Xi. Previous chromosome-wide DNAm studies have yielded contradictory reports about the extent of DNAm on Xi relative to Xa, HhaI restriction enzyme analysis suggested that the Xi was hypomethylated compared with the Xa (29), whereas in situ nick translation analysis found the Xi to be hypermethylated compared with the Xa (30), and no difference was observed through antibody staining (31). Consistent with previous genome-wide studies (13), we observed that promoter DNAm on the Xa was negatively correlated with gene expression levels and that higher CpG density was associated with lower average DNAm. On the Xi, there was less variability in DNAm, with larger effects attributable to functional chromatin state than CpG density.
To integrate chromatin marks, we used the male H1 embryonic stem cell chromatin states as these would be most reflective of the Xa chromatin status at the time of XCI; however, this male line lacks the two X chromosomes present at the time of inactivation and also lacks the Xi found in female somatic cells. Other chromatin state datasets, including from the female lymphoblast cell line GM12878, show a shift towards more active promoters, but regardless of the dataset used, the bulk of the X chromosome is comprised of states reflecting transcribed and repressed chromatin. It should also be considered that the chromatin state algorithms, developed predominantly from disomic autosomes, may not behave as robustly for the dimorphic sex chromosomes. Overall, however, while differences in chromatin state occur between cell lines, the dramatic difference in promoter DNAm from the Xa to the Xi for subject genes provides a surrogate marker for inactivated genes, whereas the majority of the X chromosome is not promoter sequences but rather gene body or intergenic sequences, which show higher levels of DNAm on the Xa than the Xi for genes subject to XCI (see Fig. 4E).
This analysis of X-linked DNAm of 1875 females, the largest to date, has provided insight into both the DNAm landscape of the X chromosome and also demonstrated remarkable stability across tissues with 62% of TSSs subject to XCI in all tissues and 9% of TSSs demonstrating escape from XCI in all 27 tissues examined. XCI status across tissues was determined to be more consistent than previously found, suggesting that deeper DNAm analysis provides more robust XCI status predictions. We have also demonstrated that the integration of CpG density, chromatin marks and DNAm can be more powerful than CpG density or histone marks alone for the assessment of XCI status. The XCI status of 63 TSSs that had not been previously examined was determined, contributing towards a chromosome-wide list of XCI statuses which will be useful both in studies of conditions that differ between males and females and also in continuing the study of how XCI is established and maintained. XCI offers a naturally occurring system in which to study the spread of silencing and more broadly it can provide insight into the general means by which epigenetic features influence silencing across the genome. We suggest that variability in XCI status likely has a genetic component but is also affected by environment. The heterochromatic Xi showed less DNAm than the euchromatin Xa in the intra- and intergenic regions of the X chromosome; however, in the gene bodies of genes that escape from XCI, equal Xa and Xi hypermethylation was observed, suggestive of a slight enrichment of DNAm owing to transcription. Beyond the importance to studying XCI, these findings highlight the differences in euchromatin and heterochromatin and the complex interplay between DNAm, chromatin modification and their ultimate relationship with expression.
Materials and Methods
GEO DNAm data
All GEO data series which used the Illumina Infinium HumanMethylation450 BeadChip platform (GPL13534) were searched through the GEO website (http://www.ncbi.nlm.nih.gov/geo/) and considered for this analysis (32). Criteria for inclusion were as follows: list date before 1 January 2014 and the publication of beta values for all 485 577 probes (thus ensuring the presence of probes on the X chromosome). A complete list of all data series used in this study can be found in Supplementary Material, Table S7. Sample information, including tissue type and the two letter codes used for each tissue type and the sex of each sample, can be found in Supplementary Material, Table S8. Additionally, the X chromosome data associated with a previously unpublished dataset were used in this study and deposited in GEO (Series ID: GSE60275; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60275). Of the 11 133 probes annotated by Illumina as being located on the X chromosome, only the 10 584 which did not also map to the autosomes (22) were used for analysis in this study. Individual samples were removed if ≥5% of X-linked CpG beta values were missing and/or if ≥1% of X-linked CpG beta values were associated with a P-value of >0.01. A total of 44 samples were removed owing to these criteria. Raw beta values for all remaining samples then underwent colour correction using the Bioconductor lumi package (51) and SWAN normalization (52).
