Abstract
Background: Lower levels of global DNA methylation in tissue and blood have been associated with increased cancer risk. Conversely, cross-sectional analyses of healthier lifestyle patterns have been associated with higher levels of global DNA methylation.
Objective: In this trial, we explored the associations between changes in lifestyle modifications (diet, weight loss), metabolic markers, and global epigenetic biomarkers in white blood cells.
Methods: Study participants were Hispanic, African American, and Afro-Caribbean overweight and sedentary female breast cancer survivors (n = 24) who participated in a larger randomized, crossover, pilot study of a 6-mo weight loss intervention and who had available blood specimens. Anthropometric measures, a food-frequency questionnaire, and peripheral blood were collected at baseline, 6 mo, and 12 mo. Plasma samples were analyzed for metabolic markers (insulin, glucose). We measured DNA methylation of long interspersed nucleotide element 1 (LINE-1) and satellite 2 by pyrosequencing and MethyLight, respectively, and global DNA methylation by the luminometric methylation assay (LUMA).
Results: DNA methylation of LINE-1 was statistically significantly elevated at 6 mo [75.5% vs. 78.5% (P < 0.0001)] and 12 mo [75.5% vs. 77.7% (P < 0.0001)], compared to baseline. Over a 12-mo period, changes in percentage body fat and plasma glucose concentrations were positively associated with LINE-1 DNA methylation (β = 0.19, P = 0.001) and LUMA DNA methylation levels (β = 0.24, P = 0.02), respectively. Similarly, 12-mo changes in dietary measures such as vegetable (β = 0.009, P = 0.048), protein (β = 0.04, P = 0.001), and total caloric (β = 0.05, P = 0.01) intake were positively associated with changes in LUMA DNA methylation, as was intake of fruit positively associated with changes in LINE-1 DNA methylation (β = 0.004, P = 0.02).
Conclusions: Our hypothesis-generating results suggest that lifestyle modifications may be associated with changes in global DNA methylation detectable at 6 and 12 mo. These biomarkers may be useful intermediate biomarkers to use in future intervention trials. This trial was registered at clinicaltrials.gov as NCT00811824.
Keywords: weight loss, epigenetics, LINE-1, Sat2, LUMA
Introduction
Despite the increase in 5-y breast cancer survival rates for all ethnic groups, African American and Hispanic women continue to have lower breast cancer survival than non-Hispanic whites, possibly because of postdiagnosis lifestyle behaviors (1). Although there is conflicting evidence, several studies have shown that changes in dietary patterns and physical activity may improve breast cancer survival (2–6). However, the biological mechanisms through which changes in lifestyle behaviors may improve breast cancer survival are not well understood.
Epigenetic modifications such as DNA methylation have been proposed as a biological mechanism by which alterations of lifestyle including diet and exercise can modify predisposition to chronic disease (7, 8). DNA methylation involves the addition of a methyl group at the 5′ position of cytosine (5mC)9 in cytosine guanine dinucleotides (CpGs). Global or genomic DNA methylation levels refer to DNA methylation levels throughout all genomic CpG sites (9). Measures of global DNA methylation biomarkers in peripheral tissue, such as blood, have been shown to respond to environmental and dietary exposures. Studies have shown that exposures to chemicals such as benzene, polycyclic aromatic hydrocarbons, persistent organic pollutants, and heavy metals can affect DNA methylation profiles (10–16). Differences in health behaviors have also been related to differences in global DNA methylation measures in a small number of studies (9). Exercise and diet were both investigated in the North Texas Healthy Heart Study, and increases in long interspersed nucleotide element 1 (LINE-1) DNA methylation was associated with higher fruit/vegetable intake and higher physical activity (17, 18). Specific dietary changes, such as fish oil intake, have been associated with the differential methylation of 27 specific CpG sites in a study of the Yup’ik Alaskan natives (19). Other studies have found that increased physical activity during childhood is associated with higher DNA methylation of LINE-1; however, increased exercise frequency in the elderly has been related to lower global methylation as measured by the luminometric methylation assay (LUMA) (20, 21). Most studies have found that current smoking is not related to changes in global DNA methylation, whereas alcohol consumption seems to be related to an increase in DNA methylation of CpG sites within CCGG sequences, but not in the LINE-1 promoter region in some studies (11, 12, 18, 22–26). Similarly, 2 studies found no associations when investigating BMI and LINE-1 DNA methylation (11, 22), whereas another study among women found that lower DNA methylation of LINE-1 was correlated with increased BMI (27). Different epigenetic biomarkers have yielded differing results when measuring the effects of environmental exposures and health behaviors, suggesting that several epigenetic markers should be investigated when assessing changes because of these environmental and behavioral factors.
In this study, we used anthropometric measures, health behavior measures, and blood samples collected in a pilot study of a dietary change and physical activity weight loss intervention among overweight and sedentary minority breast cancer survivors (28). We conducted hypothesis-generating analyses to assess the associations between changes in anthropometric measures, metabolic biomarkers, diet, and physical activity with changes in 3 separate measures of DNA methylation that have previously been associated with environmental and behavioral exposures: global levels of DNA methylation measured by LUMA and levels of DNA methylation at the repeated sequences LINE-1 and satellite 2 (Sat2). To our knowledge, this is one of the first reports examining changes in DNA methylation after a lifestyle modification intervention.
Methods
Study design and population.
