Abstract
The age of polymorphic alleles in humans is often estimated from population genetic patterns in extant human populations, such as allele frequencies, linkage disequilibrium, and rate of mutations. Ancient DNA can improve the accuracy of such estimates, as well as facilitate testing the validity of demographic models underlying many population genetic methods. Specifically, the presence of an allele in a genome derived from an ancient sample testifies that the allele is at least as old as that sample. In this study, we consider a common method for estimating allele age based on allele frequency as applied to variants from the US National Institutes of Health (NIH) Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project. We view these estimates in the context of the presence or absence of each allele in the genomes of the 5300 year old Tyrolean Iceman, Ötzi, and of the 50,000 year old Altai Neandertal. Our results illuminate the accuracy of these estimates and their sensitivity to demographic events that were not included in the model underlying age estimation. Specifically, allele presence in the Iceman genome provides a good fit of allele age estimates to the expectation based on the age of that specimen. The equivalent based on the Neandertal genome leads to a poorer fit. This is likely due in part to the older age of the Neandertal and the older time of the split between modern humans and Neandertals, but also due to gene flow from Neandertals to modern humans not being considered in the underlying demographic model. Thus, the incorporation of ancient DNA can improve allele age estimation, demographic modeling, and tests of natural selection. Our results also point to the importance of considering a more diverse set of ancient samples for understanding the geographic and temporal range of individual alleles.
Keywords: Ötzi, Neandertals, Europe
Introduction
The quality of ancient human DNA (aDNA) sequencing has steadily improved during the past decade, culminating recently in the high-coverage sequencing of two archaic hominin genomes from human skeletal remains pre-dating 40,000 years ago (Meyer et al., 2012; Prüfer et al., 2014). Additionally, Meyer et al. (2013) recently sequenced a mitochondrial genome from the Middle Pleistocene site of Sima de los Huesos in Spain, pushing the oldest sequenced human DNA beyond 300,000 years. In addition to providing information about human demographic history, the growing sample of aDNA is useful for understanding the age of mutations that segregate in extant populations and, therefore, the timing of natural selection that has shaped present-day human populations. Herein, we illustrate the importance of aDNA for addressing questions of allele age and timing of selection by first briefly reviewing related literature, and by analyzing allele age in the context of two ancient genomes from different time periods and different phylogenetic distance to present-day Europeans. By comparing with allele age estimates based on allele frequency in an extant population, we conclude that consideration of whether an allele is present or absent in aDNA can provide important information about allele age and improve on the former type of estimates.
Presence of recently selected alleles in ancient European specimens
A straightforward means of testing selective hypotheses, e.g., based on long-range haplotype methods (Sabeti et al., 2002; Voight et al., 2006), is to analyze putatively selected haplotypes and single-nucleotide variants (SNVs) in ancient human specimens. Perhaps the most successful application to date of aDNA to a potential case of recent natural selection pertains to the genetic variants underlying lactase persistence. The ability to digest lactose, a disaccharide found in milk, typically slows in most mammals around the time of weaning. In many humans the ability to produce the enzyme lactase, which digests lactose into glucose and galactose sugars, continues into adulthood, a phenotype known as ‘lactase persistence’. The timing and evolution of lactase persistence was documented first in Europeans, where the genomic region surrounding LCT, the gene encoding lactase, has been consistently reported as one of the most extreme long-range haplotype based examples of recent selection in Europe (Enattah et al., 2002, 2007, 2008; Bersaglieri et al., 2004; Tishkoff et al., 2006). The selection has been shown to be in the nearby MCM6 gene and results in downregulation of the cessation of lactase production after weaning (Enattah et al., 2002). Similar disruptive changes to the MCM6 gene have convergently evolved in both African (Tishkoff et al., 2006) and Middle Eastern (Enattah et al., 2007) populations. Selection for lactase persistence shows the importance of comparing genetic data to known cultural changes in the past, such as the timing and geographic distribution of cattle and camel pastoralism and milk consumption (Holden and Mace,1997; Gerbault et al., 2009). The age of the mutation and subsequent beginning of the selective sweep underlying lactase persistence in Europeans (C/T-13910) has been estimated between 3000 and 12,000 years, which seems to coincide with the presence of domesticated cattle (Bollongino et al., 2006) and a record of increasing pastoralism and dairying in several human populations, particularly in northern Europe. For example, Tishkoff and colleagues (2006) estimated the age, using a coalescent simulation model that incorporated selection and recombination, at approximately 8000 to 9000 years depending on the degree of dominance assumed for the allele. While consistent with the anthropological record, the confidence intervals spanning 2000 to 19,000 years points to the large uncertainty in the estimates. This is consistent with the large range of variation in coalescence times (Slatkin and Rannala, 2000).
Estimates of allele age and timing of selection based on population genetic patterns observed in extant humans depend heavily on assumptions about the demographic history of human populations and are often associated with large ranges of error (as illustrated above for the timing of C/T-13910). By testing whether specific genetic variants were absent or present in an ancient sample, aDNA can be used to test hypotheses about the timing of selective changes in past human populations (Burger et al., 2007; Malmström et al., 2010; Plantinga et al., 2012). This can lead to much more precise time estimates, though these depend on the ability to accurately date ancient skeletal materials. For example, the derived allele (−13910*T) that underlies lactase persistence in Northern Europeans was found in only one copy out of 20 (~5%) in a 5000 year old skeletal sample from Sweden (Malmström et al., 2010), in ~27% of a sample of 26 Basque individuals dating between 4500 and 5000 years ago (Plantinga et al., 2012), and was completely absent from a skeletal sample of nine individuals from eastern Europe dating between 5000 and 5800 years ago (Burger et al., 2007).
Holocene demography of Europe and ancient DNA
Archaeological evidence suggests that the transition from a hunting and gathering lifestyle to a more sedentary agricultural ‘Neolithic’ lifestyle, which began in the Near East by 10,000 years ago, spread across Europe between 8000 and 4000 years ago (Price, 2000). Archaeological and genetic evidence has traditionally been divided between two viewpoints regarding the spread of agricultural lifestyles from the Near East. The earlier and more popular viewpoint argues that this transition is characterized by a large amount of demic diffusion involving a large influx of agricultural populations across Europe (Childe, 1958). Others view this transition as being dominated by cultural diffusion, such that Mesolithic hunter-gatherers in Europe embraced Neolithic lifestyles with little genetic input from Near Eastern farmers (Zvelebil and Dolukhanov, 1991).