Sexing samples using DNAm
In many of the GEO datasets, the sex of individual samples was not provided; thus, to confirm the sex of all samples and to assign a sex to samples of unknown sex, multiple unsupervised hierarchical clusterings were performed using the hclust function (stats) in R. In the first round, hierarchical clustering was performed using only the 8527 probes which uniquely mapped to the X chromosome (22) whereas in the second round of clustering, only those five CpGs (cg03554089, cg12653510, cg05533223, cg11717280 and cg20698282) associated with the XIST IC were used. Within a cluster, samples with known sex were compared with those with unknown sex to assign a sex to samples of unknown sex. A sex was not assigned in clusters where <80% of given sexes were consistent. Ten GEO series were excluded from further analysis as a sex could not able to be assigned to the majority of samples (listed in Supplementary Material, Table S7). Of the remaining series, 2959 samples were determined to be either male (n = 1053) or female (n = 1906) based on hierarchical clustering. For 14 samples, the sex was listed as female in GEO phenotypic data; however, DNAm was consistent with the sample being male, suggesting either these samples represent individuals with Turner syndrome or that these samples were incorrectly labelled as female. For another 14 samples, the sex was listed within GEO as being male but the DNAm patterns were consistent with a female sex, suggestive of Klinefelter syndrome (47, XXY) or a misclassification of females as males. All potential Turner and Klinefelter samples were excluded from our analysis. The final sex assignment for each sample, along with the corresponding GEO sample ID, can be found in Supplementary Material, Table S8.
Broad chromatin state analysis
The ENCODE Broad Chromatin State Segmentation track from the H1-hESC cells (hg19) was downloaded through Galaxy (https://usegalaxy.org/) and an intersection performed with the basepair (hg19) location of each target CpG from the 450K array. Violin plots of Xa and Xi DNAm were generated using the vioplot and vioplot2 functions from the vioplot package in R (53). Xi DNAm was calculated using the formula [(female DNAm × 2) − (male DNAm)].
Statistical analysis
The kernel density plot in Figure 1A was generated using the kde2d function from the MASS package in R (54). All statistical analysis was performed using R (version 3.0.3) with listed P-values corrected for multiple comparisons using a standard Bonferroni correction. Statistical comparisons of means were performed using the wilcox.test function from the stats package whereas distributions were compared using the ks.test (Kolmogorov–Smirnov test) function. Determining outlier females based on level of escape from XCI was done using Iglewica and Hoaglin's outlier test (http://contchart.com/outliers.aspx), two sided with a modified Z score cut-off of 3.5 to identify females who were outliers with respect to the percentage of TSSs that showing escape from XCI. Those females (n = 30) identified as outliers are listed in Supplementary Material, Table S9 and were excluded from further analysis as the increased degree of escape from XCI appeared to occur in samples with a consistent, chromosome-wide, lower level of DNAm, which raised concerns of more severe disruptions in DNAm.
Two WB data series (GSE53128 and GSE37965) contained twin female samples and were analysed separately to create a list of genes that showed variable escape from XCI. A χ2-analysis comparing the number of concordant (subject in both twins, escape in both twins or uncallable in both twins) to discordant (subject in one twin but escape in the other) and semi-concordant (all other combinations) TSSs was then performed using GraphPad Prism.
Expression analysis
PB expression data from a publically available dataset (GSE32280, Affymetrix Human Genome U133 Plus 2.0 Array) were downloaded for eight healthy control samples (male: n = 4, female: n = 4). 1400 sites were associated with X-linked genes for which there was also 450K array DNAm data, and these were divided into quartiles based on the average level of male expression.
DNAm and the relationship to distance from the closest TSS
Previously published (1,21) analysis of X-linked expression was combined to generate two training sets used to predict XCI status. For each CpG, the XCI status associated with the closest TSS was determined and only those TSSs at which all informative studies agreed on the XCI status were included (Supplementary Material, Table S10). Over half (subject: 59%, escape: 54%) of all TSSs within both training sets had previously been examined by at least two of the three previous expression studies. After CpGs that had male DNAm of >25% were excluded (PB, n = 558) to avoid hypermethylated genes such as the cancer-testes family (33), the 2398 CpGs in the subject training set and the 148 CpGs in the escape training set were translated into a moving average of 14 CpGs (1/10 of the size of the escape training set). In both training sets, the moving average was calculated even at the seven CpGs farthest from the TSS (both up and down stream) but is shown as a dotted rather than solid line in Supplementary Material, Figure S4. All CpGs in the escape training set were located ±1 Kb from the TSS making it impossible to make any conclusions about the relationship between DNAm and distance to the TSS beyond this range. Only CpGs of −500–1000 bp surrounding the TSS were therefore used to predict XCI status.