Analyses used data and blood samples from breast cancer survivors who participated in the parent weight loss intervention trial, which has been described elsewhere (28). Briefly, a group of 42 minority breast cancer survivors participated in a randomized, crossover, waitlist-controlled pilot and feasibility study to test the effects of a 6-mo weight loss program using physical activity and dietary change. The study was targeted to Hispanic, African American, and Afro-Caribbean cancer survivors who met the following eligibility criteria: aged 21–70 y; diagnosis of stage 0–IIIa breast cancer; no evidence of recurrent or metastatic disease; completed surgery, chemotherapy, and radiation therapy at least 6 mo prior; BMI (in kg/m2) >25; sedentary (defined as physically active to the point of sweating <20 min/wk); not actively engaged in a weight loss program; nonsmoker; hemoglobin A1c <8%; blood pressure <140/90; and LDL cholesterol <150 mg/dL). Breast cancer diagnosis and treatment and all clinical measures were confirmed by medical record review. Clinical data and blood samples were collected at baseline, 6 mo, and 12 mo. The current analyses were restricted to the subset of 24 women who provided buffy coat samples at baseline and 6 and/or 12 mo. For all variables presented in Table 1, we compared values for women who did (n = 24) and did not (n = 18) provide buffy coat samples. We found no statistically significant differences other than in the proportion of African American women (33% in subset with samples, 6% in subset without samples, P = 0.03). The Columbia University Medical Center Institutional Review Board approved the study, and all participants provided written informed consent in English or Spanish.
TABLE 1.
Baseline demographic, clinical, and lifestyle characteristics of Hispanic, African American, and Afro-Caribbean breast cancer survivors participating in a dietary change and physical activity weight loss trial (n = 24)1
| Participant characteristics | Values |
| Demographic characteristics | |
| Age, y | 52.2 ± 8.7 (35–69) |
| Race/ethnicity, n (%) | |
| African descent | 8 (33.3) |
| Hispanic descent | 16 (66.7) |
| Education, n (%) | |
| <High school | 9 (37.5) |
| High school or GED | 9 (37.5) |
| Some college | 2 (8.3) |
| College and higher | 4 (16.7) |
| Clinical characteristics | |
| BMI, kg/m2 | 32.8 ± 5.5 (26.3–43.7) |
| BMI category, n (%) | |
| Overweight (25–29.9 kg/m2) | 9 (37.5) |
| Obese (30–34.9 kg/m2) | 8 (33.3) |
| Morbidly obese (≥35 kg/m2) | 7 (29.2) |
| Menopausal status at baseline, n (%) | |
| Premenopausal | 4 (16.7) |
| Postmenopausal | 20 (83.3) |
| Time since diagnosis, y | 3.4 ± 2.5 (1.2–9.4) |
| Stage at diagnosis, n (%) | |
| DCIS (0) | 4 (16.7) |
| I | 9 (37.5) |
| II | 8 (33.3) |
| III | 3 (12.5) |
| Tumor immunohistochemistry,2 n (%) | |
| ER+ and/or PR+ tumor | 15 (62.5) |
| Her2/neu+ tumor | 5 (23.8) |
| Triple negative tumor | 6 (25.0) |
| Treatments ever received,2 n (%) | |
| Surgery | 24 (100.0) |
| Mastectomy | 8 (33.3) |
| Lumpectomy | 18 (75.0) |
| Lymph nodes removed, n | 2.7 ± 4.8 |
| Radiation | 18 (75.0) |
| Chemotherapy | 19 (79.2) |
| Tamoxifen | 7 (29.2) |
| Aromatase inhibitor | 11 (45.8) |
| Premenopausal women currently on hormonal therapy,3 n (%) | |
| None | 3 (75.0) |
| Tamoxifen | 1 (25.0) |
| Postmenopausal women currently on hormonal therapy,4 n (%) | |
| None | 10 (50.0) |
| Tamoxifen | 0 (0.0) |
| Aromatase inhibitor | 10 (50.0) |
| Lifestyle characteristics | |
| Dietary intake5 | |
| Vegetables,6 servings/d | 1.4 ± 1.0 |
| Fruit intake, frequency/d | 1.4 ± 0.9 |
| Fat, % energy | 38.0 ± 4.0 |
| Total energy intake, kcal/d | 1250 ± 639 |
| Physical activity7 | |
| Occupational index | 2.6 ± 0.5 |
| Household/caregiving index | 2.3 ± 0.5 |
| Active living index | 2.7 ± 0.6 |
| Sports and exercise index | 1.5 ± 0.2 |
Unless indicated otherwise, values are means ± SDs; ranges in parentheses (n = 24). DCIS, ductal carcinoma in situ; ER, estrogen receptor; GED, graduate equivalency degree; Her2/neu, human epidermal growth factor receptor 2; PR, progesterone receptor.
Frequencies and percentages were calculated for nonmissing data.
n = 4.
n = 20.
Dietary intake measured by the Block FFQ, Hispanic version.
Approximately 70 g/serving.
Physical activity measured by the Kaiser Physical Activity Survey.
Anthropometric measures.
Weight and height were measured at baseline, 6 mo, and 12 mo with use of a calibrated electronic scale (FR Instruments) and stadiometer (Accustat). Waist and hip circumferences were measured by trained study staff using a Gulick II tape measure (Country Technology). Body composition was measured by DXA with use of the Hologic QDR 4500 densitometer (Hologic). Measurements were available at baseline, 6 mo, and 12 mo for weight, waist and hip circumference, and waist-to-hip ratio. Body composition was only measured at baseline and 6 mo.
Physical activity, dietary, and weight loss intervention.
The intervention has been described in detail elsewhere (28). Briefly, the 6-mo weight loss program encouraged increasing physical activity to 90 min/wk, reducing caloric intake (1200 kcal/d for 1–2 wk, followed by 1600 kcal/d), and distributing caloric intake as 45% protein/30% carbohydrates/25% fat. The following daily behaviors were emphasized to promote adhering to a reduced-calorie diet: eat breakfast, eat 5 small meals, eat ≥2 servings fruit, eat ≥3 servings vegetables, drink 2 L water, read food labels when choosing foods, and pay attention to intake of total calories, protein, fat, and carbohydrates.
Dietary assessment.
Dietary intake at baseline, 6 mo, and 12 mo was assessed with use of the Hispanic version of the Block FFQ, which includes foods commonly consumed by African Americans (29).
Physical activity assessment.
