Abstract
Melampomagnolide B (MMB) is a natural sesquiterpene structurally related to parthenolide (PTL). We have shown that MMB exhibits anti-leukemic properties similar to PTL. Unlike PTL, the presence of a primary hydroxyl group in the MMB molecule allows the opportunity for examining the biological activity of a variety of conjugated analogs of MMB. We have now synthesized a series of carbamate analogs of MMB and evaluated these derivatives for anti-cancer activity against a panel of sixty human cancer cell lines. Analogs 6a and 6e exhibited promising anti-leukemic activity against human leukemia cell line CCRF-CEM with GI50 values of 680 and 620 nM, respectively. 6a also showed GI50 values of 1.98 and 1.38 µM respectively, against RPMI-8226 and SR leukemia cell lines and GI50 values of 460 and 570 nM against MDA-MB-435 melanoma and MDA-MB-468 breast cancer cell lines, respectively. 6e had GI50 values of 650 nM and 900 nM against HOP-92 non-small cell lung and RXF 393 renal cancer cell lines.
Keywords: Parthenolide, Melampomagnolide B, Anti-cancer activity, Carbamates, Heterocyclic amines
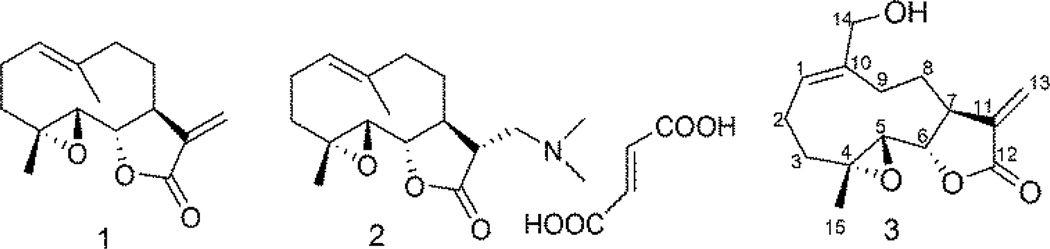
Parthenolide (PTL; 1, Fig. 1), a sesquiterpene lactone isolated from the medicinal herb Feverfew (Tanacetum parthenium), has been widely reported in the literature as an anticancer agent that is effective against both hematological and solid tumors.1,2 PTL and its analogs promote apoptosis by inhibiting the activity of the NF-κB transcription factor complex, and thereby down-regulates anti-apoptotic genes under NF-κB control.3 Recent studies also demonstrate that PTL induces robust apoptosis of primary acute myeloid leukemia (AML) stem cells in culture.4,5 AML is a clonal malignancy of the hematopoietic system characterized by accumulation of immature cell populations in the bone marrow or peripheral blood,6 and is the most common type of leukemia in adults but has the lowest survival rate of all leukemias.7
Figure 1.
Structures of PTL (1), DMAPT fumarate (2) and MMB (3)
More recently, we have shown that PTL and PTL analogs also selectively induce almost complete glutathione depletion and severe cell death in CD34+ AML cells, 8 but exhibit significantly less toxicity in normal CD34+ cells. PTL analogs perturb glutathione homeostasis by a multifactorial mechanism, including inhibition of key glutathione metabolic enzymes (GCLC and GPX1), and direct depletion of glutathione. Thus, primitive leukemia cells are uniquely sensitive to agents that target aberrant glutathione metabolism, an intrinsic property of primary human AML cells.
PTL is a major source for several novel anti-leukemic compounds arising from our research program over the past decade. The two best examples are: dimethylaminoparthenolide (DMAPT; 2, Fig. 1), and melampomagnolide B (MMB; 3, Fig 1). MMB is a melampolide originally isolated from Magnolia grandiflora.9 MMB can be synthesized from commercially available PTL via SeO2/tBuOOH oxidation.10, 11 Both of the above compounds 2 and 3 have been identified as new antileukemic sesquiterpenes with properties similar to PTL.11,12 DMAPT is currently in phase 1 clinical studies for evaluation as a treatment for acute myeloid leukemia cell (AML).12
More importantly, from a drug design point of view, MMB is a more intriguing molecule than either PTL or DMAPT because of the presence of the primary hydroxyl group at C-14, which can be structurally modified to improve potency, water solubility, bioavailability and tissue targeting of the molecule.
