Summary
Lentiviral gene transfer has a significant impact on the development of biomedical research. One of the most important features of lentiviruses is the capability to infect both dividing and nondividing cells. However, little is known whether integration preference exists, specifically in early embryos. An in-depth genome analysis on 112 independent lentiviral integration sites from 43 transgenic founder mice was performed to determine if there are preferable sites for lentiviral integration in early embryonic genome. Our results demonstrated that lentiviruses were biased in integrating within intragenic regions, especially in the introns. However, no integration preference was found associated with specific chromosomes, repetitive elements, or CpG islands, nor was there any preference for integrating at close proximity to transcription start sites. Our findings suggested that lentiviruses were biased to integrate into the intragenic regions of early embryonic genome of mouse.
Keywords: lentiviral vector, transgenesis, integration preference, genome, mouse zygote
INTRODUCTION
The applications of viral gene transfer have expanded dramatically in the past decades due to the improvement of safety features and the development of pseudotype technology (Burns et al., 1993; Temin, 1990; Yee et al., 1994). The development of a three vectors system for the production of lentivirus has greatly reduced the possibility of recombination, thus minimizing the risk of generating competent viruses (Perletti et al., 2004; Romano, 2005; Zufferey et al., 1998). Although competent retroviruses were first used to create transgenic mice in the 1970s (Jaenisch, 1976, 1983; Jaenisch et al., 1981), viral gene transfer was not widely used in biomedical research until the development of replication incompetent retrovirus in the 1980s (Shimotohno and Temin, 1981). However, the application of viral gene transfer in the creation of transgenic animals was not widely accepted until high transgenic rate was achieved by introducing pseudotyped retroviruses into the perivitelline space (PVS) of mature bovine oocytes and early embryos (Chan et al., 1998). Similar strategy has now been applied in the other species at high efficiency (Cabot et al., 2001; Chan et al., 1998, 2001; Lois et al., 2002; Yang et al., 2008). However, transgenic rate was significantly reduced when targeted zygotes instead of oocytes, which could be attributed to the presence of the nuclear envelope at the zygotic stage that delays the integration event (Chan et al., 1998). On the other hand, lentiviruses are capable of infecting dividing and nondividing cells, and are not affected by the presence of nuclear envelope (Yang et al., 2007).
Despite the high gene transfer rate, there are disadvantages that limit the application of lentiviruses. These include the capacity of lentiviral vector and the unknown integration pattern, which may affect endogenous gene functions and the expression of the trans-gene. Integration site has profound effect on the expression of transgene in animals, and is known as the position effects (Clark et al., 1994). Although copy number has been considered as an important factor on expression level, recent study in transgenic mice demonstrated that copy number of the transgene was not necessarily correlated with expression profile (Yang et al., 2007). Unlike pronuclear microinjection, a single copy of the transgene at each integration site is expected in retroviral and lentiviral gene transfer. However, the question of whether there is specific integration preference in early embryonic genome remains unknown.
Analysis on retroviral and lentiviral integration sites was mostly performed in vitro by infection of cultured cells or using sick animals caused by viral infection (Cousens et al., 2004; Lewinski et al., 2006; Nowrouzi et al., 2006). For example, human immunodeficiency virus (HIV) had shown strong integration preference for transcriptionally active genes in different cell lines (Mitchell et al., 2004); however, due to the limited number of in vivo studies in human HIV patients, no conclusive interpretation could be made. Furthermore, animal studies on ovine pulmonary adenocarcinomas have also been reported (Cousens et al., 2004; Philbey et al., 2006); however, the evidence of viral integration hotspot remains unclear.
Target cells in most of the studies were not synchronized before infection with lentiviruses. Although several reports have described the integration patterns in dividing and nondividing cells (Barr et al., 2006; Ciuffi et al., 2006), integration preference in transgenic mice merits in-depth investigation because both pronuclear microinjection and lentiviral gene transfer target early zygotic stage embryo at 11–18 h postinsemination (Hogan, 1994; Lois et al., 2002; Yang et al., 2007). Therefore, transgenic animals created by lentiviruses provide a unique model for investigating if integration preference exists at specific embryonic stage. Recently, Bryda et al. have identified the proviral integration sites in seven transgenic rats generated by lentiviruses (Bryda et al., 2006). However, detailed genome analysis was not performed. Here we presented an in-depth analysis on 112 independent integration sites from 43 transgenic founder mice generated by zygotic gene transfer using lentiviruses. Our goal was to investigate if embryonic stage influences the preference of lentiviral integration.