Physical activity was assessed with use of a self-administered adaptation of the Kaiser Physical Activity Survey. The survey provides global summary activity indexes for housework/caregiving, active living habits, sports/exercise, and occupation (score: 1–5; 1: low activity, 5: high activity) (30). All 4 indexes were assessed at baseline and sports/exercise was assessed at 6 and 12 mo.
Serum metabolic marker analyses.
Serum samples were analyzed in batches after all samples were collected. Glucose was measured via an Integra 400 Plus automated chemistry analyzer (Roche Diagnostics). RIA was used to measure insulin (Siemens). Insulin resistance was calculated with use of HOMA-IR (31).
DNA extraction and bisulfite treatment.
Blood was collected at baseline, 6-mo, and 12-mo visits for each participant. White blood cells (WBCs) were collected and stored at −80°C until they were batch analyzed at the completion of the study. Genomic DNA was extracted from the total WBC fraction by a standard salting out procedure. Aliquots of DNA (500 ng) were bisulfite-treated with the EZ DNA methylation kit (Zymo Research) following the manufacturer’s protocol. The DNA was resuspended in 20-μL distilled water and stored at −20°C until use.
DNA methylation measures.
All samples were run in the same batch for all DNA methylation assays to avoid batch effects. The LUMA method was performed as previously described (7, 32, 33), and percentage of DNA methylation was expressed as:
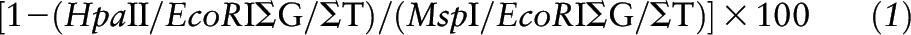 |
The interassay coefficient of variation was 1.5%.
Pyrosequencing for LINE-1 methylation levels was performed with use of PCR and sequencing primers as previously described (34). Pyrosequencing was conducted with use of a PyroMark Q24 instrument (Qiagen) with subsequent quantitation of methylation levels determined with the PyroMark Q24 1.010 software. Three CpG sites were included in the analysis. Each set of amplifications included bisulfite-converted CpGenome universal methylated, unmethylated, and nontemplate controls. The interassay coefficient of variation was 0.67%.
The MethyLight assay was used to determine Sat2 methylation levels. The MethyLight real-time PCR reactions were performed with use of sequences of probes and forward and reverse primers of Sat2-M1, as described by Weisenberger et al. (35). Universal methylated DNA served as a methylated reference, and an Alu-based control reaction was used to measure the concentrations of input DNA to normalize the signal for each methylation reaction also described by Weisenberger et al. (35). Fully methylated DNA to be used as the fully methylated reference sample, was obtained by enzymatically methylating DNA extracted from human DNA methyl transferases double knock-out (DKO) human cell line HCT116 DKO (Zymo Research). Each MethyLight reaction was performed in duplicate, and the percent of methylated reference (PMR) values represent the mean. The interassay coefficient of variation was 1.2%. MethyLight data for Sat2 was expressed as PMR values. The PMR is a relative quantitative measure, and obtained percentages can be above 100% considering the variable number of the consensus sequence used as control and the number of repeats in individual samples.
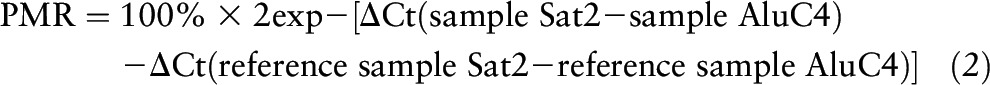 |
Statistical analysis.
Given the modest changes in health behaviors and metabolic markers in both arms of the parent study (28), and the relatively small number of women for whom baseline and follow-up buffy coat samples were available (n = 24), participant data were analyzed as a cohort. Paired t tests were performed to compare the baseline values to 6- and 12-mo values for anthropometric measures, health behaviors, metabolic biomarkers, and DNA methylation measures. For each of the anthropometric measures, health behaviors, and metabolic biomarkers, a generalized estimated equation (GEE) model with repeated measures nested with participants was used to examine the effect of its percent change on the percent change of DNA methylation from baseline to 6 mo and 12 mo. Each of the GEE models was adjusted by using the baseline value of the predictor of interest and the randomization arm as covariates. We determined that the randomization arm was a confounder between percent change of some, but not all, of the predictors of interest and the percent change of DNA methylation. Confounding was defined as changing the β estimate for the predictor by >10%. GEE model β estimates are based on associations with a 1% change in the specified variable. To increase the interpretability of results, in the results section we discuss changes in anthropometric, metabolic biomarker, and dietary measures in terms of 10% changes in the specified variable. Statistical significance was set at P < 0.05. Analyses were conducted with use of SAS 9.2.
Results
Baseline characteristics.
Demographic, lifestyle, and clinical characteristics of study participants (n = 24) at baseline are shown in Table 1. At study enrollment, the mean age of participants was 52 y (range: 35–69 y) and 83% were postmenopausal. Participants were 67% Hispanic and 33% African American. On average, women were 3.4 y postdiagnosis (range: 1.2–9.4 y). Mean BMI was 32.8 (range: 26.3–43.7). On average, reported caloric intake was 1250 kcal/d, which is less than the recommended 1600 kcal/d intake for women over age 50 (36), and is likely an underreporting given that all participants were overweight or obese at baseline. On average, 38% of reported caloric intake was derived from fat, which is higher than the recommended <30% of intake from fat (37). Daily intake of vegetables averaged 1.4 servings/d (∼70 g/serving), and, on average, participants ate fruit 1.4 times/d, indicating that the combined intake of vegetables and fruit was well below the recommended 5–9 daily servings (37).
Change in anthropometric measures, dietary intake, and metabolic biomarkers.