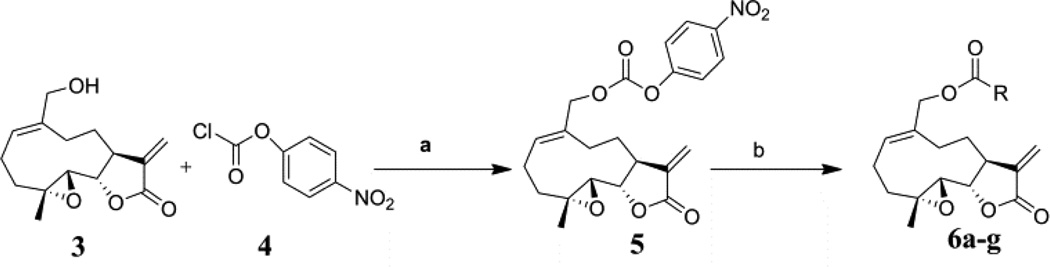
In the current study, we have prepared a series of novel carbamate analogs of MMB. These compounds were initially designed as potential prodrugs of MMB. However, we have found that on examining the anticancer activity of these compounds, several of the molecules exhibited significant growth inhibition properties in a panel of sixty human cancer cell lines. Two of these compounds exhibited GI50 values of ≤10 µM against the majority of the human cancer cell lines in the panel.
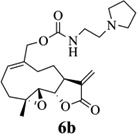
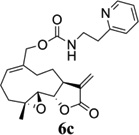
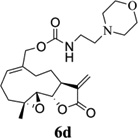
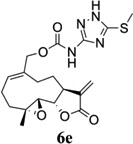
Carbamate analogs of MMB were prepared by reaction of the p-nitrophenyloxycarbonyl ester of MMB13 with a variety of primary and secondary heterocyclic amines containing pyrrolidine, morpholine, piperidine, imidazole, triazole and pyridine moieties, to afford carbamate products 6a–6g14 with generally improved water-solubility (Scheme 1, Table 1) compared to MMB. The key p-nitrophenyloxycarbonyl ester of MMB was prepared by the reaction of MMB with p-nitrophenylchloroformate in the presence of triethylamine. All conjugation reactions were carried out at ambient temperature in dichloromethane. We have reported previously that the reaction of sesquiterpenes containing an exocyclic double bond attached to the 13-position of the 5-membered lactone ring with primary and secondary amines leads to the facile formation of Michael addition products.15 However, under the reaction conditions employed in Scheme 1, the rate of O-carbamoylation appears to be much faster than the rate of C-13 Michael addition, and only in a few cases, with amines such as 2-morpholinoethylamine, 2-piperidinoethylamine and 3-aminopropylimidazole, were Michael addition byproducts observed (usually in low yields of 5–10%), due likely to the high nucleophilicity of these amines.
Scheme 1.
Synthesis of carbamoylated MMB analogs 6a–6g: (a) CH2Cl2, triethylamine, rt, 24 h; (b) CH2Cl2, heterocyclic amines, rt, 5–12 h.
Table 1.
Structures, reaction conditions, yields, and melting points for carbamate analogs of melampomagnolide B
| Amine | Product | Yield (%) |
Time (hr) |
Mp (°C) |
|---|---|---|---|---|
 |
50 | 12 | 150 | |
 |
67 | 12 | 50 | |
 |
72 | 8 | 150 | |
 |
65 | 6 | 80 | |
 |
70 | 8 | 107 | |
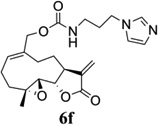 |
75 | 5 | 70 | |
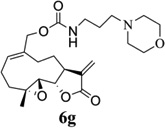 |
68 | 8 | 60 |
All compounds were purified by column chromatography (silica gel; methanol/dichloromethane) to afford pure compounds in 50–75% yield. The synthesized compounds were fully characterized by 1H NMR, 13C NMR and high resolution mass spectral analysis14.
The above carbamate analogs were evaluated for growth inhibition properties against a panel of 60 human cancer cell lines derived from nine human cancer cell types, grouped into disease sub-panels that represent leukemia, lung, colon, central nervous system (CNS), melanoma, renal, ovary, breast, and prostate cancer cells. Growth inhibitory (GI50) effects were measured as a function of the variation of optical density as a percentage of control.16, 17 Initial screening assays were carried out at a single concentration of 10 µM. Five analogs, 6a–6e, were identified as hits based on their ability to inhibit by 60% the growth of at least 8 of the 60 tumor cell lines in the panel. These five analogs were then evaluated in 5-dose assays over the concentration range 10−4–10−8 µM, and their GI50 values against the tumor cell lines in the panel determined (Table 2). Two analogs, 6a and 6e, were identified as lead compounds. Compound 6a exhibited potency against leukemia cell line CCRF-CEM, melanoma cell line MDA-MB-435 and breast cancer cell line MDA-MB-468 in the nanomolar range with GI50 values of 680 nM, 460 nM and 570 nM, respectively. Compound 6e was found to possess potent anti-leukemic activity against leukemia cell line CCRF-CEM, non-small cell lung cancer cell line HOP-92 and renal cancer cell line RXF 393 with GI50 values of 620 nM, 650 nM and 900 nM, respectively (Table 2).