RESULTS
Integration sites of lentiviruses carrying the GFP gene were cloned and sequenced from 43 transgenic founders using the specific primer located at the transgene (see Fig. 1). These sequences were then used to map the position of the integration sites relative to the mouse genome assembly (mm8). Redundant sequences and those shorter than 100 bp were excluded from further analysis. As a result, we identified at least one integration site in each animal, and a total of 112 integration sites were successfully mapped within the mouse genome. The matched sequences were ranging from 100 to 850 bp, and the average size of the fragments was 431.99 bp. To determine if these integration sites were randomly distributed throughout the genome, we have generated a set of 10,000 simulated integration sites created using a random number generator in the mouse genome to serve as controls. The chromosomal positions of the 112 authentic sites and the control integration sites were then analyzed and compared. In the following text and tables, the numbers and P values refer to comparisons between the authentic integration sites and the set of 10,000 simulated integration sites.
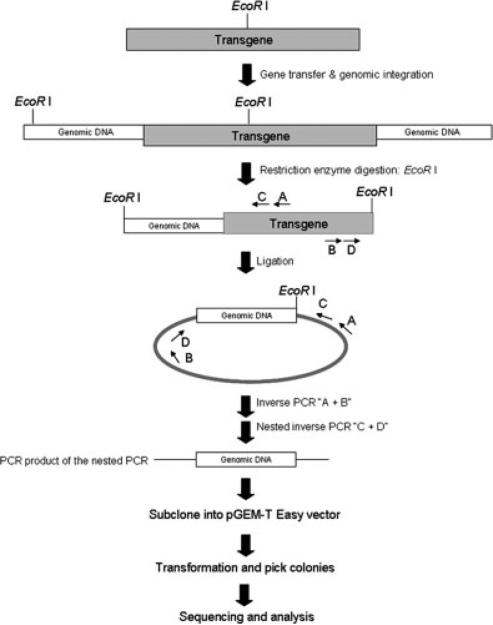
FIG. 1.
The flow chart for identifying the lentiviral integration sites in this study. Genome DNA from transgenic founder mice was digested with the EcoR I restriction enzyme, and conducted the self-ligation of enzyme digested genome DNA. Two rounds of PCR were performed for amplifying circularized genomic DNA using two different sets of primers sequentially. A and B primers were used for the first round of inverse PCR, and C and D primers were used for the second round of nested PCR. After two rounds of PCR, the nested PCR products were subcloned into the pGEM-T Easy vector, and transformed into the bacterial competent cells. Colonies with different sizes were picked up for plasmid DNAs extraction, sequencing, and then analyzed the sequence data in UCSC website and self-generated software.
Chromosomal Distribution of Lentiviral Integration Sites
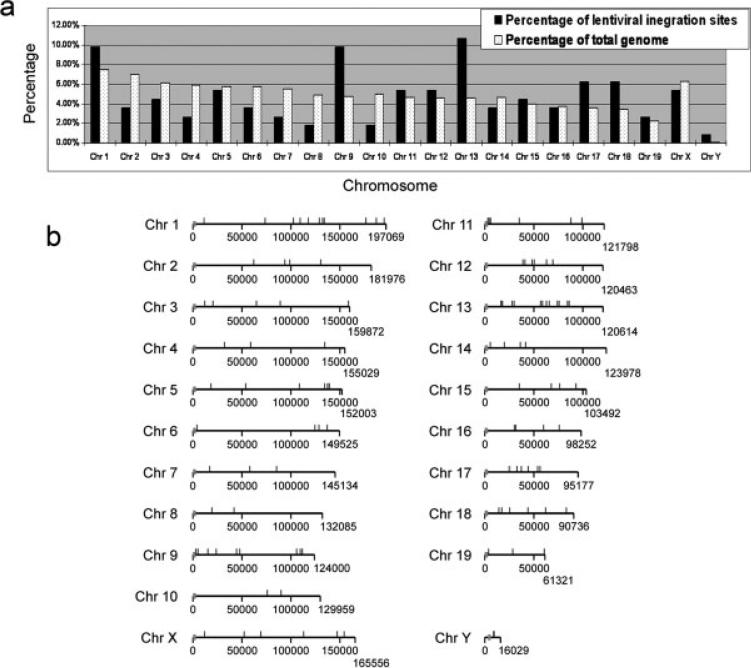
Spatial arrangement of chromosomes may alter the accessibility of chromosomes, and influent subsequent integration events. Our first step was to determine if integration preference exists in early embryo. Our results revealed that all integration sites were broadly distributed among autosomes and sex chromosomes (Fig. 2a,b). On chromosome 9 and 13, a higher percentage of integration events was observed; whereas lower than expected number of events was found on chromosomes 8 and 10. Although variations on integration events and chromosome sizes were observed, the distribution of integration events was not significantly differed among chromosomes (Fig. 2a; P > 0.05).
FIG. 2.
Chromosome distribution of lentiviral integration sites in transgenic mice. (a) Lentiviral vector integration sites from transgenic mice were plotted as the percentage of all integration sites in different chromosomes, and compared with the percentage of the mouse genome included in each chromosome. The integration frequency of integration sites in different chromosomes was not significantly different (P > 0.05) when compared with the related sizes of various chromosomes. (b) The 112 integration sites were plotted into the related positions in the individual chromosomes. The chromosome locations of the viral integration sites in the mouse genome were illustrated using the Parasight program (http://eichlerlab.gs.washington.edu/jeff/parasight/index.html). Gray lines indicated integration sites and gray boxes indicated the location of the centromeres. Positions of the chromosomes were given in kb.