Mean changes in anthropometric measures, dietary intake, and plasma metabolic biomarkers from baseline to 6 and 12 mo have been reported elsewhere for all study participants. In Table 2, we report results for this study sample for objective measures including anthropometric and metabolic markers and subjective measures including diet and physical activity. During the course of this study, on average, participants lost ∼2% of their body weight from baseline to 6 and 12 mo (both P = 0.01; Table 2). There were corresponding changes in waist, but not hip, circumference. At 6 mo, there was a mean percentage loss in body fat of 2.4% (P = 0.03). At 12 mo, we observed a 10.6% decrease in plasma insulin concentration (P < 0.01) and an 11.4% decrease in insulin resistance as assessed by HOMA-IR (P < 0.01). In addition, there was an increase in the sports/exercise index at 6 and 12 mo (P < 0.001).
TABLE 2.
Anthropometric measures, metabolic markers, diet, physical activity, and global DNA methylation at baseline, 6 mo, and 12 mo among Hispanic, African American, and Afro-Caribbean breast cancer survivors participating in a dietary change and physical activity weight loss trial1
| Month 6 (n = 24) |
Month 12 (n = 22) |
||||||
| Baseline values (n = 24) | Values | Mean change, % | P2 | Values | Mean change, % | P3 | |
| Objective measures | |||||||
| Anthropometry | |||||||
| Weight, kg | 86.4 ± 14.3 | 84.7 ± 14.6 | −1.9 | 0.01 | 84.5 ± 14.0 | −2.1 | 0.01 |
| Waist circumference, cm | 103 ± 12.2 | 99.7 ± 9.8 | −2.7 | <0.01 | 99.1 ± 8.5 | −2.7 | 0.01 |
| Hip circumference, cm | 116 ± 11.4 | 116 ± 12.2 | −0.2 | 0.78 | 115 ± 11.0 | −0.6 | 0.55 |
| Waist-to-hip ratio | 0.9 ± 0.1 | 0.9 ± 0.1 | −2.2 | 0.14 | 0.9 ± 0.1 | −0.4 | 0.63 |
| Body fat,4 % | 41.6 ± 5.4 | 40.2 ± 5.4 | −2.4 | 0.03 | — | — | — |
| Metabolic markers | |||||||
| Plasma glucose concentration, mg/dL | 100 ± 16.9 | 102 ± 19.1 | 0.7 | 0.73 | 97.6 ± 13.9 | −2.2 | 0.16 |
| Plasma insulin concentration, uIU/mL | 18.2 ± 10.7 | 17.5 ± 11.8 | 4.8 | 0.52 | 14.6 ± 6.5 | −10.6 | <0.01 |
| HOMA-IR, units | 4.7 ± 3.1 | 4.7 ± 3.8 | 6.7 | 0.78 | 3.6 ± 2.0 | −11.4 | <0.01 |
| DNA methylation | |||||||
| LUMA, % | 69.6 ± 4.0 | 70.7 ± 4.1 | 2.5 | 0.14 | 70.7 ± 3.7 | 1.9 | 0.18 |
| LINE-1, % | 75.5 ± 1.6 | 78.5 ± 1.7 | 4.2 | <0.0001 | 77.7 ± 1.3 | 3.0 | <0.0001 |
| Sat2, % | 91.3 ± 58.6 | 103 ± 48.4 | 26.6 | 0.53 | 66.6 ± 37.4 | 0.7 | 0.073 |
| Subjective measures | |||||||
| Diet | |||||||
| Total caloric intake, kcal/d | 1250 ± 639 | 1120 ± 402 | 8.0 | 0.43 | 1050 ± 219 | 1.0 | 0.17 |
| Vegetable intake,5 servings/d | 1.4 ± 1.0 | 1.7 ± 1.7 | 67.6 | 0.49 | 1.3 ± 0.5 | 42.0 | 0.68 |
| Fruit intake, frequency/d | 1.4 ± 0.9 | 1.6 ± 1.1 | 85.9 | 0.29 | 1.6 ± 0.7 | 66.9 | 0.12 |
| Protein intake, g | 45.1 ± 22.7 | 45.3 ± 17.6 | 22.3 | 0.97 | 40.8 ± 7.5 | 9.9 | 0.40 |
| Fat intake, % total calories | 12.0 ± 6.3 | 10.7 ± 3.2 | 4.7 | 0.35 | 3.0 ± 0.9 | −3.4 | 0.14 |
| Physical activity | |||||||
| Sports and exercise index | 1.5 ± 0.2 | 2.6 ± 1.4 | 81.5 | <0.001 | 2.2 ± 1.1 | 51.2 | <0.001 |
Values are means ± SDs unless otherwise indicated. LINE-1, long interspersed nucleotide element 1; LUMA, luminometric methylation assay; Sat2, satellite 2.
P values calculated from paired t tests comparing baseline to 6 mo.
P values calculated from paired t tests comparing baseline to 12 mo.
Body fat percentage data were not collected at 12 mo.
Approximately 70 g/serving.
Change in DNA methylation biomarkers.
DNA methylation levels for each marker at baseline, 6 mo, and 12 mo are shown in Table 2. Increases in percent methylation of the repetitive element LINE-1 were observed from baseline to 6 and 12 mo (4.2% and 3.0%, respectively; both P < 0.0001). There were no statistically significant differences observed in percent methylation measured by LUMA or percent methylation of the tandem repeat Sat2.
Associations between changes in anthropometric measures, metabolic markers, diet, and physical activity and changes in markers of DNA methylation.
GEE models were used to estimate the individual effects of changes in anthropometric measures, metabolic biomarkers, diet, and physical activity on measures of DNA methylation (Table 3). Weight loss itself was not associated with changes in any of the global DNA methylation markers. Of the objective measures, changes in body fat and glucose were associated with changes in methylation. A 10% decrease in body fat was associated with a 1.9% (95% CI: 0.8%, 3.1%) decrease in DNA methylation of the repetitive element LINE-1. We also observed an association between a 10% increase in glucose with a 2.4% (95% CI: 0.4%, 4.4%) increase in global DNA methylation, as measured by LUMA. No other associations were observed between changes in anthropometric or metabolic markers and changes in other epigenetic biomarkers.
TABLE 3.