Table 2.
Growth inhibition (GI50; µM)b data for PTL (1), DMAPT (2) and carbamoylated MMB analogs 6a–6e against a panel of human cancer cell lines
| Panel/cell line | 1a | 2a | 6a | 6b | 6c | 6d | 6e |
|---|---|---|---|---|---|---|---|
| Leukemia | GI50 | GI50 | GI50 | GI50 | GI50 | GI50 | GI50 |
| CCRF-CEM | 7.94 | 1.99 | 0.68 | 2.49 | 2.65 | 3.03 | 0.62 |
| HL-60(TB) | 5.01 | 1.58 | 2.04 | 4.15 | NDb | 3.59 | NDb |
| K-562 | 19.9 | 2.51 | 3.45 | 3.26 | NDb | 3.37 | NDb |
| MOLT-4 | 15.8 | 3.16 | 2.05 | 3.48 | 5.54 | 5.00 | 2.32 |
| RPMI-8226 | 7.94 | 2.51 | 1.98 | 8.71 | 8.20 | 5.72 | 2.57 |
| SR | NDb | ND | 1.38 | 10.2 | 4.10 | 3.65 | 2.36 |
| Non-Small cell lung cancer | |||||||
| HOP-92 | 12.5 | 10.0 | 1.45 | 2.25 | 2.25 | 2.30 | 0.65 |
| NCI-H522 | 5.01 | 2.51 | 1.25 | 1.78 | 1.64 | 2.13 | 1.26 |
| Colon cancer | |||||||
| COLO 205 | 15.8 | 31.6 | 2.06 | 4.78 | 3.54 | 8.89 | 1.79 |
| HCT-116 | 10.0 | 5.01 | 1.41 | 3.18 | 1.89 | 2.87 | 1.13 |
| SW-620 | 15.8 | 3.98 | 1.46 | 3.46 | 3.13 | 3.48 | 1.12 |
| CNS cancer | |||||||
| SF-539 | 19.9 | 2.51 | 1.98 | 15.0 | 5.77 | 14.4 | 1.76 |
| SNB-75 | 50.1 | NDb | 6.13 | 18.0 | 3.78 | 19.5 | 1.71 |
| Melanoma | |||||||
| LOX IMVI | 7.94 | 10.0 | 2.23 | 7.88 | 4.96 | 4.84 | 1.95 |
| MALME-3M | 12.5 | NDb | 1.90 | 4.18 | 7.52 | 6.12 | 2.32 |
| M14 | NDb | 15.8 | 2.84 | 9.65 | 5.58 | 7.97 | 1.59 |
| MDA-MB-435 | NDb | 7.94 | 0.46 | 6.56 | 6.01 | 5.89 | 2.24 |
| Ovarian cancer | |||||||
| IGROV1 | 19.9 | 19.9 | 2.31 | 4.09 | 14.4 | 3.49 | 3.66 |
| OVCAR-3 | 19.9 | 12.5 | 1.69 | 8.41 | NDb | 6.20 | NDb |
| Renal cancer | |||||||
| ACHN | NDb | 15.8 | 1.79 | 3.80 | 2.75 | 3.74 | 1.75 |
| CAKI-1 | 10.0 | 12.5 | 2.03 | 6.86 | 2.88 | 4.31 | 1.99 |
| RXF 393 | 12.5 | 15.8 | 1.20 | 4.08 | 2.22 | 3.00 | 0.90 |
| TK-10 | NDb | 3.16 | 2.60 | 3.11 | 3.78 | 3.93 | 2.51 |
| Prostate Cancer | |||||||
| DU-145 | NDb | 5.01 | 2.44 | 7.42 | 4.59 | 3.74 | 3.49 |
| Breast cancer | |||||||
| MCF7 | 15.8 | 5.01 | 1.62 | 3.91 | 2.85 | 3.54 | 1.33 |
| BT-549 | NDb | 5.01 | 2.69 | 4.75 | 2.60 | 4.99 | 1.47 |
| T-47D | NDb | 39.8 | 3.07 | 4.86 | 6.19 | 6.32 | 2.23 |
| MDA-MB-468 | NDb | NDb | 0.57 | 2.30 | 3.22 | 3.26 | 1.29 |
GI50 values obtained from NCI database
GI50, concentration of analog (µM) that halves cellular growth.
ND, not determined.
GI50 values < 1µM are bolded
Compounds 6a and 6e also exhibited significant growth inhibition against the following sub-panels of human cancer cell lines: non-small cell lung cancer (GI50 values 0.65–1.45 µM); colon cancer (GI50 values 1.12–2.06 µM); melanoma (GI50 values 0.46–2.84 µM); renal cancer (GI50 values 0.90–2.60 µM); and breast cancer (GI50 values 0.57–3.07 µM) (Table 2).