Intra- and Intergenic Distribution of Lentiviral Integration Sites
Transcriptionally active regions are structurally vulnerable, and high DNA repair activity is expected, which may favor exogenous gene integration (Milutinovic et al., 2002). Unwinding of the two DNA strands during transcription increases the accessibility of chromosomes and exposes to viral preintegration complex, thus increases the likelihood of successful integration. In order to determine whether viral integration sites were predominantly located within actively transcribed region of the genome, we classified each integration site as intra- or intergenic based on the position of the annotated mouse RefGenes, which are protein-coding genes as described by the National Center for Biotechnology Information (NCBI) mRNA reference sequences collection. In contrast to the 31.73% of simulated integration sites, a much higher number (41.96%; 47/112, P < 0.05) of the lentiviral integration sites were mapped to the intragenic portion of the mouse genome (Table 1). Among these intragenic lentiviral integration sites, majority (95.75%; 45/47) of them were located within the introns and were overrepresented when compared with the simulated integration sites (Table 1; P < 0.05). Specifically, only two integration sites were mapped within the exons. These results suggested that lentiviruses prefer to integrate within intragenic regions and specifically within the introns.
Table 1.
The Characteristics of Lentiviral Integration Sites Within the Intragenic and Intergenic Regions
| No. of lentiviral integration sites (%) | No. of 10,000 simulated integration sitesa (%) | P value | |
|---|---|---|---|
| Intragene | 47 (41.96) | 3,173 (31.73) | 0.021* |
| Codingb | 1 (0.89) | 124 (1.24) | 0.741 |
| UTRc | 1 (0.89) | 79 (0.79) | 0.903 |
| Intron | 45 (40.18) | 2,970 (29.70) | 0.016* |
| Intergene | 65 (58.04) | 6,827 (68.27) | 0.021* |
| Total | 112 (100.00) | 10,000 (100.00) |
Simulated 10,000 integration sites were generated randomly.
Represented the coding region which can translate to amino acid.
Represented the untranslated region.
Showed the significant difference (P < 0.05) for χ2 test comparing with the simulated data.
Distribution of Lentiviral Integration Sites Within Repetitive Elements
Repetitive elements comprise over 42% of the mouse genome (Waterston et al., 2002). Therefore, it was logical to envision that integration preference might exist within these elements. We have compared the positions of the integration sites with the annotated positions of the long interspersed nuclear elements (LINEs), the short interspersed nuclear elements (SINEs), the long terminal repeat elements (LTRs), the low complexity repeats, the DNA repeat elements (DNA), the simple repeats (Simple), and the satellite repeats. Among the 112 integration sites, 44.64% (50/112) were located within the repetitive elements (Table 2). The integration frequency within LINE, SINE, LTR, Low complexity, DNA, Simple and Satellite repeats were 25.00% (28/ 112), 7.14% (8/112), 8.04% (9/112), 0.00% (0/112), 0.00% (0/112), 4.46% (5/112), and 0.00% (0/112), respectively (Table 2). However, there was no difference when compared with the simulated integration sites (P > 0.05).
Table 2.
The Characteristics of Lentiviral and Simulated Integration Sites Within the Repetitive Elements
| Repetitive elements | No. of the integration sites (%)a | No. of the 10,000 simulated integration sites (%)b | P valuec |
|---|---|---|---|
| LINE | 28 (25.00) | 1,994 (19.94) | 0.183 |
| SINE | 8 (7.14) | 771 (7.71) | 0.823 |
| LTR | 9 (8.04) | 1,010 (10.10) | 0.470 |
| Low complexity | 0 (0.00) | 42 (0.42) | 0.492 |
| DNA | 0 (0.00) | 84 (0.84) | 0.330 |
| Simple | 5 (4.46) | 284 (2.84) | 0.305 |
| Satellite | 0 (0.00) | 1 (0.01) | 0.916 |
| Total | 50 (44.64) | 4,186 (41.86) | 0.553 |
Percentage of 112 integration sites.
Percentage of 10,000 simulated integration sites.
P value was determined by the χ2 test comparing the number of lentiviral integration sites and simulated integration sites.