GEE estimates for the associations between anthropometric measures, dietary intake, and metabolic measures and global and repetitive element DNA methylation biomarkers of Hispanic, African American, and Afro-Caribbean breast cancer survivors participating in a dietary change and physical activity weight loss trial1
| LUMA |
LINE-1 |
Sat2 |
||||
| β2 (95% CI) | P3 | β2 (95% CI) | P3 | β2 (95% CI) | P3 | |
| Objective measures | ||||||
| Anthropometry | ||||||
| Weight, kg | 0.14 (−0.45, 0.74) | 0.64 | 0.01 (−0.24, 0.27) | 0.93 | 0.35 (−6.79, 7.49) | 0.92 |
| Waist circumference, cm | 0.21 (−0.32, 0.74) | 0.43 | −0.03 (−0.24, 0.18) | 0.77 | −4.99 (−10.63, 0.65) | 0.08 |
| Hip circumference, cm | 0.18 (−0.13, 0.49) | 0.26 | −0.01 (−0.11, 0.10) | 0.87 | 2.11 (−0.21, 4.43) | 0.07 |
| Waist-to-hip ratio | 0.33 (−0.03, 0.69) | 0.07 | −0.07 (−0.24, 0.10) | 0.41 | −1.49 (−5.85, 2.87) | 0.50 |
| Body fat,4 % | 0.13 (−0.22, 0.49) | 0.46 | 0.19 (0.08, 0.31) | 0.001 | −2.80 (−7.15, 1.56) | 0.21 |
| Metabolic markers | ||||||
| Plasma glucose concentration, mg/dL | 0.24 (0.04, 0.44) | 0.02 | −0.004 (−0.08, 0.07) | 0.91 | 1.68 (−1.27, 4.62) | 0.26 |
| Plasma insulin concentration, uIU/mL | 0.03 (−0.01, 0.07) | 0.13 | 0.01 (−0.005, 0.03) | 0.15 | 0.07 (−0.41, 0.56) | 0.76 |
| HOMA-IR, units | 0.03 (−0.01, 0.07) | 0.10 | 0.01 (−0.01, 0.03) | 0.28 | 0.09 (−0.36, -0.55) | 0.69 |
| Subjective measures | ||||||
| Diet | ||||||
| Total caloric intake, kcal/d | 0.05 (0.01, 0.09) | 0.01 | 0.01 (−0.02, 0.03) | 0.50 | 0.29 (−0.35, 0.92) | 0.38 |
| Vegetable intake,5 servings/d | 0.009 (0.0001, 0.02) | 0.048 | 0.002 (−0.005, 0.009) | 0.58 | 0.05 (−0.07, 0.18) | 0.42 |
| Fruit intake, frequency/d | 0.01 (−0.004, 0.03) | 0.13 | 0.004 (0.0006, 0.008) | 0.02 | 0.05 (−0.02, 0.12) | 0.16 |
| Protein intake, g | 0.04 (0.01, 0.07) | 0.01 | 0.01 (−0.009, 0.02) | 0.39 | 0.23 (−0.24, 0.71) | 0.34 |
| Fat intake, % total calories | 0.05 (0.0007, 0.09) | 0.05 | 0.01 (−0.02, 0.04) | 0.52 | 0.18 (−0.63, 1.00) | 0.66 |
| Physical activity | ||||||
| Sports and exercise index | 0.01 (−0.001, 0.02) | 0.07 | 0.002 (−0.005, 0.01) | 0.56 | 0.12 (−0.05, 0.29) | 0.16 |
GEE, generalized estimated equation; LINE-1, long interspersed nucleotide element 1; LUMA, luminometric methylation assay; Sat2, satellite 2.
β coefficients were adjusted for the baseline value of the predictor of interest and randomization arm.
P values were calculated using Wald chi-square statistics from a GEE model.
Body fat percentage was ascertained at 6 mo only.
Approximately 70 g/serving.
Of the subjective diet and physical activity measures, only changes in diet were associated with changes in LUMA and LINE-1, and no other associations were observed. A decrease in caloric intake of 10% was associated with a 0.48% (95% CI: 0.10%, 0.86%) decrease in DNA methylation. A 10% increase in servings of vegetables and protein was associated with an increase in LUMA DNA methylation of 0.85% (95% CI: 0.01%, 1.7%) and 0.41% (95% CI: 0.12%, 0.70%), respectively. A 10% increase in the frequency of eating fruit was associated with an increase in LINE-1 DNA methylation of 0.42% (95% CI: 0.06%, 0.77%).
Discussion
In this study, we evaluated 6- and 12-mo changes in global DNA methylation in Hispanic, African American, and Afro-Caribbean breast cancer survivors who were part of a larger study examining the effects of a specific dietary change and physical activity weight loss program among sedentary and overweight/obese minority breast cancer survivors (28). A decrease in percentage of body fat, an increase in plasma glucose concentrations, and changes in diet were associated with changes in global markers of DNA methylation. Although the magnitude of these associations was small, these results are hypothesis generating and suggest that changes in diet, body fat mass, and the subsequent metabolic changes may affect epigenetic biomarkers. Furthermore, results suggest that DNA methylation biomarkers may be suitable intermediate biomarkers for lifestyle intervention trials.
A decrease in global DNA methylation is associated with increased genomic instability and chromosomal rearrangements (38, 39), a common biological mechanism in several diseases including cancer. DNA methylation is a mechanism to suppress the expression of repetitive and viral DNA sequences that can affect cell functioning (40). Reduced DNA methylation in WBCs may be an indicator of systemic hypomethylation and of cumulative environmental impacts. Conversely, an increase in DNA methylation levels of repetitive elements may have a protective effect against genomic instability and unwanted chromosomal rearrangements (41).