We have determined the hydrolytic stability of the above five carbamate derivatives in human plasma and have shown that compounds 6b–6e have half-lives in the range 100–180 min, while compound 6a has a much longer half-life of 8 h in human plasma. Thus, we consider compounds 6b–6d to be anticancer agents that are also metabolized by plasma esterases to the active parent compound MMB, while compound 6a would be considered a more potent anticancer agent than MMB that is likely not metabolized to MMB in vivo.
The above results are interesting for a number of reasons: first, the antileukemic activities of 6a and 6e against the sub-panel of human leukemia cells indicates that these carbamate analogs of MMB are more potent than the parent compound. Second, the potent growth inhibition properties of 6a and 6e against human solid tumor cell lines is the first report of such activities for MMB analogs. Third, these interesting results indicate that structural modification of the MMB molecule through appropriate carbamoylation of the primary hydroxyl group can lead to an improvement in the anticancer properties of MMB.
In summary, we have reported on a series of novel carbamate derivatives of MMB derived from heterocyclic and heteroaromatic amines. Among these derivatives, compounds 6a and 6e have been identified as potent anticancer agents with growth inhibition activities in the nanomolar range against a variety of hematological and solid tumor cell lines. Analogs 6a and 6e exhibit promising anti-leukemic activity against human leukemia cell line CCRF-CEM with GI50 values of 680 and 620 nM, respectively. Compound 6a also exhibits GI50 values of 460 and 570 nM against MDA-MB-435 melanoma and MDA-MB-468 breast cancer cell lines, respectively, and 6e has GI50 values of 650 nM and 900 nM against HOP-92 non-small cell lung and RXF 393 renal cancer cell lines, respectively. Further structure-activity relationship studies will focus on the structural optimization of these interesting lead analogs and on the molecular basis for their mechanism of action.
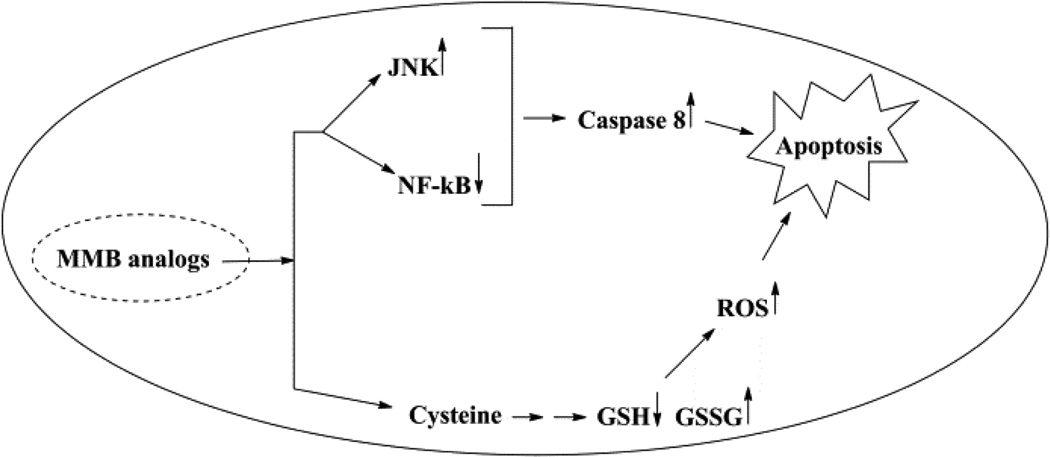
Figure 2.
Mechanism of action of MMB analogs
Acknowledgements
We are grateful to the NIH/National Cancer Institute (grant # R01 CA158275) for supporting this research, to the Arkansas Research Alliance for an Arkansas Scholar award and to the NCI drug screening program.