DNAm ranges used to predict XCI status
To predict XCI status, individual CpGs associated with the subject training set or escape training set were averaged together to create a single DNAm average associated with each TSS. Within the escape training set, a mean of seven CpGs were combined for each TSS average, whereas on average, six CpGs were combined in the subject training set. The SD of this average DNAm for females and males was then calculated along with a sex DNAm delta (difference between female and male DNAm). Two criteria were used to call a TSS subject to XCI. First, average female DNAm needed to be within a range defined by the subject training set. The minimum boundary was set as the mean of all subject training TSS minus 2 SD, and the maximum boundary was set as the mean of all subject training TSS plus 2 SD plus a constant of 25% beta value. Second, the sex DNAm delta had to be within a range defined by the average sex DNAm delta in the subject training set, with the boundaries set at ±2 SD of the average of all informative sex DNAm deltas in the subject training set.
Before establishing escape DNAm ranges for TSSs that escaped from XCI, the subject criteria defined earlier were first applied to TSSs in the escape training set. Any escape training set TSSs that fell within the subject DNAm range for both female and sex DNAm delta (note the three pink squares in Supplementary Material, Fig. S4C) were excluded from the establishment of escape DNAm ranges. Escape criteria were 2-fold. First, the average female DNAm needed to be within a range defined by the escape training set, with the minimum boundary set at a constant 0% and the maximum boundary set as the mean of all escape training TSSs plus 2 SD. Second, the sex DNAm delta had to be within a range defined by the average sex DNAm delta in the escape training set, with the boundaries set at ±2 SD of the average sex DNAm deltas in the escape training set.
Using the subject and escape criteria defined earlier, 100% of TSSs within the subject training were predicted to be subject to XCI in addition to three TSSs that had been included in the escape training set. These three TSSs suggest tissue-specific XCI status between PBs and the tissues used to generate the training sets. Eighty-nine per cent (n = 17) of the remaining TSSs in the escape training set were predicted to escape from XCI, whereas 11% (n = 2) were uncallable. Supplementary Material, Table S2 lists the DNAm threshold ranges used to call XCI for each tissue. If the female DNAm fell in the uncallable range, an XCI status of subject or escape was called based on the XCI of the sex DNAm delta alone. This was also true if the sex DNAm delta was uncallable but the female DNAm had an XCI status. In extremely rare cases (0.01% in PBs), the female DNAm level and the sex DNAm delta predicted different XCI statuses; we considered these cases to be uncallable.
Supplementary Material
Funding
This work was supported by the Canadian Institutes of Health Research (MOP-13690 and MOP-119586 to C.J.B.). E.M.P. is funded by a Canadian Institutes of Health Research Doctoral Frederick Banting and Charles Best Canada Graduate Scholarship. M.J.J. is supported by a Mining for Miracles post-doctoral fellowship from the Child and Family Research Institute. Funding to pay the Open Access publication charges for this article was provided by grants from the Canadian Institutes of Health Research (MOP-13690 and MOP-119586 to C.J.B.).
Supplementary Material
Acknowledgements
We thank members of the Kobor lab Sarah Goodman, Lucia Lam, Sarah Newmann, Julia MacIsaac and Sarah Mah for the running of the 450 K arrays, Rachel Edgar and Elodie Portales-Casamar for initial discussion of GEO data analysis and members of the Brown lab for additional insight and thoughtful discussion.
Conflict of Interest statement. None declared.