Our finding that LINE-1 DNA methylation levels increased at the end of our intervention is supported by recently published data of a statistically significant increase in LINE-1 methylation resulting from a dietary intervention (42). In the study by Scoccianti et al. (42), all groups experienced a moderate increase in LINE-1 DNA methylation levels after 4 wk. However, the authors found no relation between LINE-1 DNA methylation and specific diets, such as one with enriched flavonoids and isothiocyanates (particularly a cruciferous vegetable-based diet), or one supplemented with flavonoids (green tea and soy products). A study by Zhang et al. (17) also found that a high fruit and vegetable intake was associated with high DNA methylation of LINE-1 in blood. In addition, higher LINE-1 DNA methylation was found in individuals with higher exercise levels in the same population (18) and in other studies (20, 21), suggesting an increase in LINE-1 DNA methylation might be associated with healthy lifestyle habits. Future studies are needed to confirm these associations and to understand the importance of the biological mechanisms at play.
Results of previous studies suggest changes in glucose concentration can induce epigenetic changes. Increased LINE-1 and global DNA methylation have been associated with higher glucose concentrations and HOMA insulin resistance, respectively (43–45). In addition, chromatin modifications (46) and altered DNA methylation at specific sequences such as the leptin and cyclin-dependent kinase inhibitor 2A (CDKN2A) promoters (47, 48) have also been shown to be related to changes in glucose concentrations. A study that investigated DNA methyltransferase levels in a colon cancer cell line found that hypoglycemia lead to a decrease in the concentrations of this enzyme, and conversely, studies in mouse cells have shown higher DNA methyltransferase activity and higher DNA methylation of cytosines as a result of exposure to increased glucose concentrations, proposing a biological mechanism through which decreases in glucose can lead to a decrease in overall DNA methylation (48, 49). Our study adds to the growing evidence that suggests glucose concentrations may result in increases in global DNA methylation levels; however, unlike previous studies, our findings were restricted to measurements by LUMA and not DNA methylation at repetitive sequences such as LINE-1. This might be due to the fact that we observed only a small change in glucose concentrations over our intervention period, which might not be enough to trigger a change in LINE-1 DNA methylation levels. Previous research investigated DNA methylation of LINE-1 and Alu repetitive elements (43–45). To our knowledge, our study is the first to explore a global measure of DNA methylation in association with variations in glucose concentrations. Our current results and the published evidence suggest glucose concentrations and metabolism might play a role in epigenetic changes. However, additional studies are needed to further explore this association.
In this study we assessed 3 different global epigenetic biomarkers to investigate differential effects of our intervention on blood DNA methylation levels. The epigenetic biomarkers used here target different genomic sequences representing different biological constructs that possibly respond differently to environmental and behavioral changes. We measured global DNA methylation by the LUMA assay, which targets CCGG sequences throughout the genome, and DNA methylation of the repetitive element LINE-1 and tandem repeat Sat2. CCGG sequences measured by LUMA account for ∼7% of all CpG sites on the genome and are used frequently as a surrogate measure of global DNA methylation levels (50–53). However, because it detects such a small fraction of the overall 5mC content, this method may underestimate changes in 5mC levels, possibly explaining why we did not observe an overall change in LUMA levels at the different time points but detected an association with individual dietary measures and glucose concentrations. However, although data are available on the effect of vegetable-based chemicals increasing DNA methylation markers such as LINE-1 (42), no data to date support caloric intake changing DNA methylation or epigenetic modifications. A previous study highlighted the high number of promoter sequences covered by LUMA (54). Therefore, LUMA might provide an indication of changes in the regulation of expression of specific genes. The estimates of the associations between changes in dietary factors and LUMA measures are statistically significant, but quite small, which raises two additional concerns. First, although we took precautions when performing laboratory measurements, we might be observing an effect related to the technical variation of the assay, which is higher than the observed change. Second, the biological significance of very small changes in DNA methylation is not known, and, even if these associations exist, their contribution to the epigenome may be negligible. Overall, we are cautious in our interpretation of these findings and believe they should be investigated in future studies.
Our study has several limitations. The sample size is small, which limits our ability to detect associations between these epigenetic marks and lifestyle modifications. It is also unclear how the duration of the intervention affects changes in epigenetic biomarkers. Increases in LINE-1 DNA methylation were observed at 6 mo after the initiation of the intervention and persisted to month 12, suggesting stability after the initial change. However, a longer intervention could provide information about the sustained effect of lifestyle changes in blood DNA methylation levels. An additional limitation is the assessment of dietary information, which was obtained from a self-reported questionnaire. As we previously noted about this particular study, given that the majority of the participants were overweight or obese and the mean reported caloric intake was 1250 kcal/d, it is likely that study participants underestimated their dietary intake (28). Inaccurate reporting at baseline for total caloric intake could result in an underestimation or an overestimation of mean percent change at 12 mo, which would affect the association with DNA methylation. Lastly, a common challenge using total WBCs is the heterogeneity in cell type composition (55). The relative amounts of the different blood cell fractions vary across individuals and might affect global DNA methylation biomarker levels (22). Therefore, care should be taken when comparing results to other studies.
Future studies should include a larger sample, with a potentially extended intervention and intermediate monitoring points to better assess the rate at which changes to epigenetic marks take place in blood. In addition, it is important to consider that the changes we expect to observe as a result of dietary interventions will not be large and most likely would be cumulative as a result of additional interactions with environmental, lifestyle, and demographic factors (56). Despite its limitations, our study contributes to the growing evidence that suggests that the health impacts of changes in dietary habits and other lifestyle factors may be mediated through epigenetic modifications. Our study provides preliminary evidence for the use of WBC DNA methylation biomarkers to monitor lifestyle interventions. However, larger and longer lifestyle interventions are needed to clearly assess the effects on global DNA methylation levels.
Acknowledgments
LD-C, WYT, KDC, RMS, DLH, and HG designed the research; LD-C, QW, KDC, DLH, and HG conducted the research; WZ, JAM, WYT, CV, and LF analyzed the data; LD-C, WZ, JAM, KDC, and HG wrote the paper; and HG had primary responsibility for the final content. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: CDKN2A, cyclin-dependent kinase inhibitor 2A; CpG, cytosine guanine dinucleotide; GEE, generalized estimated equation; LINE-1, long interspersed nucleotide element 1; LUMA, luminometric methylation assay; PMR, percent of methylated reference; Sat2, satellite 2; WBC, white blood cell; 5mC, 5-methylcytosine.