Considering the two viewpoints, using phylogeographic analyses of present human DNA samples has led to contrasting results (Sokal and Menozzi, 1982; Ammerman and Cavalli-Sforza, 1984; Sokal et al., 1991; Puit et al., 1994; Richards et al., 2000; Rosser et al., 2000; Simoni et al., 2000; Belle et al., 2006; Balaresque et al., 2010). It is important to note that estimates based solely on phylogeographic analysis of genetic variation in extant humans can be biased by several factors. Importantly, recent demographic events, such as gene-flow and back-migration between Europe and the Near East can result in the geography of extant human populations misrepresenting that of their perceived ancestors (Richards et al., 2000; Balaresque et al., 2010). Additionally, when considering a limited number of genetic loci, clines of genetic variation can be sensitive to stochastic processes such as isolation by distance (Novembre and Stephens, 2008).
Ancient DNA is an ideal data source to circumvent these issues and settle this debate because genetic differences between skeletal samples associated with Mesolithic and Neolithic technologies can be directly assayed and correlated with cultural differences. A model of cultural diffusion predicts little to no major genetic differences associated with culture change in Holocene samples, while the demic diffusion model predicts substantial influx of novel genetic variation. The majority of aDNA studies of Europe have involved mtDNA analysis, primarily from hunter-gatherer and farming populations of north and central Europe. These studies have revealed that approximately ~83% of pre-Neolithic peoples of Europe carried mtDNA haplogroup U and none belong to haplogroup H, a composition that is markedly different from present samples in which haplogroup H is dominant (Haak et al., 2005, 2008, 2010; Bramanti, 2008; Bramanti et al., 2009; Guba et al., 2011; Fu et al., 2012; Lee et al., 2012; Nikitin et al., 2012). On the other hand, ~12% from early farming populations belong to haplogroup U, while haplogroup H is present in between 25 and 37% of mtDNA from early farming samples, both consistent with the haplotype frequencies of extant Europeans. Combined, these results from ancient mtDNA analyses suggest that pre-Neolithic hunter-gatherers contributed at most 20% to the mtDNA genetic composition of present European populations (Fu et al., 2012). These ancient mtDNA results were recently combined with a dataset of 1151 complete mtDNAs from across Europe (Fu et al., 2012). Fu and colleagues (2012) found evidence for a population expansion between 15,000 and 10,000 years ago in mtDNA typical of pre-Neolithic hunter-gatherers and a subsequent contraction of these haplotypes between 10,000 and 5000 years ago, consistent with the expansion of mtDNA from agricultural populations from the Near East.
In addition to ancient mtDNA data, results from whole genome sequencing of ancient Holocene-aged individuals have been applied to the question of Neolithic population replacement in Europe. Within the past two years, substantial amounts of autosomal DNA have been reported for seven ancient individuals in Europe. These include Ötzi (the Tyrolean Iceman) dating to roughly 5300 years ago (Keller et al., 2012), three hunter-gatherers associated with the Pitted Ware culture and one farmer associated with the Funnel Beaker culture from Scandinavia dating to approximately 5000 years ago (Skoglund et al., 2012), and two 7000 year old Iberian hunter-gatherers (Sánchez-Quinto et al., 2012). Genome-wide analysis of the Iceman’s genome revealed that of extant populations, the Iceman appears most closely related to Sardinians, which has been argued to reflect the common ancestry between the Sardinian and Alpine populations of ~5000 years ago (Keller et al., 2012). The study of the four ancient Scandinavians surprisingly revealed that the hunter-gatherers of this sample are more similar to (though still distinct from) Northern Europeans, while the farmer is more similar to present-day Southern Europeans, with a high degree of genetic similarity to present-day individuals in Greece and Cyprus. Finally, analysis of the two Iberian individuals showed no close relationship between these individuals and present-day populations in Iberia or southern Europe. The general picture emerging from analysis of aDNA is that the spread of farming technology during the Neolithic was associated with significant population movements from the Near East across Europe. This population replacement should be taken under consideration in utilizing data from aDNA to address questions about recent natural selection in Europe, particularly in studies of a single locus.
Ancient DNA, demography, and selection
To what extent should the evidence of demographic population replacement affect the application of aDNA in testing hypotheses of recent natural selection? As a hypothetical example, consider the analysis by Malmström et al. (2010), which revealed that the −13910*T lactase persistence allele was relatively rare (5%) in a middle Neolithic sample of hunter-gatherers associated with the Pitted Ware culture. This result seemingly strengthens the argument for the recent origin and rapid increase in frequency of the lactase persistence allele in populations of Europe. However, given that these mid-Neolithic hunter-gatherer populations may not be closely related to present day populations of northern Europe, the rapid shift in allele frequency may reflect the demographic shift rather than natural selection. In light of this, it is important that applications of aDNA to questions related to selection consider a wider geographic range of aDNA samples. For example, the frequency of the −13910*T allele in ancient populations of the Near East can shed important additional light on the timing and geography of selection for lactase persistence.
Estimating allele age
One of the exciting outcomes of the development of aDNA technologies is the use of ancient samples to improve allele age estimation from modern samples, as recently demonstrated by Malaspinas et al. (2012). To examine uncertainties—due to demographic events—associated with using aDNA to assess allele age, be it a neutral allele or one under recent selection, we aim to first determine how closely estimates of allele age from extant populations conform to ancient specimens with known geographic and temporal provenience. Lactase persistence provides one example where the absence of the derived allele in ancient samples is consistent with a recent mutation. However, discovery of the allele in additional ancient populations could potentially push back the timing of the allele. We focus here on the information that can be gleaned from the presence or absence of derived alleles in ancient samples on the genome-wide distribution of allele age. We further compare the information gleaned from aDNA with estimates of allele age based on extant population genetic information, which can provide insight into the accuracy of the latter.
Several methods have been developed to estimate allele age using allele frequency (Kimura and Ohta, 1973; Griffiths and Tavaré, 1998), intra-allelic variation (Serre et al., 1990; Risch et al., 1995; Slatkin and Rannala, 2000), patterns of linkage disequilibrium (Rannala and Reeve, 2001), or shared haplotypes (Genin et al., 2004). We chose to utilize a set of allele age estimates generated from allele frequencies (using the approach developed by Griffiths and Tavaré, 1998) as part of the US National Institutes of Health (NIH) Heart, Lung, and Blood Institute (NHLBI)-sponsored Exome Sequencing Project (ESP). We focus specifically on the set of allele ages for approximately 1.15 million exonic SNVs estimated from a sample of 4298 European Americans (Fu et al., 2013).