Distribution of Lentiviral Integration Sites With Respect to the CpG Islands
CpG islands are common features of mammalian promoters (Saxonov et al., 2006). Approximately, 40% and 70% of mouse and human promoters are associated with CpG islands (Saxonov et al., 2006). Therefore, the insertion of exogenous genes, viral genome, or transposon elements at the CpG islands may affect the expression of the endogenous genes. In order to determine if CpG islands influence lentiviral integration events, we have determined the distance between each integration site and the closest CpG islands. After analyzed the integration pattern regarding to the location of CpG islands, majority of the integration sites (>95%) were mapped at least 5 kb from the nearest CpG islands, and the number of integration sites located within 0–5 kb, 5–10 kb, 10– 25 kb, 25–50 kb, and 50–100 kb to the middle region of CpG islands were 3.57% (4/112), 8.04% (9/112), 9.82% (11/112), 11.61% (13/112), and 18.75% (21/112), respectively (Table 3). However, no difference from the simulated integration sites (P > 0.05) was found, suggesting that lentiviruses may not have preference integrating at close proximity to the CpG islands. Although there was a slight increase in the number of integration sites at 5–10 kb from the CpG islands, there was no difference when compared with the simulated sites (Table 3; P > 0.05).
Table 3.
Lentiviral Integration Profile vs. CpG Islands in Transgenic Mice
| Distance to center of the CpG islands (kb) | No. of integration sites (%) | No. of ~10,000 simulated integration sites (%) | P value |
|---|---|---|---|
| 0 ~ 5 | 4 (3.57) | 584 (5.84) | 0.37 |
| 5 ~ 10 | 9 (8.04) | 435 (4.35) | 0.059 |
| 10 ~ 25 | 11 (9.82) | 1077 (10.78) | 0.745 |
| 25 ~ 50 | 13(11.61) | 1169 (11.70) | 0.976 |
| 50 ~ 100 | 21 (18.75) | 1359 (13.60) | 0.114 |
| >100 | 54 (48.32) | 5369 (53.73) | 0.245 |
| Total | 112(100.00) | 9,993 (100.00) |
P value was determined by the χ2 test comparing the number of lentiviral integration sites and simulated integration sites.
Distribution of Lentiviral Integration Sites Near the Transcription Start Sites
Prior studies suggested that some lentiviruses preferred to integrate near the transcription start sites (Lewinski et al., 2006; MacNeil et al., 2006; Mitchell et al., 2004). In order to determine the correlation between integration sites and transcription units, the distance between each integration site and the annotated position of the nearest transcription start site for a mouse RefSeq gene was determined. Among the 112 integration sites, 4.46% (5/112) of them were located within 0–5 kb from the transcription start sites (Table 4). The number of integration sites located within 5–10 kb, 10–25 kb, 25–50 kb, and 50–100 kb to the transcription start sites of the RefSeq genes were 8.93% (10/112), 13.39% (15/ 112), 14.29% (16/112), and 15.18% (17/112), respectively. 43.75% (49/112) integration sites were located at more than 100 kb from the transcription start sites of the RefSeq genes (Table 4). On the basis of our analysis, we have found no difference between the integration sites and the simulated set (Table 4; P > 0.05).
Table 4.
Lentiviral Integration Profile vs. Transcription Start Sites in Transgenic Mice
| Distance to transcription start sites (kb) | No. of integration sites (%) | No. of 10,000 simulated integration sites (%) | P value |
|---|---|---|---|
| 0 ~ 5 | 5 (4.46) | 726 (7.26) | 0.256 |
| 5 ~ 10 | 10 (8.93) | 568 (5.68) | 0.141 |
| 10 ~ 25 | 15 (13.39) | 1,248 (12.48) | 0.771 |
| 25 ~ 50 | 16 (14.29) | 1,355 (13.55) | 0.821 |
| 50 ~ 100 | 17 (15.18) | 1,492 (14.92) | 0.939 |
| >100 | 49 (43.75) | 4,611 (46.11) | 0.618 |
| Total | 112 (100.00) | 10,000 (100.00) |
P value was determined by the χ2 test comparing the number of lentiviral integration sites and simulated integration sites.
DISCUSSION
Pseudotyped lentiviral vector is a highly efficient gene transfer method for the generation of transgenic animals. Unlike retroviruses, lentiviruses are capable of infecting nondividing cells such as neurons, which greatly increase their application in gene therapy and the development of transgenic animal models. However, one of the potential risks of viral gene transfer is the unknown integration preference that may disrupt endogenous gene functions, and these potential side effects have raised concerns in clinical applications. Thus, there is a tremendous interest in determining if lentiviruses have integration preference in live animals.
Most of the viral integration studies were performed in tissue culture cells (Lewinski et al., 2006; Mitchell et al., 2004; Nowrouzi et al., 2006). Bryda et al. have determined the chromosomal positions of lentiviral pro-virus in seven transgenic founder rats; however, detailed analysis on integration pattern was not performed (Bryda et al., 2006). Compared with previous studies, our interest was focused on determining if integration hot spots exist in 43 transgenic founder mice produced by lentiviral infection at the zygotic stage. These transgenic mice were unique models for investigating if embryo stage influents the integration preference of lentiviruses. Unlike the transgenic rat study, we have a relatively large sample size of 112 integration sites from 43 founder animals. An in-depth genome analysis was performed to determine if there is integration preference for unique chromosomal regions.