References
- 1.Pulte D, Redaniel MT, Brenner H, Jeffreys M. Changes in survival by ethnicity of patients with cancer between 1992–1996 and 2002–2006: is the discrepancy decreasing? Ann Oncol 2012;23:2428–34. [DOI] [PubMed]
- 2.Rock CL, Flatt SW, Natarajan L, Thomson CA, Bardwell WA, Newman VA, Hollenbach KA, Jones L, Caan BJ, Pierce JP. Plasma carotenoids and recurrence-free survival in women with a history of breast cancer. J Clin Oncol 2005;23:6631–8. [DOI] [PubMed]
- 3.Holmes MD, Chen WY, Feskanich D, Kroenke CH, Colditz GA. Physical activity and survival after breast cancer diagnosis. JAMA 2005;293:2479–86. [DOI] [PubMed] [Google Scholar]
- 4.Holmes MD, Stampfer MJ, Colditz GA, Rosner B, Hunter DJ, Willett WC. Dietary factors and the survival of women with breast carcinoma. Cancer 1999;86:826–35. [DOI] [PubMed] [Google Scholar]
- 5.Pierce JP, Stefanick ML, Flatt SW, Natarajan L, Sternfeld B, Madlensky L, Al-Delaimy WK, Thomson CA, Kealey S, Hajek R, et al. . Greater survival after breast cancer in physically active women with high vegetable-fruit intake regardless of obesity. J Clin Oncol 2007;25:2345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rock CL, Demark-Wahnefried W. Nutrition and survival after the diagnosis of breast cancer: a review of the evidence. J Clin Oncol 2002;20:3302–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barres R, Zierath JR. DNA methylation in metabolic disorders. Am J Clin Nutr 2011;93:897S–900S. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Xu X. Diet, epigenetic, and cancer prevention. Adv Genet 2010;71:237–55. [DOI] [PubMed]
- 9.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics 2011;6:828–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, et al. . Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res 2007;67:876–80. [DOI] [PubMed] [Google Scholar]
- 11.Kim KY, Kim DS, Lee SK, Lee IK, Kang JH, Chang YS, Jacobs DR, Steffes M, Lee DH. Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy Koreans. Environ Health Perspect 2010;118:370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect 2008;116:1547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing C, Wang QF, Li B, Tian H, Ni Y, Yin S, Li G. Methylation and expression analysis of tumor suppressor genes p15 and p16 in benzene poisoning. Chem Biol Interact 2010;184:306–9. [DOI] [PubMed] [Google Scholar]
- 14.Pilsner JR, Hu H, Ettinger A, Sanchez BN, Wright RO, Cantonwine D, Lazarus A, Lamadrid-Figueroa H, Mercado-Garcia A, Tellez-Rojo MM, et al. . Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect 2009;117:1466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pilsner JR, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, Factor-Litvak P, Graziano JH, Gamble MV. Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Bangladeshi adults. Am J Clin Nutr 2007;86:1179–86. [DOI] [PubMed] [Google Scholar]
- 16.Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK, Hu H, Sparrow D, Vokonas P, Baccarelli A. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect 2010;118:790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang FF, Morabia A, Carroll J, Gonzalez K, Fulda K, Kaur M, Vishwanatha JK, Santella RM, Cardarelli R. Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population. J Nutr 2011;141:1165–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang FF, Cardarelli R, Carroll J, Zhang S, Fulda KG, Gonzalez K, Vishwanatha JK, Morabia A, Santella RM. Physical activity and global genomic DNA methylation in a cancer-free population. Epigenetics 2011;6:293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aslibekyan S, Wiener HW, Havel PJ, Stanhope KL, O'Brien DM, Hopkins SE, Absher DM, Tiwari HK, Boyer BB. DNA methylation patterns are associated with n–3 fatty acid intake in Yup’ik people. J Nutr 2014;144:425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luttropp K, Nordfors L, Ekstrom TJ, Lind L. Physical activity is associated with decreased global DNA methylation in Swedish older individuals. Scand J Clin Lab Invest 2013;73:184–5. [DOI] [PubMed] [Google Scholar]
- 21.White AJ, Sandler DP, Bolick SC, Xu Z, Taylor JA, DeRoo LA. Recreational and household physical activity at different time points and DNA global methylation. Eur J Cancer 2013;49:2199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, Yang AS, Vokonas P, Lissowska J, Fustinoni S, et al. . Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol 2012;41:126–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, Kelsey KT. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 2007;16:108–14. [DOI] [PubMed] [Google Scholar]
- 24.Figueiredo JC, Grau MV, Wallace K, Levine AJ, Shen L, Hamdan R, Chen X, Bresalier RS, McKeown-Eyssen G, Haile RW, et al. Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev 2009;18:1041–9. [DOI] [PMC free article] [PubMed]
- 25.Hillemacher T, Frieling H, Moskau S, Muschler MA, Semmler A, Kornhuber J, Klockgether T, Bleich S, Linnebank M. Global DNA methylation is influenced by smoking behaviour. Eur Neuropsychopharmacol 2008;18:295–8. [DOI] [PubMed]
- 26.Wilhelm CS, Kelsey KT, Butler R, Plaza S, Gagne L, Zens MS, Andrew AS, Morris S, Nelson HH, Schned AR, et al. Implications of LINE1 methylation for bladder cancer risk in women. Clin Cancer Res 2010;16:1682–9. [DOI] [PMC free article] [PubMed]
- 27.Piyathilake CJ, Badiga S, Alvarez RD, Partridge EE, Johanning GL. A lower degree of PBMC L1 methylation is associated with excess body weight and higher HOMA-IR in the presence of lower concentrations of plasma folate. PLoS ONE 2013;8:e54544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenlee HA, Crew KD, Mata JM, McKinley PS, Rundle AG, Zhang W, Liao Y, Tsai WY, Hershman DL. A pilot randomized controlled trial of a commercial diet and exercise weight loss program in minority breast cancer survivors. Obesity (Silver Spring) 2013;21:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Block G, Hartman AM, Naughton D. A reduced dietary questionnaire: development and validation. Epidemiology 1990;1:58–64. [DOI] [PubMed] [Google Scholar]
- 30.Ainsworth BE, Sternfeld B, Richardson MT, Jackson K. Evaluation of the Kaiser physical activity survey in women. Med Sci Sports Exerc 2000;32:1327–38. [DOI] [PubMed] [Google Scholar]
- 31.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and B-cell function from fasting glucose and insulin concentrations in man. Diabetologia 1985;28:412–9. [DOI] [PubMed] [Google Scholar]
- 32.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekstrom TJ, Harris TB, et al. . Intra-individual change over time in DNA methylation with familial clustering. JAMA 2008;299:2877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karimi M, Johansson S, Ekstrom TJ. Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics 2006;1:45–8. [DOI] [PubMed] [Google Scholar]
- 34.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 2004;32:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005;33:6823–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.United States Department of Agriculture [Internet]. [cited 2014 Aug 29]. Available from: www.choosemyplate.gov/weight-management-calories/calories/empty-calories-amount.html.