We considered two whole-genomes of skeletal samples with known geographic and temporal provenience, namely Ötzi, the Tyrolean Iceman, estimated to be approximately 5300 years old (Keller et al., 2012) and an approximately 50,000 year old Nean-dertal from the Altai Mountains of Siberia (Prüfer et al., 2014). The accuracy of population genetic estimates of derived allele age depends both on the details of the method applied for their estimation, as well as on selection operating on the allele and the global effects of demography. For example, an allele has been previously reported (Sams and Hawks, 2013) that is present in Ötzi but estimated to be younger than 5000 using an LD-based method. However, we hypothesize that derived allele ages estimated from the extant European American population will be much older on average when they are present in an ancient genome, compared with derived alleles that are not observed in the ancient individual. Empirical testing and quantification of this hypothesis can weigh into how commonly-used methods for timing selection and age of mutations can be biased by demographic factors (such as population replacements, bottlenecks, and population growth) as well as by other factors, and how accurate they are when considering variability across loci in coalescence times, mutation and recombination rates, etc.
Materials and methods
Allele-ages
We extracted text-based (.txt) Exome-Sequencing Project data from the NHLBI Exome Sequencing Project’s Exome Variant Server (http://evs.gs.washington.edu/EVS/, accessed December 2013). We filtered the data to exclude duplicate SNVs, SNVs representing transition polymorphisms, and any SNVs that did not include an allele age estimate for the European sub-sample of the ESP.
The ESP data files do not include the project’s calls for the ancestral state used to determine derived allele-age. Therefore, we used our own inferred ancestral state (described below) to polarize the frequency at each resulting SNV. During this process, we noticed that a small proportion (~0.04%) of SNVs had listed ages clearly corresponding to the frequency of our inferred ancestral allele, which likely results from differences in calling the ancestral state. To alleviate this problem, we removed these miscalled sites from our analysis. In total, we analyzed a sample of 119,421 polymorphic sites.
Inference of ancestral state
We inferred ancestral state by alignment of the human reference (hg19) to three primates [chimpanzee (pantro2), orangutan (ponabe1), and rhesus macaque (rhemac2)]. Alignment gaps and non-syntenic regions were removed. Ancestral state was called by the chimpanzee allele as long as that allele was also present in a second primate outgroup.
PhyloP data
PhyloP scores generated from the EPO 36 eutherian mammal alignment with human sequence masked were generated and provided by Wenqing Fu and Joshua Akey. We used the 95th percentile to differentiate highly conserved sites (top 5%) and not highly conserved sites.
Iceman genome data
We obtained aligned genome reads from the Tyrolean Iceman (Ötzi) from the European Short Read Archive. At the time of our download, the Ötzi data were provided as three BAM files; we used samtools (http://samtools.org) to merge these into a single dataset and extracted the SNV sites for each allele with a valid age estimate from the European portion of the ESP data. In order to align the Ötzi reads (hg18) to the HapMap/1000 Genomes data (hg19), we first had to perform a genome build liftover using the UCSC Genome Browser Batch Coordinate Conversion utility (http://genome.ucsc.edu/cgi-bin/hgLiftOver).
The extracted set of reads were filtered as in previous studies to include only reads with base quality >40 and mapping quality >37. Additionally, we filtered out sites with collapsed coverage greater than 15. We chose not to use a minimum read depth for the Iceman genome in order to maximize the number of available sites represented. Because of this, we chose a single random read from each SNV and joined the data from Ötzi with the ESP allele ages.
Neandertal genome data
The Altai Neandertal VCF files were downloaded from the Max Planck Institute’s Department of Evolutionary Genetics server (http://cdna.eva.mpg.de/neandertal/altai/AltaiNeandertal/VCF/). We filtered the data using minimum quality filters, which included the following filters: removal of repeat sites from the Tandem Repeat Finder, removal of sites with mapping quality <30, inclusion of only sites that pass a genome alignability filter that requires that 50% of 35-mers overlap a position map uniquely allowing up to one mismatch, and removal of sites with read depth in the most extreme 5% of the genome-wide coverage distribution. In order to compare the presence-absence distributions of the Iceman and Neandertal, we extracted only SNVs that were also ascertained in the Iceman. Rather than choosing a single read as in the Iceman, we simply scored (as 0 or 1) each genotype for the presence or absence of the derived allele.
Block bootstrap tests
We used a block bootstrap approach (Lahiri, 2003; Keinan et al., 2007) to assess the significance of the median difference in allele age between presence/absence categories from zero and to assess the significance of deviations of odds-ratios from one (after dividing the sample into four young/old, present/absent bins). We resampled the ESP dataset (with replacement) in blocks of 200 Kb a total of 10,000 times and used the mean and standard error of these bootstrap distributions to calculate Z-scores and p-values.
Results and discussion
We analyzed, in total, 119,421 derived allele age estimates as estimated by the ESP for exonic single-nucleotide variants (Fu et al., 2013). These estimates are based on coalescent simulations using a model developed by Tennessen et al. (2012) and allele frequencies from a sample of 4298 European Americans. For each SNV, we considered whether the derived allele is observed (present) or not (absent) in each of two ancient genomes, the approximately 5300 year old Tyrolean Iceman (Ötzi) (Keller et al., 2012) and an approximately 50,000 year old Neandertal from the Altai Mountains of Siberia (Prüfer et al., 2014).
Ascertainment in the Iceman genome shows reasonable fit to allele age estimation
The distribution of ESP age estimates for derived alleles that are present in the Iceman genome exhibits a significant shift towards older ages compared with that for derived alleles present in the Iceman genome (Fig. 1; median difference, p < 10−10). This result holds also after removing the low frequency variants (derived allele count below 10), which make up a majority of the dataset (data not shown). The overall proportion of derived alleles estimated to be younger than 5000 years old and present in the Iceman genome is very small (Table 1; 0.024%). Stated differently, alleles estimated to be younger than 5000 years old are associated with 67-fold decreased odds (OR = 0.0015) of being observed in the Iceman genome relative to alleles estimated to be older than 5000 years.