Several approaches were used for analyzing chromosomal integration sites. We first surveyed the distribution of integration sites in different chromosomes. Our results suggested that lentiviruses integrated randomly among chromosomes in the transgenic mice (Fig. 2a,b). In fact, our finding was consistent with previous studies on avian sarcoma virus (ASV) in human HeLa cells (Narezkina et al., 2004) and feline immunodeficient virus (FIV) in human HepG2 cells (Kang et al., 2006) that no global bias was found in different chromosomes.
We then determined if lentiviruses have biased on integrating between the intragenic and intergenic regions of the mouse genome. By analyzing the distribution of the lentiviral integration sites, a tendency toward the intragenic region, especially the introns, was observed (Table 1). Over 95% of the intragenic integration sites were located in the introns instead of the exons, which suggested that the likelihood of disrupting normal gene function may be low. Previous studies have also shown strong preference of integration at the active genes regions in MLV- and HIV-based vector (Mitchell et al., 2004), whereas FIV preferred to integrate into actively transcribed genes in human hepatoma cells (Kang et al., 2006). Similar to ASV, HIV, and adeno-associated virus (AAV); lentivirus and FIV also had strong preference integrating into the intragenic regions (Liu et al., 2006; Trobridge et al., 2006). Furthermore, integration events in nondividing cells were mostly found at close proximity of the transcription units (Barr et al., 2006; Ciuffi et al., 2006). In fact, there are advantages for targeting an active gene region, which include the unwinding of DNA strands for subsequent transcription events (Milutinovic et al., 2002), thus creating a more accessible region for lentiviral integration. In addition to favorable chromosomal structures, necessary components for subsequent transcription were also localized at the active gene regions (Milutinovic et al., 2002), which allows immediate production of viral gene products upon integration. Because of the high tendency of integrating at the euchromatin regions instead of the heterochromatin regions, these suggested the potential influence of chromosomal structure on viral integration events.
Based on the repetitive elements study, lentiviral integration has no preference in specific repetitive elements (Table 2). Our results did not support previous studies that foamy virus (FV) has less integration sites located in the LINE repeats (Trobridge et al., 2006), HIV-1 preferred to integrate at close proximity to LINE-1 or Alu elements in human genome (Stevens and Griffith, 1994, 1996), and HIV-based vector favored the Alu region when targeted the IMR-90 cells synchronized at G1 stage (Ciuffi et al., 2006). The disparities could be related to the host cell types and the accessibility of the repetitive elements. Despite the differences, repetitive elements in mouse zygotes seem to be less favorable for lentiviral integration, which may or may not relate to the cell phases while infection was achieved. Thus, the role of cell cycle on viral integration merits further investigation, whereas metaphase II arrested oocytes prior to fertilization, and zygotic stage embryos could be a good model because of their naturally synchronized cell phases.
CpG islands are usually associated with active genes in vertebrate and are key regulators on gene expression at epigenetic level (Saxonov et al., 2006). Therefore, it is our interest to determine if lentiviruses have any preference integrating into the CpG islands. Our results suggested that lentiviruses had no preference integrating into regions apart from the CpG islands (Table 3). In fact, conflicting results on viral integration preference regarding to the CpG islands have been reported. Wu et al. have shown the association between the integration sites of murine leukemia virus (MLV) and the CpG islands, whereas no association was found in HIV (Wu et al., 2003). Their findings were consistent with the other studies on HIV and MLV integration preference regarding to the CpG islands (Lewinski et al., 2006; Mitchell et al., 2004). Furthermore, FV, MLV, and AAV preferred to integrate within 1 kb of the CpG islands (Trobridge et al., 2006), which were differed from lentiviral vectors and HIV. Although we could not predict the outcome in transgenic mice generated by the other vector systems such as MLV, our transgenic mice were generated by lentiviral infection at the zygotic stage and have shown no specific preference regarding to the CpG islands, which was consistent with prior studies using lentiviral based vector (Lewinski et al., 2006; Mitchell et al., 2004). Despite no integration preference at close proximity of the CpG islands was observed, high CG content at the region of CpG islands may limit the accessibility of restriction enzyme for effective digestion, which may reduce the possibility in identifying proviral sequence at this region. It is also important to note that only ~25% of the integration sites, compared with Southern blotting result, were identified as authentic sites based on our stringent criteria. Most of the integration sites were not able to be identified, which suggested the limitation of the techniques as well as the complexity of integration events in vivo.