- 37.American Institute for Cancer Research [Internet]. [cited 2014 Aug 29]. Available from: www.aicr.org.
- 38.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010;70:27–56. [DOI] [PubMed] [Google Scholar]
- 39.Toyota M, Suzuki H. Epigenetic drivers of genetic alterations. Adv Genet 2010;70:309–23. [DOI] [PubMed] [Google Scholar]
- 40.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 1997;13:335–40. [DOI] [PubMed] [Google Scholar]
- 41.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007;8:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scoccianti C, Ricceri F, Ferrari P, Cuenin C, Sacerdote C, Polidoro S, Jenab M, Hainaut P, Vineis P, Herceg Z. Methylation patterns in sentinel genes in peripheral blood cells of heavy smokers: influence of cruciferous vegetables in an intervention study. Epigenetics 2011;6:1114–9. [DOI] [PubMed] [Google Scholar]
- 43.Ulrich CM, Toriola AT, Koepl LM, Sandifer T, Poole EM, Duggan C, McTiernan A, Issa JP. Metabolic, hormonal and immunological associations with global DNA methylation among postmenopausal women. Epigenetics 2012;7:1020–8. [DOI] [PMC free article] [PubMed]
- 44.Pearce MS, McConnell JC, Potter C, Barrett LM, Parker L, Mathers JC, Relton CL. Global LINE-1 DNA methylation is associated with blood glycaemic and lipid profiles. Int J Epidemiol 2012;41:210–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J, Goldberg J, Bremner JD, Vaccarino V. Global DNA methylation is associated with insulin resistance: a monozygotic twin study. Diabetes 2012;61:542–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pirola L, Balcerczyk A, Tothill RW, Haviv I, Kaspi A, Lunke S, Ziemann M, Karagiannis T, Tonna S, Kowalczyk A, et al. . Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells. Genome Res 2011;21:1601–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouchard L, Thibault S, Guay SP, Santure M, Monpetit A, St-Pierre J, Perron P, Brisson D. Leptin gene epigenetic adaptation to impaired glucose metabolism during pregnancy. Diabetes Care 2010;33:2436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skowronski K, Dubey S, Rodenhiser D, Coomber B. Ischemia dysregulates DNA methyltransferases and p16INK4a methylation in human colorectal cancer cells. Epigenetics 2010;5:547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiang EP, Wang YC, Chen WW, Tang FY. Effects of insulin and glucose on cellular metabolic fluxes in homocysteine transsulfuration, remethylation, S-adenosylmethionine synthesis, and global deoxyribonucleic acid methylation. J Clin Endocrinol Metab 2009;94:1017–25. [DOI] [PubMed] [Google Scholar]
- 50.Deneberg S, Grovdal M, Karimi M, Jansson M, Nahi H, Corbacioglu A, Gaidzik V, Dohner K, Paul C, Ekstrom TJ, et al. . Gene-specific and global methylation patterns predict outcome in patients with acute myeloid leukemia. Leukemia 2010;24:932–41. [DOI] [PubMed] [Google Scholar]
- 51.Lee JJ, Geli J, Larsson C, Wallin G, Karimi M, Zedenius J, Hoog A, Foukakis T. Gene-specific promoter hypermethylation without global hypomethylation in follicular thyroid cancer. Int J Oncol 2008;33:861–9. [PubMed] [Google Scholar]
- 52.Römermann D, Hasemeier B, Metzig K, Schlegelberger B, Langer F, Kreipe H, Lehmann U. [Methylation status of LINE-1 sequences in patients with MDS or secondary AML.] Verh Dtsch Ges Pathol 2007;91:338–42(in German). [PubMed] [Google Scholar]
- 53.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat Rev Genet 2004;5:446–55. [DOI] [PubMed] [Google Scholar]
- 54.Xu X, Gammon MD, Hernandez-Vargas H, Herceg Z, Wetmur JG, Teitelbaum SL, Bradshaw PT, Neugut AI, Santella RM, Chen J. DNA methylation in peripheral blood measured by LUMA is associated with breast cancer in a population-based study. FASEB J 2012;26:2657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu HC, Delgado-Cruzata L, Flom JD, Kappil M, Ferris JS, Liao Y, Santella RM, Terry MB. Global methylation profiles in DNA from different blood cell types. Epigenetics 2011;6:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martínez JA, Milagro FI, Claycombe KJ, Schalinske KL. Epigenetics in adipose tissue, obesity, weight loss, and diabetes. Adv Nutr 2014;5:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]