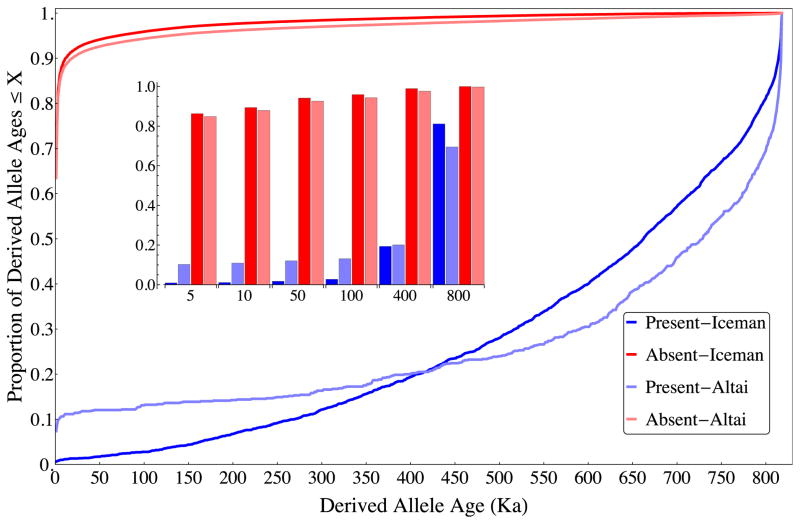
Figure 1.
Empirical cumulative distributions of mean allele age divided by its presence or absence in the Iceman and the Altai Neandertal. Empirical distributions of derived allele age estimates from ESP for each subset of alleles based on their presence/absence in the Iceman and the Altai Neandertal. As expected, alleles observed in ancient genomes are older than those not observed, and the distribution of the latter conforms to observations from extant allele frequencies of most alleles being young. Inset bar chart summarizes the distributions at several time points (in ka). Note in particular the excess proportion of ‘young present’ alleles in the Altai genome.
Table 1.
Number of alleles for each combination of presence (P) or absence (A) in the Iceman and older (O) or younger (Y) age (in ka) compared to the threshold indicated in each row.
| Meana | SDa | Y + P | Y + A | O + P | O + A | ORb | SE | Z | p-valuec |
|---|---|---|---|---|---|---|---|---|---|
| 5 | 0 | 24 | 99,573 | 2770 | 17,054 | 0.0015 | 0.00032 | 4.64 | 3.5 × 10−6 |
| 5 | 2 | 0 | 0 | 2794 | 116,627 | N/A | N/A | N/A | N/A |
| 8 | 0 | 27 | 102,670 | 2767 | 13,957 | 0.0013 | 0.00026 | 4.9 | 9.8 × 10−7 |
| 8 | 2 | 17 | 75,282 | 2777 | 41,345 | 0.0034 | 0.00078 | 3.85 | 0.0001 |
Mean plus standard deviation (in ka) of coalescent age from ESP used as age cutoff.
Mean odds-ratio from block-bootstrap test.
p-value for difference of odds-ratio from 0.
While only 0.024% of alleles were estimated as younger than 5000 years and present in the Iceman, this represents a significant deviation from the expectation of no such alleles (p = 3.5 × 10−6). Therefore, we must explain the presence of the small fraction of young alleles discovered in the Iceman. Assuming unbiased age estimations, the problem must lie either in statistical error of age estimation or with error in the aDNA, including deamidation of cytosine residues, sequencing error, and contamination. The estimated fraction of the Iceman sequence subject to these sorts of errors has been estimated to be approximately 2.5% of the genome (Keller et al., 2012). Our analysis should be confounded by these to a lesser extent since we only considered sites with known polymorphisms in present-day populations and removed transition polymorphisms (C-T, A-G) to reduce the impact of cytosine deamidation errors. Only 0.86% of derived alleles that are present in the Iceman are estimated to be younger than 5000 years, and they show a different distribution, shifted towards older ages, compared with absent alleles that are also estimated to be younger than 5000 years (Fig. 1). Combined, these results support that the vast majority of derived alleles observed in the Iceman are genuine.
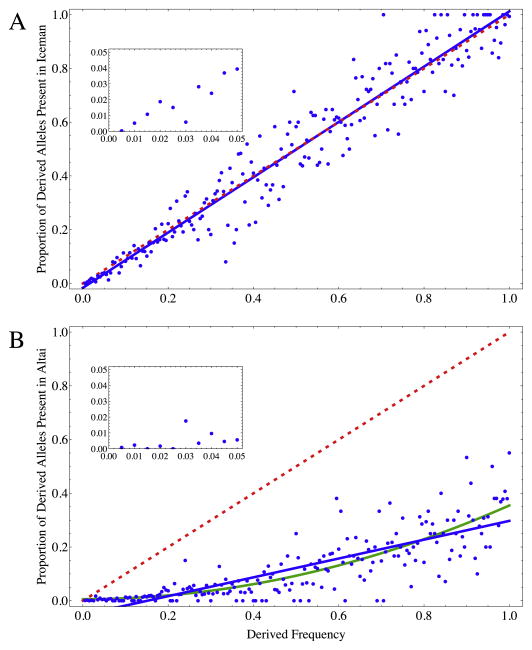
Another piece of evidence supporting these conclusions stems from the frequency spectrum of present alleles: Under neutrality, the probability that an allele is observed in a single ancient genome from an ancestral population can be approximated by the present-day frequency of the allele. Fig. 2A shows the correlation of present-day frequency against the fraction of derived alleles that are observed in the Iceman. The slope of the best-fit regression line is 1.03, reflecting a slight and insignificant increase compared with the neutral expectation, potentially due to the averaging of allele frequencies that fit into a certain frequency bin or the small number of total sites in the higher frequency bins.
Figure 2.