References and notes
- 1.Knight DW. Nat. Prod. Rep. 1995;12:271. doi: 10.1039/np9951200271. [DOI] [PubMed] [Google Scholar]
- 2.(a) Skalska J, Brookes PS, Nadtochiy SM, Hilchey SP, Jordan CT, Guzman ML, Maggirwar SB, Briehl MM, Bernstein SH. PLoS ONE. 2009;4:e8115. doi: 10.1371/journal.pone.0008115. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shama N, Crooks PA. Bioorg. Med. Chem. Lett. 2008;18:3870. doi: 10.1016/j.bmcl.2008.06.050. [DOI] [PubMed] [Google Scholar]; (c) Hewamana S, Alghazal S, Lin TT, Clement M, Jenkins C, Guzman ML, Jordan CT, Neelakantan S, Crooks PA, Burnett AK, Pratt G, Fegan C, Rowntree C, Brennan P, Pepper C. Blood. 2008;111:4681. doi: 10.1182/blood-2007-11-125278. [DOI] [PubMed] [Google Scholar]; (d) Oka D, Nishimura K, Shiba M, Nakai Y, Arai Y, Nakayama M, Takayama H, Inoue H, Okuyama A, Nonomura N. Int. J. Cancer. 2007;120:2576. doi: 10.1002/ijc.22570. [DOI] [PubMed] [Google Scholar]
- 3.(a) Bork PM, Schmitz ML, Kuhnt M, Escher C, Heinrich M. FEBS Lett. 1997;402:85. doi: 10.1016/s0014-5793(96)01502-5. [DOI] [PubMed] [Google Scholar]; (b) Wen J, You KR, Lee SY, Song CH, Kim DG. J. Biol. Chem. 2002;277:38954. doi: 10.1074/jbc.M203842200. [DOI] [PubMed] [Google Scholar]; (c) Hehner SP, Heinrich M, Bork PM, Vogt M, Ratter F, Lehmann V, Schulze-Osthoff K, Dröge W, Schmitz ML. J. Biol. Chem. 1998;273:1288. doi: 10.1074/jbc.273.3.1288. [DOI] [PubMed] [Google Scholar]; (d) Sweeney CJ, Li L, Shanmugam R, Bhat-Nakshatri PB, Jayaprakasan V, Baldridge LA, Gardner T, Smith M, Nakshatri H, Cheng L. Clin. Cancer Res. 2004;10:5501. doi: 10.1158/1078-0432.CCR-0571-03. [DOI] [PubMed] [Google Scholar]; (e) Yip-Schneider MT, Nakshatri H, Sweeney CJ, Marshall MS, Wiebke EA, Schmidt CM. Mol. Cancer Ther. 2005;4:587. doi: 10.1158/1535-7163.MCT-04-0215. [DOI] [PubMed] [Google Scholar]; (f) Nozaki S, Sledge GW, Nakshatri H. Oncogene. 2001;20:2178. doi: 10.1038/sj.onc.1204292. [DOI] [PubMed] [Google Scholar]
- 4.Guzman ML, Rossi RM, Karnischky L, Li X, Peterson DR, Howard DS, Jordan CT. Blood. 2005;105:4163. doi: 10.1182/blood-2004-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Guzman ML, Jordan CT. Expert Opin. Biol. Ther. 2005;5:1147. doi: 10.1517/14712598.5.9.1147. [DOI] [PubMed] [Google Scholar]; (b) Dai Y, Guzman ML, Chen S, Wang L, Yeung SK, Pei XY, Dent P, Jordan CT, Grant S. Br. J. Haematol. 2010;151:70. doi: 10.1111/j.1365-2141.2010.08319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kim YR, Eom JI, Kim SJ, Jeung HK, Cheong JW, Kim JS, Min YH. J. Pharmacol. Exp. Ther. 2010;335:389. doi: 10.1124/jpet.110.169367. [DOI] [PubMed] [Google Scholar]
- 6.Deschler B, Lubbert M. Cancer. 2006;107:2099. doi: 10.1002/cncr.22233. [DOI] [PubMed] [Google Scholar]