Many studies suggested that most viruses had strong preference integrating at close proximity to the transcription start sites (Mitchell et al., 2004). HIV favors integration at chromosomal region of high gene density, especially in a cluster of active genes (Lewinski et al., 2006; MacNeil et al., 2006; Mitchell et al., 2004). Narezkina et al. (2004) also reported that ASV preferred to integrate at the region where RNA polymerase II binds, whereas FV preferred to integrate at close proximity to the transcription start sites (Trobridge et al., 2006). However, our study on the distribution of lentiviral integration sites in regard to the transcription start sites has no difference from that of the simulated integration sites (Table 4). The conflicting result with the HIV study was not expected because integration mechanism of HIV and HIV-based vectors should be comparable if not identical. The disparity on integration preference regarding to transcription units and active genes suggested two possible explanations: (i) HIV has similar but not identical integration mechanism with HIV-based vector, which is not likely the case and (ii) integration preference is influenced by the target cells. The inconsistent results found between the HIV studies (Schroder et al., 2002) and our studies on transgenic mice could be related to the target cells while the integration events occurred. Based on our findings, integration events in transgenic mice generated by lentiviruses may be influenced by embryonic stage, thus zygotic stage mouse embryo may serve as a model for in-depth investigation of lentiviral integration.
One may also concern whether integration occurs at the zygotic stage or at more advanced embryonic stage. Although we could not eliminate such possibility, we have investigated the effect of temperature on viral infectivity (Chan, 1997). Viral infectivity decreased dramatically in the first few hours of incubation at 37°C (Chan, 1997). Furthermore, only a small volume of the viral solution was injected into the PVS of early embryos, approximately 10–100 infectious viral particles were expected to be delivered. Thus the likelihood of continuous infection for a prolong period of time during early development was not expected (Chan et al., 1998), instead, integration is expected to occur before the first embryonic division.
This study was evolved based on prior studies of HIV-1 integration pattern in cultured cells (MacNeil et al., 2006; Nowrouzi et al., 2006). Single restriction enzyme was commonly used for genomic digestion followed by cloning and sequence analysis (MacNeil et al., 2006; Nowrouzi et al., 2006), however, whether such restriction site is randomly distributed throughout the genome was not clearly defined. Thus digestion using a single restriction enzyme such as EcoR I may be nonrandomly distributed in the mouse genome, which may limit the likelihood of identifying all integration sites. In order to determine if EcoR I sites were randomly distributed throughout the genome we have generated a set of 10,000 randomly selected EcoR I sites in the mouse genome. A nonrandom distribution of EcoR I site was observed in the mouse genome (data not shown). However, we have found less EcoR I sites that were randomly distributed among intra-/intergenic integration sites when compared with the 10,000 simulated sites. This result suggests that lentiviral integration sites identified in this study are much more than that of the simulated control. Because of the likelihood of nonrandom distribution of a single restriction site such as EcoR I, additional restriction enzymes might be considered to minimize potential bias in future studies.
Here we reported a detailed analysis on integration sites of transgenic mice generated by lentiviruses at the zygotic stage. Our transgenic mouse model provides a unique tool for delineating if integration preference of lentiviruses exists at specific cell phase. Understanding the integration pattern of lentiviruses will not only benefit the future development of transgenic technology but also the clinical application of lentiviral vector.
MATERIALS AND METHODS
Generation of VSVG-LVU-GFP and Envelope-Free LVU-GFP
A self-inactivated lentiviral vector expressing the GFP gene under the control of ubiquitin promoter (VSVGLVU-GFP) was used in this study (Gift from C. Lois). VSVG-LVU-GFP was generated by transfection of the 293FT packaging cells (Invitrogen Inc.) with lentiviral vector, which was cotransfected with plasmid pD8.9 and pVSVG. Supernatant was collected and concentrated by ultracentrifugation to approximately 1 × 109 infectious units/ml.
Determining the Titer of VSVG-LVU-GFP
The concentration of infectious vector particles (titer) was determined by the expression of GFP. In a 6-well plate, 2.5 × 105 of 293FT cells were plated and used as target cells. The following day, target cells were cocultured with a serially diluted VSVG-LVU-GFP in the presence of 8 μg/ml of polybrene and the titer was determined by multiplying the number of GFP-positive colonies by the dilution factor and presented by colony forming units (cfu)/ml.
Preparation of Mouse Zygotes for Perivitelline-Space Injection
Female ICR mice of 6- to 8-week-old were superovulated by an intraperitoneal injection of 7.5 IU pregnant mares’ serum gonadotropin (PMSG). Mouse zygotes were recovered from superovulated ICR females after 18 h of 7.5 IU human chorionic gonadotropin (hCG) injection and mating with ICR male mice. The zygotes were cultured in human tubal fluid medium until virus injection was performed. A micropipette of 1 to 2 μm (inner diameter) was used, and viruses were injected into the pronuclear stage zygotes. Mouse zygotes were then implanted into the oviducts of pseudopregnant ICR females, and carried to term. Four weeks after birth, tail snips were recovered for PCR analysis to determine their transgenic status followed by chromosomal integration analysis.