Proportion of derived alleles present in the Iceman and the Altai Neandertal by derived frequency. Plots illustrate the fraction of derived alleles observed in the Iceman (A) and Altai Neandertal (B) at each 0.5% frequency bin. The dashed red line (y = x) illustrates the expectation from purely neutral evolution, assuming the ancient individual is from a population directly ancestral to the modern sample. Note the extreme skew from this expectation for the Neandertal in panel B. Solid blue lines in both panels indicate linear regression ‘best-fit’ of the data. The linear regression slope for the Iceman is not significantly different from the neutral expectation (B = 1.03, SE = 0.02). The linear regression slope for Altai is significantly different from 1 (B = 0.35, SE = 0.018). The green line in panel B illustrates nonlinear (quadratic) regression, which better fits the data (R2 = 0.85 versus 0.66) and illustrates a more complex population history with the present sample. Insets in both panels show the lowest ten frequency bins. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Another explanation for the small surplus of alleles estimated at younger than 5000 years and present in the Iceman genome involved statistical error in allele age estimation. Indeed, when we considered the mean age plus two standard deviations as a conservative upper bound estimate of allele age, we lose all power to differentiate alleles by the age of the Iceman sample since that upper bound is lower than 5000 years for no alleles in our sample (Table 1). As the Iceman is unlikely to be directly ancestral to any individuals in the modern sample, the age of this sample (and other ancient samples) may be significantly younger than the time of the population split between the present-day population and the population of the Iceman. In this context, the dating of the skeletal sample is a conservative lower bound for the minimum age estimates we should expect to find in the ancient sample. Hence, we repeated analyses with a slightly less conservative age of 8000 years (Table 1).
Ascertainment in a Neandertal genome provides a poorer fit to allele age estimation
To bring more perspective to the results from the Iceman genome, we additionally examined patterns of presence-absence in the recently published (Prüfer et al., 2014) high-coverage genome of a Neandertal discovered in Denisova Cave, Russia. As with the Iceman, the distribution of ages for alleles present in the Altai Neandertal is significantly shifted towards older ages compared with that of alleles that are absent (Fig. 1; median difference, p < 10−10). For this genome, we used a conservatively young estimated age of 50,000 years. In comparing with age estimates from ESP bases on allele frequencies, it is important to recognize that they are based on a model that does not include a contribution of Neandertal ancestry to present-day European populations (Fu et al., 2013); therefore, we might expect to see a larger bias in the distribution of presence-absence by age than observed in the much younger Iceman genome. Table 2 illustrates that the model still performs relatively well, given that the fraction of derived alleles observed in modern humans and the Neandertal genome is expected to be small, since the majority of derived alleles in the ESP European sample are expected to have arisen since the population divergence of ancestral modern humans and Neandertals (Coventry et al., 2010; Keinan and Clark, 2012; Tennessen et al., 2012; Fu et al., 2013; Kiezun et al., 2013). Additionally, the distribution of ages relative to whether an allele is observed in the Neandertal genome suggests that the age estimates are relatively accurate (Fig. 1). Similarly, the overall fraction of derived alleles observed in the Neandertal genome (0.7%) is substantially reduced compared with the Iceman (2.3%), in line with its older age.
Table 2.
Number of alleles for each combination of presence (P) or absence (A) in the Altai Neandertal and older (O) or younger (Y) age (in ka) compared to the threshold indicated in each row.
| Meana | SDa | Y + P | Y + A | O + P | O + A | ORb | SE | Z | p-valuec |
|---|---|---|---|---|---|---|---|---|---|
| 50 | 0 | 93 | 109,707 | 763 | 8858 | 0.01 | 0.0015 | 6.9 | 5.35 × 10−12 |
| 50 | 2 | 79 | 100,573 | 777 | 17,992 | 0.018 | 0.0029 | 6.31 | 2.74 × 10−10 |
| 100 | 0 | 101 | 111,767 | 755 | 6798 | 0.008 | 0.001 | 7.24 | 4.38 × 10−13 |
| 100 | 2 | 86 | 104,775 | 770 | 13,790 | 0.015 | 0.0023 | 6.45 | 1.11 × 10−10 |
Mean plus standard deviation (in ka) of coalescent age from ESP used as age cutoff.
Mean odds-ratio from block-bootstrap test.
p-value for difference of odds-ratio from 0.
However, the fraction of sites estimated to be younger than 50,000 years that are present in the Neandertal genome is 0.08% and the overall odds of observing a derived allele younger than 50,000 years old relative to those older than 50,000 years old (OR = 0.01) are less reduced compared with the Iceman. Compared with the Iceman, this represents an even greater excess of ‘young present’ alleles above zero (p = 5.35 × 10−12). One explanation for this pattern is the fact that statistical uncertainty in allele age increases with allele age. Therefore, the pattern of presence/absence in the Altai genome could be explained by this increased statistical uncertainty. Consequently, we also measured the same statistics using an upper bound of mean age plus two standard deviations (Table 2), holding the age cutoff constant at 50,000 years and the odds-ratio remains highly significant (OR = 0.018, p = 2.74 × 10−10 for difference from zero). This excess of ‘young present’ alleles in the Altai genome is visually evident in Fig. 1.
The distribution of ages of alleles that are present in the Altai genome before the 400 ka (thousands of years) cross over is essentially flat, reflecting the fact that the majority of ‘present’ alleles in this genome are lower frequency compared with those ‘present’ in the Iceman. We should not expect the Neandertal genome to carry a much higher proportion of truly ancient alleles compared with the Iceman for alleles that are of very low frequency. This excess of lower frequency alleles likely reflects Neandertal ancestry in the present-day sample.
We conclude, therefore, that the observation of derived alleles in the Neandertal genome given ages estimated from present-day samples is more biased. The amount of sequencing error/contamination necessary to explain the fraction of incorrectly estimated ages (0.11) is 11 times greater than the 0.01 inferred amount of error in this high-coverage genome, which is further reduced by genotype calling (Prüfer et al., 2014), making errors in the Neandertal sequence itself an unlikely explanation for this bias. This skew in the presence-absence distribution for the Altai genome should not be surprising, as the Neandertals have a mostly separate population history from our African ancestors after at least 300 ka. This different population history is strongly reflected in the frequency distribution of ‘present’ Altai alleles (Fig. 2B). If more recent admixture from this archaic population is not explicitly modeled to produce the allele age estimates, we should expect the low-frequency variants contributed to modern populations by Neandertals to be estimated to be much younger than they truly are.