- 7.(a) Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]; (b) Lowenberg B, Suciu S, Archimbaud E, Haak H, Stryckmans P, De Cataldo R, Dekker AW, Berneman ZN, Thyss A, Van der Lelie J, Sonneveld P, Visani G, Fillet G, Hayat M, Hagemeijer A, Solbu G, Zittoun R. J Clin Oncol. 1998;16:872. doi: 10.1200/JCO.1998.16.3.872. [DOI] [PubMed] [Google Scholar]; (c) Tazzari PL, Cappellini A, Ricci F, Evangelisti C, Papa V, Grafone T, Martinelli G, Conte R, Cocco L, McCubrey JA, Martelli AM. Leukemia. 2007;21:427. doi: 10.1038/sj.leu.2404523. [DOI] [PubMed] [Google Scholar]
- 8.Pei S, Minhajuddin M, Callahan KP, Balys M, Ashton JM, Neering SJ, Lagadinou ED, Corbett C, Ye H, Liesveld JL, O’Dwyer KM, Li Z, Shi L, Greninger P, Settleman J, Benes C, Hagen FK, Munger J, Crooks PA, Becker MW, Jordan CT. J. Biol. Chem. 2013;288:33542. doi: 10.1074/jbc.M113.511170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Feraly FS. Phytochemistry. 1984;23:2372. [Google Scholar]
- 10.Macias FA, Galindo JCG, Massanet GM. Phytochemistry. 1992;31:1969. [Google Scholar]
- 11.Shama N, ShanShan P, Fred KH, Craig TJ, Peter AC. Bioorg. Med. Chem. 2011;19:1515. [Google Scholar]
- 12.Guzman ML, Rossi RM, Neelakantan S, Li X, Corbett CA, Hassane DC, Becker MW, Bennett JM, Sullivan E, Lachowicz JL, Vaughan A, Sweeney CJ, Matthews W, Carroll M, Liesveld JL, Crooks PA, Jordan CT. Blood. 2007;110:4427. doi: 10.1182/blood-2007-05-090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Synthetic procedure and analytical data for the p-nitrophenyloxycarbonyl ester of MMB (5): To the reaction mixture of MMB (100 mg, 0.378 mmol) and triethylamine (45.8 mg, 0.454 mmol) in dichloromethane (2 mL), p-nitrophenyl chloroformate (76.3 mg, 0.378 mmol) was added at 0 °C. The reaction mixture was stirred for 24 h at ambient temperature. When the reaction was completed (monitored by TLC), water was added to the reaction mixture and the aqueous mixture was extracted with dichloromethane. The organic layer was washed with water, followed by brine solution, dried over anhydrous Na2SO4 and concentrated to afford the crude product. The crude product was purified by column chromatography (silica gel, 2% methanol in dichloromethane) to afford compound 5 as a pale yellow solid. 1H NMR (CDCl3, 400 MHz): δ 8.26 (d, J = 9.6 Hz, 2H), 7.37(d, J = 9.8 Hz, 2H), 6.25 (s, 1H), 5.83 (t, J = 8.4 Hz, 1H), 5.56 (s, 1H), 4.81(d, J = 12.8 Hz, 1H), 4.72 (d, J = 12.4 Hz, 1H), 3.86 (m, 1H), 2.85 (m, 2H), 2.56 (m, 7H), 1.77 (m, 2H), 1.55 (s, 1H), 1.16 (t, J = 13.2 Hz, 1H). 13C NMR (CDCl3, 100 MHz): δ 169.1, 155.2, 152.2, 145.4, 138.6, 133.6, 132.8, 125.3, 121.5, 120.3, 80.8, 71.5, 63.1, 59.8, 42.7, 36.4, 25.6, 24.9, 23.9, 17.9 ppm.
- 14.General synthetic procedure and analytical data for carbamate derivatives of MMB: To the p-nitro phenyloxycarbonyl ester derivative of MMB (5) (70 mg, 0.16 m mol) in dichloromethane (2 mL), the appropriate amine (0.16 m mol) was added at 0°C. The reaction mixture was stirred for 18 h at ambient temperature. When the reaction was completed (monitored by TLC), water was added to the reaction mixture and the aqueous mixture was extracted with dichloromethane. The organic layer was washed with water, followed