References
- 1.Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 2.Cotton A.M., Ge B., Light N., Adoue V., Pastinen T., Brown C.J. Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol. 2013;14:R122. doi: 10.1186/gb-2013-14-11-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellott D.W., Hughes J.F., Skaletsky H., Brown L.G., Pyntikova T., Cho T.J., Koutseva N., Zaghlul S., Graves T., Rock S., et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng X., Berletch J.B., Nguyen D.K., Disteche C.M. X chromosome regulation: diverse patterns in development, tissues and disease. Nat. Rev. Genet. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J., Deng X., Disteche C.M. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS ONE. 2008;3:e2553. doi: 10.1371/journal.pone.0002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peeters S.B., Cotton A.M., Brown C.J. Variable escape from X-chromosome inactivation: identifying factors that tip the scales towards expression. BioEssays. 2014;36:746–756. doi: 10.1002/bies.201400032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotton A.M., Lam L., Affleck J.G., Wilson I.M., Penaherrera M.S., McFadden D.E., Kobor M.S., Lam W.L., Robinson W.P., Brown C.J. Chromosome-wide DNA methylation analysis predicts human tissue-specific X inactivation. Hum. Genet. 2011;130:187–201. doi: 10.1007/s00439-011-1007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen D.K., Disteche C.M. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 9.Sharp A.J., Stathaki E., Migliavacca E., Brahmachary M., Montgomery S.B., Dupre Y., Antonarakis S.E. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011;21:1592–1600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michels K.B., Binder A.M., Dedeurwaerder S., Epstein C.B., Greally J.M., Gut I., Houseman E.A., Izzi B., Kelsey K.T., Meissner A., et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat. Methods. 2013;10:949–955. doi: 10.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- 11.Saxonov S., Berg P., Brutlag D. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 13.Wagner J.R., Busche S., Ge B., Kwan T., Pastinen T., Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014;15:R37. doi: 10.1186/gb-2014-15-2-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrlich M., Gama-Sosa M.A., Huang L.-H., Midgett R.M., Kuo K.C., McCune R.A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucl. Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hackenberg M., Previti C., Luque-Escamilla P.L., Carpena P., Martinez-Aroza J., Oliver J.L. CpGcluster: a distance-based algorithm for CpG-island detection. BMC Bioinf. 2006;7:446. doi: 10.1186/1471-2105-7-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lokk K., Modhukur V., Rajashekar B., Martens K., Magi R., Kolde R., Kolt Ina M., Nilsson T.K., Vilo J., Salumets A., et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014;15:R54. doi: 10.1186/gb-2014-15-4-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huh I., Zeng J., Park T., Yi S.V. DNA methylation and transcriptional noise. Epigenet. Chrom. 2013;6:9. doi: 10.1186/1756-8935-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lou S., Lee H.M., Qin H., Li J.W., Gao Z., Liu X., Chan L.L., Lam V., So W.Y., Wang Y., et al. Whole-genome bisulfite sequencing of multiple individuals reveals complementary roles of promoter and gene body methylation in transcriptional regulation. Genome Biol. 2014;15:408. doi: 10.1186/s13059-014-0408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothbart S.B., Strahl B.D. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta. 2014;839:627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M., et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cotton A.M., Chen C.Y., Lam L.L., Wasserman W.W., Kobor M.S., Brown C.J. Spread of X-chromosome inactivation into autosomal sequences: role for DNA elements, chromatin features and chromosomal domains. Hum. Mol. Genet. 2014;23:1211–1223. doi: 10.1093/hmg/ddt513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price M.E., Cotton A.M., Lam L.L., Farre P., Emberly E., Brown C.J., Robinson W.P., Kobor M.S. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenet. Chrom. 2013;6:4. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irizarry R.A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P., Cui H., Gabo K., Rongione M., Webster M., et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doi A., Park I.H., Wen B., Murakami P., Aryee M.J., Irizarry R., Herb B., Ladd-Acosta C., Rho J., Loewer S., et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zykovich A., Hubbard A., Flynn J.M., Tarnopolsky M., Fraga M.F., Kerksick C., Ogborn D., MacNeil L., Mooney S.D., Melov S. Genome-wide DNA methylation changes with age in disease-free human skeletal muscle. Aging Cell. 2014;13:360–366. doi: 10.1111/acel.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber M., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 27.Joo J.E., Novakovic B., Cruickshank M., Doyle L.W., Craig J.M., Saffery R. Human active X-specific DNA methylation events showing stability across time and tissues. Eur. J. Hum. Genet. 2014;22:1376–1381. doi: 10.1038/ejhg.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellman A., Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 29.Viegas-Pequignot E., Dutrillaux B., Thomas G. Inactive X chromosome has the highest concentration of unmethylated Hha I sites. Proc. Natl. Acad. Sci. USA. 1988;85:7657–7660. doi: 10.1073/pnas.85.20.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prantera G., Ferraro M. Analysis of methylation and distribution of CpG sequences on human active and inactive X chromosomes by in situ nick translation. Chromosoma. 1990;99:18–23. doi: 10.1007/BF01737285. [DOI] [PubMed] [Google Scholar]