General performance of allele age estimation relative to ancient genomes
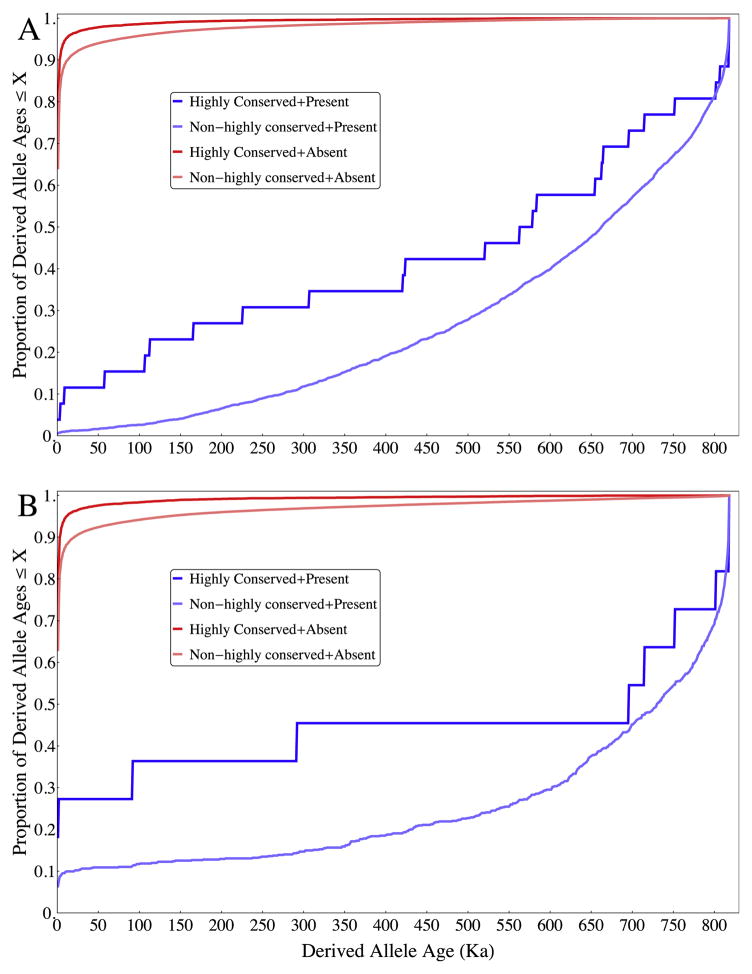
Our results suggest that, overall, the frequency- and coalescent-based allele age estimation employed by Fu et al. (2013) performs relatively well compared to empirical observations in aDNA. We note that our results described above and in the tables and figures in this manuscript refer to the mean allele ages (and mean + 2SD where noted) estimated by Fu et al. (2013). In reality, a small proportion of rare derived alleles (proportional to the frequency) are expected to be much older than the average age for that frequency under neutrality (Kimura and Ohta, 1973), which may explain a portion of our observed ‘young present’ alleles. Additionally, age estimation can be biased for alleles in which frequency has been affected by natural selection. In fact, we observe in the Iceman that alleles at conserved sites (high phyloP scores) have systematically younger age estimates than at non-conserved sites (Fig. 3), a result that is consistent with purifying selection pushing alleles towards low frequencies, and with previous empirical observations (Fu et al., 2013; Kiezun et al., 2013).
Figure 3.
Empirical cumulative distributions of age divided by presence-absence in Iceman and high-low phyloP scores. Empirical distributions of derived alleles in our sample divided into four categories based on presence-absence in the Iceman (A) and Altai Neandertal (B), and an estimate of high base conservation (as measured by the sample 95th percentile of phyloP scores). More highly conserved sites on average are lower in frequency and, therefore, have younger estimated ages. More conserved sites also contain a higher proportion of ‘young present’ alleles in the ancient genomes, which likely reflects deviations of these sites as a group from neutral expectations.
Considering the large range of error implicit in the coalescent process, none of the 24 young observed alleles are significantly younger than 5000 years (mean + 2SD). It is also worthy of note that the true number of independent loci is 20 (several of the 24 SNPs are in LD). Similarly, 79 of 93 of the young alleles observed in the Neandertal are significantly younger than 50,000 years (Tables 1 and 2). This highlights an important problem with allele-age estimation for which aDNA will be especially useful in the future. Extensions of our allele presence-absence procedure to larger samples of high quality and geologically well-dated samples of aDNA will facilitate the reduction of these large ranges of error by empirically documenting the geographic and temporal locality of specific alleles.
Towards improved demographic models
In addition to utilizing ancient genomes to narrow the range of allele ages estimated from population genetic data, we envision a process by which allele presence-absence in ancient genomes can be used to create and fit more realistic demographic models. The importance of using an ancient genome in this context is that if age estimations were absolutely accurate, demography could not produce our observed results. We should never observe alleles that arose in the past 5000 years in a 5300 year-old genome. Therefore, we must account for the potential sources of bias in these age estimates.
A major source of bias on allele age estimates generated from frequency and coalescent simulations is the demographic model used to generate the coalescent trees. Fu et al. (2013) rely on a demographic model of the history of European and African populations generated by Tennessen et al. (2012), which has several key parameters that affect the average age of alleles, most importantly the timing of recent population expansion, but also the timing of the Out-of-Africa event and the rate of growth before and during the recent population expansion. The effects of the different parameterization of several recently published models that include recent population growth (Coventry et al., 2010; Nelson et al., 2012; Tennessen et al., 2012) leads to as much as a two-fold difference in average allele age (Fu et al., 2013). However, several events that have not been included in any recently published models may also substantially bias allele age when not included in the underlying model used for simulation. For example, recent expansion of the European population is typically modeled as in situ growth when, in all likelihood, this expansion event included or was preceded by an influx of gene flow from agricultural populations in the Near East, which themselves may have experienced substantial gene flow from Africa (Lazaridis et al., 2013). Fu et al., 2013 have examined the effects of modest migration rates on allele age during recent population expansion (between 0 and 15 × 10−5 per chromosome, per generation), the effective admixture rate from populations outside of ancient Europe may have been much larger.
Our observations of alleles in the Neandertal genome highlights another major issue with these demographic models, which is that they do not include the contribution of archaic human populations to present-day human ancestry. By the most recent estimates, these ancient gene flow events contributed, on average, ~2% of the ancestry of present-day Europeans (Prüfer et al., 2014). This suggests that a small but substantial fraction of low frequency variants, introduced by gene flow from Neandertals, will have true ages that are much older than the average age of alleles with similar frequency under a model without Neandertal admixture. While models have been developed that include archaic introgression (for example, see Harris and Nielsen, 2013), none have been explicitly applied to the allele age problem. The inclusion of more aDNA in the way that we have illustrated here will allow improving the parameterization of human demographic models.