by brine solution, dried over anhydrous Na2SO4 and concentrated to afford the crude product. The crude product was purified by column chromatography (silica gel, 5% methanol in dichloromethane) to afford the carbamate analogs (6a–g) as white solids. ((1aR,7aS,10aS,10bS,E)-1a-methyl-8-methylene-9-oxo-1a,2,3,6,7,7a,8,9, 10a,10b-decahydrooxireno[2',3':9,10]cyclodeca[1,2-b]furan-5-yl)methyl-4,4-difluoropiperidine-1-carboxylate (6a) 1H NMR (CDCl3), 400 MHz): δ 6.27 (d, J = 2.8 Hz, 1H), 5.67 (t, J = 8.4 Hz, 1H), 5.56 (s, 1H), 4.69 (d, J = 12.4 Hz, 1H), 4.52 (d, J = 12 Hz, 1H), 3, 87 (t, J = 9.6 Hz, 1H), 3.60 (brs, 4H), 2.87 (d, J = 9.2 Hz, 2H), 2.50-2.16 (m, 6H), 1.96 (brs, 4H), 1.71(t, J = 10 Hz, 1H), 1.55 (s, 3H), 1.14 (t, J = 12 Hz, 1H). 13C NMR (CDCl3, 100 MHz): δ 169.4, 154.8, 138.7, 135.3, 129.9, 121.5, 120.5 (t, JCF = 5.3 Hz, 1C), 81.1, 67.7, 63.4, 60.0, 42.7, 41.0, 36.7, 34.0, 25.8, 24.4, 23.9, 18.1 ppm. HRMS (ESI) m/z calcd for C21H28F2NO5 (M + H)+ 412.1930, found 412.1933. ((1aR,7aS,10aS,10bS,E)-1a-methyl-8-methylene-9-oxo-1a,2,3,6,7,7a,8,9, 10a,10b-decahydrooxireno[2',3':9,10]cyclodeca[1,2-b]furan-5-yl)methyl-(2-(pyrrolidin-1-yl)ethyl) carbamate (6b) 1H NMR (CDCl3, 400 MHz): δ 6.22 (d, J = 3.2 Hz, 1H), 5.65 (t, J = 7 Hz, 1H), 5.55 (s, 1H), 4.62 (d, J = 11.6 Hz, 1H), 4.47 (d, J = 12.4 Hz, 1H), 3.82 (t, J = 9.6 Hz, 1H), 3.35 (s, 2 H), 2.90-2.83 (m, 2H), 2.70 (s, 4H), 2.42 (d, J = 9.6 Hz, 2H), 2.38-2.13 (m, 7H) 1.84 (s, 4H), 1.66 (t, J = 12 Hz, 1H), 1.52 (s, 3H), 1.13 (t, J = 11.6 Hz, 1H). 13C NMR (CDCl3, 100 MHz): δ 169.3, 156.1, 138.6, 135.3, 129.8, 120.1, 80.9, 67.0, 63.1, 59.8, 55.1, 53.8, 42.5, 38.9, 36.5, 25.6, 24.3, 23.6, 23.2, 17.8 ppm. HRMS (ESI) m/z calcd for C22H33N2O5 (M + H)+ 405.2384, found 405.2390. ((1aR,7aS,10aS,10bS,E)-1a-methyl-8-methylene-9-oxo-1a,2,3,6,7,7a,8,9, 10a,10b-decahydrooxireno[2',3':9,10]cyclodeca[1,2-b]furan-5-yl)methyl-(2-(pyridin-2-yl)ethyl) carbamate (6c) 1H NMR (CDCl3, 400 MHz): δ 8.50 (s, 1H), 7.62 (t, J = 7.2 Hz, 1H), 7.15 (d, J = 7.6 Hz, 2H), 6.19 (s, 1H), 5.64 (t, J = 8 Hz, 1H), 5.56-5.56 (m, 2H), 4.62 (d, J = 12.4 Hz, 1H), 4.44 (d, J = 12.8 Hz, 1H), 3.84 (t, J = 9.2 Hz, 1H), 3.60 (d, J = 6 Hz, 2H), 2.99-2.82 (m, 4H), 2.41-2.12 (m, 5H), 1.75 (s, 1H), 1.64 (d, J = 10.4 Hz, 1H), 1.52 (s, 3H), 1.07 (t, J = 13.6 Hz, 1H). 13C NMR (CDCl3, 100 MHz): δ 169.3, 159.1, 156.0, 149.1, 138.6, 136.6, 135.5, 129.9, 123.4, 121.6, 120.2, 81.0, 67.0, 63.2, 59.8, 42.5, 40.1, 37.1, 36.6, 25.8, 24.4, 23.7, 17.9 ppm. HRMS (ESI) m/z calcd for C23H29N2O5 (M + H)+ 413.2071, found 413.2073. ((1aR,7aS,10aS,10bS,E)-1a-methyl-8-methylene-9-oxo-1a,2,3,6,7,7a,8,9,10a,10b-decahydrooxireno[2',3':9,10]-cyclodeca[1,2-b]furan-5-yl)-methyl(2-morpholino ethyl) carbamate (6d) 1H NMR (CDCl3, 400 MHz): δ 6.11(d, J = 3.6 Hz, 1H), 5.58 (t, J = 8 Hz, 1H), 5.43 (d, J = 2.8 Hz, 1H), 5.06 (s, 1H), 4.53 (d, J = 12.4 Hz, 1H), 4.34 (d, J = 12 Hz, 1H), 3.74 (t, J = 8.8 Hz, 1H), 3.56 (s, 4 H), 3.16 (t, J = 5.2 Hz, 2H), 2.78-2.72 (m, 2H), 2.32 (s, 8H), 2.21-2.00 (m, 4H), 1.55(t, J = 10.4 Hz, 1H), 1.41 (s, 3H), 1.01 (t, J = 12 Hz, 1H). 