- 31.Miller D.A., Okamoto E., Erlanger B.F., Miller O.J. Is DNA methylation responsible for mammalian X chromosome inactivation? Cytogenet. Cell Genet. 1982;33:345–349. doi: 10.1159/000131782. [DOI] [PubMed] [Google Scholar]
- 32.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucl. Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Smet C., Lurquin C., Lethe B., Martelange V., Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol. Cell. Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tinker A.V., Brown C.J. Induction of XIST expression from the human active X chromosome in mouse/human somatic cell hybrids by DNA demethylation. Nucl. Acids Res. 1998;26:2935–2940. doi: 10.1093/nar/26.12.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D., Liu X., Zhou Y., Xie H., Hong X., Tsai H.J., Wang G., Liu R., Wang X. Individual variation and longitudinal pattern of genome-wide DNA methylation from birth to the first two years of life. Epigenet. 2012;7:594–605. doi: 10.4161/epi.20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alisch R.S., Barwick B.G., Chopra P., Myrick L.K., Satten G.A., Conneely K.N., Warren S.T. Age-associated DNA methylation in pediatric populations. Genome Res. 2012;22:623–632. doi: 10.1101/gr.125187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bocklandt S., Lin W., Sehl M.E., Sanchez F.J., Sinsheimer J.S., Horvath S., Vilain E. Epigenetic predictor of age. PLoS ONE. 2011;6:e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talebizadeh Z., Simon S.D., Butler M.G. X chromosome gene expression in human tissues: male and female comparisons. Genomics. 2006;88:675–681. doi: 10.1016/j.ygeno.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin S.G., Wu X., Li A.X., Pfeifer G.P. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucl. Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lingenfelter P.A., Adler D.A., Poslinski D., Thomas S., Elliot R.W., Chapman V.M., Disteche C.M. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat. Genet. 1998;18:212–213. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- 41.Lock L.F., Takagi N., Martin G.R. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- 42.Shen L., Kondo Y., Guo Y., Zhang J., Zhang L., Ahmed S., Shu J., Chen X., Waterland R.A., Issa J.P. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mould A.W., Pang Z., Pakusch M., Tonks I.D., Stark M., Carrie D., Mukhopadhyay P., Seidel A., Ellis J.J., Deakin J., et al. Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenet. Chrom. 2013;6:19. doi: 10.1186/1756-8935-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gendrel A.V., Apedaile A., Coker H., Termanis A., Zvetkova I., Godwin J., Tang Y.A., Huntley D., Montana G., Taylor S., et al. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev. Cell. 2012;23:265–279. doi: 10.1016/j.devcel.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bondy C.A., Hougen H.Y., Zhou J., Cheng C.M. Genomic imprinting and Turner syndrome. Ped. Endocrin. Rev. 2012;9:728–732. [PubMed] [Google Scholar]
- 46.Lepage J.F., Hong D.S., Hallmayer J., Reiss A.L. Genomic imprinting effects on cognitive and social abilities in prepubertal girls with Turner syndrome. J. Clin. Endocrin. Metab. 2012;97:E460–E464. doi: 10.1210/jc.2011-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raefski A.S., O'Neill M.J. Identification of a cluster of X-linked imprinted genes in mice. Nat. Genet. 2005;37:620–624. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- 48.Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D.G., Sauvageau M., Kelley D.R., et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheffield N.C., Thurman R.E., Song L., Safi A., Stamatoyannopoulos J.A., Lenhard B., Crawford G.E., Furey T.S. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res. 2013;23:777–788. doi: 10.1101/gr.152140.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinf. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 52.Maksimovic J., Gordon L., Oshlack A. SWAN: subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hintze J.L., Nelson R.D. Violin plots: a box plot-density trace synergism. Amer. Stat. 1998;52:181–184. [Google Scholar]
- 54.Venables W.N., Ripley B.D. Modern Applied Statistics with S. New York: Springer; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.