Towards estimates of recent positive natural selection
Coalescent/frequency based estimates of allele age typically assume an absence of positive selection. However, in many cases we are most interested in understanding whether a particular allele has been subject to recent positive selection. Moreover, it has been proposed that the amount of positive selection increased during the recent epoch of population expansion (Hawks et al., 2007). Can ancient genomes be used to systematically identify cases of recent positive selection? There is a quickly growing literature on inferring natural selection from genetic time-series data (Bollback et al., 2008; Malaspinas et al., 2012; Feder et al., 2014). These approaches are based on the availability of several samples with genetic continuity through time. Similar to some approaches based on extant population genetic data, these time-series approaches assume a model of demographic history, including population sizes at each sampled time point. Consider the case of lactase persistence described in the Introduction. In this case, current sampling of aDNA supports selection time as estimated from present-day samples and points to the lactase persistence allele having risen to present frequency rapidly in situ in Europe. However, at present, no DNA from ancient agricultural or pre-agricultural populations of the Near East has been recovered. The lack of observation of the lactase persistence allele in ancient northern Europe does not, by itself, establish just how recent selection for lactase persistence has been. This case is, therefore, a perfect illustration of the need for larger, more geographically diverse samples of aDNA. With moderately sized samples of aDNA from northern Europe, southern Europe, and the Near East from approximately 8000 years ago we could empirically test hypotheses about positive selection on alleles genome-wide in a probabilistic framework. Failure to observe the lactase persistence allele in such larger samples from different locales will go a long way in solidifying the observation of very recent, strong positive selection on this allele.
Conclusions
We have demonstrated the utility of ancient human genomes for examining the accuracy of allele age estimates derived from extant samples and a coalescent-based framework under an assumed demographic model. Our results illustrate that such allele age estimates in European Americans, based on recently published models of European demographic history, fit well with expectations based on the observation or lack thereof of these alleles in a 5300 year old European genome (Fig. 2A). However, comparison to a high-coverage Neandertal genome illustrates at least one major gap in the models of European demographic history that have been used for allele age estimation (Fig. 2B). Our results and discussion paint a picture of improved demographic modeling, allele age estimation, and tests of natural selection via increased inclusion of the growing sample of ancient human genomes. They also point to the importance of considering a more diverse set of aDNA samples—both geographically and temporally—for making significant improvements in these areas.
Acknowledgments
We would like to thank Karen Strier, Bret Payseur, Henry Bunn, and Travis Pickering for comments on an earlier version of this paper. We would also like to thank Sriram Sankararaman and David Reich for providing a set of quality filters for the Altai Neandertal VCFs and Wenqing Fu and Joshua Akey for providing phyloP data for the ESP dataset. Finally, we would like to thank Leonardo Arbiza for providing alignments of the inferred ancestral genome. This work was supported in part by NIH grant R01GM108805 (A.K.) and by an award from the Ellison Medical Foundation (A.K.).
References
- Ammerman AJ, Cavalli-Sforza LL. The Neolithic Transition and the Genetics of Populations in Europe. Princeton University Press; Princeton: 1984. [Google Scholar]
- Balaresque P, Bowden GR, Adams SM, Leung HY, King TE, Rosser ZH, Goodwin J, Moisan JP, Richard C, Millward A, Demaine AG, Barbujani G, Previderè C, Wilson IJ, Tyler-Smith C, Jobling MA. A predominantly Neolithic origin for European paternal lineages. PLoS Biol. 2010;8:e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle EMS, Landry PA, Barbujani G. Origins and evolution of the Europeans’ genome: evidence from multiple microsatellite loci. Proc R Soc B. 2006;273:1595–1602. doi: 10.1098/rspb.2006.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri T, Sabeti P, Patterson N, Vanderploeg T, Schaffner S, Drake J, Rhodes M, Reich D, Hirschhorn J. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollback JP, York TL, Nielsen R. Estimation of 2Nes from temporal allele frequency data. Genetics. 2008;179:497–502. doi: 10.1534/genetics.107.085019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollongino R, Edwards CJ, Alt KW, Burger J, Bradley D. Early history of European domestic cattle as revealed by ancient DNA. Biol Lett. 2006;2:155–159. doi: 10.1098/rsbl.2005.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramanti B. Ancient DNA: genetic analysis of aDNA from sixteen skeletons of the Vedrovice. Anthropologie. 2008;46:153–160. [Google Scholar]
- Bramanti B, Thomas MG, Haak W, Unterlaender M, Jores P, Tambets K, Antanaitis-Jacobs I, Haidle MN, Jankauskas R, Kind CJ, Lueth F, Terberger T, Hiller J, Matsumura S, Forster P, Burger J. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science. 2009;326:137–140. doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG. Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci. 2007;104:3736–3741. doi: 10.1073/pnas.0607187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childe VG. The Dawn of European Civilization. 6. Knopf; New York: 1958. [Google Scholar]