Provirus Integration Sites Cloning
Lentiviral integration sites were identified by modifying the ligation-mediated PCR method as described in Figure 1 (Schroder et al., 2002). In brief, 10-μg genomic DNA was digested by EcoR I for 3 h and purified by Qia-Quick PCR purification kit (Qiagen). The purified DNA was then self-ligated by T4 ligase (Promega) at 4°C overnight followed by purification. Circular DNA was then amplified by nested PCR using two sets of inverse primer subsequently. The first round of inverse PCR was conducted by using specific primers (A: TTCAAGGAC GACGGCAACTAC and B: TGTCTGAAGGGATGGTTG TAG). The second round of nested PCR was used to amplify the inward region of the first inverse PCR product by using nested primers (C: CTGCTGCCCGACAAC CACTA and D: TCTGGTTTCCCTTTCGCTTTCA) (see Fig. 1). Prior to PCR amplification, circular DNA was denatured at 94°C for 5 min. PCR was then performed at 94°C for 30 sec, 63°C for 30 sec, and 72°C for 5 min for 35 times followed by 72°C for 7 min. The PCR products/ fragments were then cloned into the pGEM-T easy vector (Promega) and were subsequently used for sequencing.
Mapping the Integration Sites
The sequenced integration sites were mapped back to the mouse genome assembly (mm8) using BLAT in UCSC website (http://www.genome.ucsc.edu) (Kent, 2002). Sequences were considered as authentic integration sites if they: (i) included the terminal region of the lentiviral vector and (ii) matched with the mouse genome for more than 90% of at least 100 bp. A total of 112 sequences were considered authentic and used in this study.
Analysis of the Integration Sites
Ten thousand simulated integration sites were randomly generated in the mouse genome (excluding sequencing gaps) using a random number generator and was used as a control. To account for a possible nonrandom distribution of EcoR I sites in the mouse genome, a set of the positions of 10,000 randomly selected EcoR I sites were compiled. Annotation tables corresponding to the mouse genome assembly (mm8) and including positional information on repetitive elements, CpG islands and RefSeq genes were downloaded from the UCSC Genome Browser (Karolchik et al., 2003). The CpG island was defined according to the UCSC website: (i) GC contents were greater than 50%; (ii) the length was longer than 200bp; and (iii) the ratio of the observed number of CG dinucleotides to the expected number on the basis of the number of Cs and Gs in the segment was greater than 0.6. Custom database queries and programs were then used to classify the integration sites based on the mouse genome annotation and calculate the distance to the nearest annotation feature of interest, i.e. CpG islands or transcription start sites. Chi-square analysis and SAS for multiple comparisons were then used to compare the distribution of authentic versus control integration sites. Difference of P < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
The authors thank Dr. Carlos Lois for providing lentiviral vector backbone, Jin-Jing Yang and Tina Huang for their technical assistance. The authors also thank Katherine Larkin and Leslee Sinclair for the proofreading; Fang-Chueh Liu and Chanchao Lorthongpanich for statistic analysis, and Grey Tharp for computer support. The procedures were approved by the Emory University Animal Care and Biosafety Committees.
Contract grant sponsor: NIH (Yerkes National Primate Research Center), Contract grant number: RR-00165. Contract grant sponsor: NCRR/NIH, Contract grant number: RR018827-04.
LITERATURE CITED
- Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, Bushman FD. HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Mol Ther. 2006;14:218–225. doi: 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Bryda EC, Pearson M, Agca Y, Bauer BA. Method for detection and identification of multiple chromosomal integration sites in transgenic animals created with lentivirus. Biotechniques. 2006;41:715–719. doi: 10.2144/000112289. [DOI] [PubMed] [Google Scholar]
- Burns JC, Friedmann T, Driever W, Burrascano M, Yee JK. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: Concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabot RA, Kuhholzer B, Chan AW, Lai L, Park KW, Chong KY, Schatten G, Murphy CN, Abeydeera LR, Day BN, Prather RS. Transgenic pigs produced using in vitro matured oocytes infected with a retroviral vector. Anim Biotechnol. 2001;12:205–214. doi: 10.1081/ABIO-100108347. [DOI] [PubMed] [Google Scholar]
- Chan AW, Chong KY, Martinovich C, Simerly C, Schatten G. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science. 2001;291:309–312. doi: 10.1126/science.291.5502.309. [DOI] [PubMed] [Google Scholar]
- Chan AW, Homan EJ, Ballou LU, Burns JC, Bremel RD. Transgenic cattle produced by reverse-transcribed gene transfer in oocytes. Proc Natl Acad Sci USA. 1998;95:14028–14033. doi: 10.1073/pnas.95.24.14028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffi A, Mitchell RS, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman FD. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol Ther. 2006;13:366–373. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Clark AJ, Bissinger P, Bullock DW, Damak S, Wallace R, Whitelaw CB, Yull F. Chromosomal position effects and the modulation of transgene expression. Reprod Fertil Dev. 1994;6:589–598. doi: 10.1071/rd9940589. [DOI] [PubMed] [Google Scholar]