13C NMR (CDCl3, 100 MHz): δ 169.2, 155.9, 138.7, 135.4, 130.2, 120.0, 80.9, 67.0, 66.7, 63.1, 59.8, 57.2, 53.1, 42.5, 37.0, 36.5, 25.7, 24.4, 23.7, 17.8 ppm. HRMS (ESI) m/z calcd for C22H33N2O6 (M + H)+ 421.2333, found 421.2331. ((1aR,7aS,10aS,10bS,E)-1a-methyl-8-methylene-9-oxo-1a,2,3,6,7,7a,8,9, 10a,10b-decahydrooxireno[2',3':9,10]cyclodeca[1,2-b]furan-5-yl)methyl-(5-(methylthio)-1H-1,2,4-triazol-3-yl)carbamate (6e) 1H NMR (CDCl3, 400 MHz): δ 6.22 (s, 2H), 5.82 (t, J = 8 Hz, 1H), 5.50 (s, 1H), 4.90 (d, J = 12.4 Hz, 1H), 4.81(d, J = 12.4 Hz, 1H), 3.85 (t, J = 9.6 Hz, 1H), 2.95 (s, 1H), 2.85 (d, J = 9.2 Hz, 1H), 2.48-2.15 (m, 8H), 1.70-1.53 (m, 6H), 1.13 (t, J = 12.4 Hz, 1H) ppm. 13C NMR (CDCl3, 100 MHz): δ 169.1, 163.1, 157.4, 149.9, 138.5, 133.3, 132.7, 120.2, 80.7, 70.1, 62.9, 59.7, 42.4, 36.2, 25.6, 24.3, 23.7, 17.8, 13.5 ppm. HRMS (ESI) m/z calcd for C19H25N4O5S (M + H)+ 421.1540, found 421.1524. ((1aR,7aS,10aS,10bS,E)-1a-methyl-8-methylene-9-oxo-1a,2,3,6,7,7a,8,9, 10a,10b-decahydrooxireno[2',3':9,10]cyclodeca[1,2-b]furan-5-yl)methyl-(3-(1H-imidazol-1-yl)-propyl) carbamate (6f) 1H NMR (CDCl3, 400 MHz): δ 7.56 (s, 1H), 7.09 (s, 1H), 6.95 (s, 1H), 6.26 (d, J = 3.6 Hz, 1H), 5.69 (t, J = 8 Hz, 1H), 5.56 (d, J = 3.2 Hz, 1H), 4.85 (s, 1H), 4.64 (d, J = 12.4 Hz, 1H), 4.51(d, J = 12.4 Hz, 1H), 4.04 (t, J = 7.2 Hz, 2H), 3.88 (t, J = 9.2 Hz, 1H), 3.21 (d, J = 6 Hz, 2H), 2.93-2.85 (m, 2H), 2.47-2.16 (m, 7H), 2.03 (t, J = 6.4 Hz, 1H) 1.70 (t, J = 10.8 Hz, 1H), 1.55 (s, 3H), 1.15 (t, J = 12.4 Hz, 1H). 13C NMR (CDCl3, 100 MHz): δ 169.5, 156.3, 138.9, 137.1, 135.3, 130.4, 129.6, 120.2, 118.8, 81.1, 67.3, 63.3, 60.0, 44.4, 42.7, 38.3, 36.7, 31.5, 25.8, 24.6, 23.8, 18.0 ppm. HRMS (ESI) m/z calcd for C22H30N3O5 (M + H)+ 416.2180, found 416.2183. ((1aR,7aS,10aS,10bS,E)-1a-methyl-8-methylene-9-oxo-1a,2,3,6,7,7a,8,9, 10a,10b-decahydrooxireno[2',3':9,10]-cyclodeca[1,2-b]furan-5-yl)methyl-(3-morpholinopropyl) carbamate (6g) 1H NMR (CDCl3, 400 MHz): δ 6.23 (s, 1H), 5.74 (s, 1H), 5.65 (t, J = 8 Hz, 1H), 5.54 (s, 1H), 4.58 (d, J = 12 Hz, 1H), 4.48 (d, J = 16 Hz, 1H), 3.85 (t, J = 9.2 Hz, 1H), 3.72 (s, 4 H), 3.27 (s, 2H), 2.89-2.83 (m, 2H), 2.46-2.11 (m, 12H), 1.69 (t, J = 12 Hz 3H), 1.52 (s, 3H), 1.11 (t, J = 12 Hz, 1H). 13C NMR (CDCl3, 100 MHz): δ 169.5, 156.3, 138.9, 135.7, 129.9, 120.4, 81.2, 67.0, 66.9, 63.4, 60.1, 57.4, 53.6, 42.7, 36.8, 25.9, 25.5, 24.6, 23.9, 18.1 ppm. HRMS (ESI) m/z calcd for C23H35N2O6 (M + H)+ 435.2490, found 435.2482.
- 15.Neelakantan S, Shama N, Guzman ML, Jordan CT, Crooks PA. Bioorg. Med. Chem. Lett. 2009;19:4346. doi: 10.1016/j.bmcl.2009.05.092. [DOI] [PubMed] [Google Scholar]
- 16.Boyd MR, Paull KD. Drug Dev. Res. 1995;34:91. [Google Scholar]
- 17.Acton EM, Narayanan VL, Risbood PA, Shoemaker RH, Vistica DT, Boyd MR. J. Med. Chem. 1994;37:2185. doi: 10.1021/jm00040a010. [DOI] [PubMed] [Google Scholar]