- Cousens C, Bishop JV, Philbey AW, Gill CA, Palmarini M, Carlson JO, DeMartini JC, Sharp JM. Analysis of integration sites of Jaagsiekte sheep retrovirus in ovine pulmonary adenocarcinoma. J Virol. 2004;78:8506–8512. doi: 10.1128/JVI.78.16.8506-8512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B. Manipulating the mouse embryo: A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; Plainview, NY: 1994. p. 497. [Google Scholar]
- Jaenisch R. Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proc Natl Acad Sci USA. 1976;73:1260–1264. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R. Retroviruses and mouse embryos: A model system in which to study gene expression in development and differentiation. Ciba Found Symp. 1983;98:44–62. doi: 10.1002/9780470720790.ch4. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Jahner D, Nobis P, Simon I, Lohler J, Harbers K, Grotkopp D. Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell. 1981;24:519–529. doi: 10.1016/0092-8674(81)90343-3. [DOI] [PubMed] [Google Scholar]
- Kang Y, Moressi CJ, Scheetz TE, Xie L, Tran DT, Casavant TL, Ak P, Benham CJ, Davidson BL, McCray PB., Jr Integration site choice of a feline immunodeficiency virus vector. J Virol. 2006;80:8820–8823. doi: 10.1128/JVI.00719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: Viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:0611–0622. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Dow EC, Arora R, Kimata JT, Bull LM, Arduino RC, Rice AP. Integration of human immunodeficiency virus type 1 in untreated infection occurs preferentially within genes. J Virol. 2006;80:7765–7768. doi: 10.1128/JVI.00542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- MacNeil A, Sankale JL, Meloni ST, Sarr AD, Mboup S, Kanki P. Genomic sites of human immunodeficiency virus type 2 (HIV-2) integration: similarities to HIV-1 in vitro and possible differences in vivo. J Virol. 2006;80:7316–7321. doi: 10.1128/JVI.00604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinovic S, Zhuang Q, Szyf M. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J Biol Chem. 2002;277:20974–20978. doi: 10.1074/jbc.M202504200. [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:1127–1137. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA. Genome-wide analyses of avian sarcoma virus integration sites. J Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrouzi A, Dittrich M, Klanke C, Heinkelein M, Rammling M, Dandekar T, von Kalle C, Rethwilm A. Genome-wide mapping of foamy virus vector integrations into a human cell line. J Gen Virol. 2006;87:1339–1347. doi: 10.1099/vir.0.81554-0. [DOI] [PubMed] [Google Scholar]
- Perletti G, Osti D, Marras E, Tettamanti G, de Eguileor M. Generation of VSV-G pseudotyped lentiviral particles in 293T cells. J Cell Mol Med. 2004;8:142–143. doi: 10.1111/j.1582-4934.2004.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philbey AW, Cousens C, Bishop JV, Gill CA, DeMartini JC, Sharp JM. Multiclonal pattern of Jaagsiekte sheep retrovirus integration sites in ovine pulmonary adenocarcinoma. Virus Res. 2006;117:254–263. doi: 10.1016/j.virusres.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Romano G. Current development of lentiviral-mediated gene transfer. Drug News Perspect. 2005;18:128–134. doi: 10.1358/dnp.2005.18.2.886481. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Shimotohno K, Temin HM. Formation of infectious progeny virus after insertion of herpes simplex thymidine kinase gene into DNA of an avian retrovirus. Cell. 1981;26:67–77. doi: 10.1016/0092-8674(81)90034-9. [DOI] [PubMed] [Google Scholar]
- Stevens SW, Griffith JD. Human immunodeficiency virus type 1 may preferentially integrate into chromatin occupied by L1Hs repetitive elements. Proc Natl Acad Sci USA. 1994;91:5557–5561. doi: 10.1073/pnas.91.12.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SW, Griffith JD. Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration. J Virol. 1996;70:6459–6462. doi: 10.1128/jvi.70.9.6459-6462.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin HM. Safety considerations in somatic gene therapy of human disease with retrovirus vectors. Hum Gene Ther. 1990;1:111–123. doi: 10.1089/hum.1990.1.2-111. [DOI] [PubMed] [Google Scholar]
- Trobridge GD, Miller DG, Jacobs MA, Allen JM, Kiem HP, Kaul R, Russell DW. Foamy virus vector integration sites in normal human cells. Proc Natl Acad Sci USA. 2006;103:1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Yang SH, Agca Y, Cheng PH, Yang JJ, Agca C, Chan AW. Enhanced transgenesis by intracytoplasmic injection of envelope-free lentivirus. Genesis. 2007;45:177–183. doi: 10.1002/dvg.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Cheng PH, Banta H, Piotrowska-Nitsche K, Yang JJ, Cheng EC, Snyder B, Larkin K, Liu J, Orkin J, Fang ZH, Smith Y, Bachevalier J, Zola SM, Li SH, Li XJ, Chan AW. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature. 2008;453:921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee JK, Friedmann T, Burns JC. Generation of high-titer pseudo-typed retroviral vectors with very broad host range. Methods Cell Biol. 1994;43(Part A):99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]