- Coventry A, Bull-Otterson LM, Liu X, Clark AG, Maxwell TJ, Crosby J, Hixson JE, Rea TJ, Muzny DM, Lewis LR, Wheeler DA, Sabo A, Lusk C, Weiss KG, Akbar H, Cree A, Hawes AC, Newsham I, Varghese RT, Villasana D, Gross S, Joshi V, Santibanez J, Morgan M, Chang K, Hale W, IV, Templeton AR, Boerwinkle E, Gibbs R, Sing CF. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat Commun. 2010;1:131. doi: 10.1038/ncomms1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enattah N, Sahi T, Savilahti E, Terwilliger J. Identification of a variant associated with adult-type hypolactasia. Nature. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- Enattah N, Trudeau A, Pimenoff V, Maiuri L. Evidence of still-ongoing convergence evolution of the lactase persistence T-13910 alleles in humans. Am J Hum Genet. 2007;81:615–625. doi: 10.1086/520705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enattah NS, Jensen TGK, Nielsen M, Lewinski R, Kuokkanen M, Rasinperä H, El-Shanti H, Seo JK, Alifrangis M, Khalil IF. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am J Hum Genet. 2008;82:57–72. doi: 10.1016/j.ajhg.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder AF, Kryazhimskiy S, Plotkin JB. Identifying signatures of selection in genetic time series. Genetics. 2014;196:509–522. doi: 10.1534/genetics.113.158220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Rudan P, Pääbo S, Krause J. Complete mitochondrial genomes reveal Neolithic expansion into Europe. PLoS One. 2012;7:e32473. doi: 10.1371/journal.pone.0032473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, Akey JM NHLBI Exome Sequencing Project. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin E, Tullio-Pelet A, Begeot F, Lyonnet S, Abel L. Estimating the age of rare disease mutations: the example of Triple-A syndrome. J Med Genet. 2004;41:445–449. doi: 10.1136/jmg.2003.017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbault P, Moret C, Currat M, Sanchez-Mazas A. Impact of selection and demography on the diffusion of lactase persistence. PLoS One. 2009;4:e6369. doi: 10.1371/journal.pone.0006369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RC, Tavaré S. The age of a mutation in a general coalescent tree. Communications in Statistics. Stoch Models. 1998;14:273–295. [Google Scholar]
- Guba Z, Hadadi É, Major Á, Furka T, Juhász E, Koós J, Nagy K, Zeke T. HVS-I polymorphism screening of ancient human mitochondrial DNA provides evidence for N9a discontinuity and East Asian haplogroups in the Neolithic Hungary. J Hum Genet. 2011;56:784–796. doi: 10.1038/jhg.2011.103. [DOI] [PubMed] [Google Scholar]
- Haak W, Forster P, Bramanti B, Matsumura S, Brandt G, Tänzer M, Villems R, Renfrew C, Gronenborn D, Alt KW. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310:1016–1018. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- Haak W, Brandt G, Jong HND, Meyer C, Ganslmeier R, Heyd V, Hawkesworth C, Pike AWG, Meller H, Alt KW. Ancient DNA, Strontium isotopes, and osteological analyses shed light on social and kinship organization of the Later Stone Age. Proc Natl Acad Sci. 2008;105:18226–18231. doi: 10.1073/pnas.0807592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W, Balanovsky O, Sanchez JJ, Koshel S, Zaporozhchenko V, Adler CJ, Sarkissian Der CSI, Brandt G, Schwarz C, Nicklisch N, Dresely V, Fritsch B, Balanovska E, Villems R, Meller H, Alt KW, Cooper A The Genographic Consortium. Ancient DNA from European early Neolithic farmers reveals their Near Eastern affinities. PLoS Biol. 2010;8:e1000536. doi: 10.1371/journal.pbio.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K, Nielsen R. Inferring demographic history from a spectrum of shared haplotype lengths. PLoS Genet. 2013;9:e1003521. doi: 10.1371/journal.pgen.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawks J, Wang ET, Cochran GM, Harpending HC, Moyzis RK. Recent acceleration of human adaptive evolution. Proc Natl Acad Sci. 2007;104:20753–20758. doi: 10.1073/pnas.0707650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden C, Mace R. Phylogenetic analysis of the evolution of lactose digestion in adults. Hum Biol. 1997;69:605–628. [PubMed] [Google Scholar]
- Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Graefen A, Ball M, Matzas M, Boisguerin V, Maixner F, Leidinger P, Backes C, Khairat R, Forster M, Stade B, Franke A, Mayer J, Spangler J, McLaughlin S, Shah M, Lee C, Harkins TT, Sartori A, Moreno-Estrada A, Henn B, Sikora M, Semino O, Chiaroni J, Rootsi S, Myres NM, Cabrera VM, Underhill PA, Bustamante CD, Vigl EE, Samadelli M, Cipollini G, Haas J, Katus H, O’Connor BD, Carlson MRJ, Meder B, Blin N, Meese E, Pusch CM, Zink A. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 2012;3:698. doi: 10.1038/ncomms1701. [DOI] [PubMed] [Google Scholar]
- Kiezun A, Pulit SL, Francioli LC, van Dijk F, Swertz M, Boomsma DI, van Duijn CM, Slagboom PE, van Ommen GJB, Wijmenga C, Consortium GOTN, De Bakker PIW, Sunyaev SR. Deleterious alleles in the human genome are on average younger than neutral alleles of the same frequency. PLoS Genet. 2013;9:e1003301. doi: 10.1371/journal.pgen.1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Ohta T. The age of a neutral mutant persisting in a finite population. Genetics. 1973;75:199–212. doi: 10.1093/genetics/75.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri SN. Resampling Methods for Dependent Data. Springer; Dordrecht: 2003. [Google Scholar]
- Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Sudmant PH, Schraiber JG, Castellano S, Kirsanow K, Economou C, Bollongino R, Fu Q, Bos K, Nordenfelt S, de Filippo C, Prüfer K, Sawyer S, Posth C, Haak W, Hallgren F, Fornander E, Ayodo G, Babiker HA, Balanovska E, Balanovsky O, Ben-Ami H, Bene J, Berrada F, Brisighelli F, Busby GBJ, Cali F, Churnosov M, Cole DEC, Damba L, Delsate D, van Driem G, Dryomov S, Fedorova SA, Francken M, Romero IG, Gubina M, Guinet JM, Hammer M, Henn B, Helvig T, Hodoglugil U, Jha AR, Kittles R, Khusnutdinova E, Kivisild T, Kucinskas V, Khusainova R, Kushniarevich A, Laredj L, Litvinov S, Mahley RW, Melegh B, Metspalu E, Mountain J, Nyambo T, Osipova L, Parik J, Platonov F, Posukh OL, Romano V, Rudan I, Ruizbakiev R, Sahakyan H, Salas A, Starikovskaya EB, Tarekegn A, Toncheva D, Turdikulova S, Uktveryte I, Utevska O, Voevoda M, Wahl J, Zalloua P, Yepiskoposyan L, Zemunik T, Cooper A, Capelli C, Thomas MG, Tishkoff SA, Singh L, Thangaraj K, Villems R, Comas D, Sukernik R, Metspalu M, Meyer M, Eichler EE, Burger J, Slatkin M, Pääbo S, Kelso J, Reich D, Krause J. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2013;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Makarewicz C, Renneberg R, Harder M, Krause-Kyora B, Müller S, Ostritz S, Fehren-Schmitz L, Schreiber S, Müller J, Wurmb-Schwark N, von Nebel A. Emerging genetic patterns of the European Neolithic: Perspectives from a late Neolithic Bell Beaker burial site in Germany. Am J Phys Anthropol. 2012;148:571–579. doi: 10.1002/ajpa.22074. [DOI] [PubMed] [Google Scholar]
- Malaspinas AS, Malaspinas O, Evans SN, Slatkin M. Estimating allele age and selection coefficient from time-serial data. Genetics. 2012;192:599–607. doi: 10.1534/genetics.112.140939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmström H, Linderholm A, Lidén K, Storå J, Molnar P, Holmlund G, Jakobsson M, Gotherström A. High frequency of lactose intolerance in a prehistoric hunter-gatherer population in northern Europe. BMC Evol Biol. 2010;10:89. doi: 10.1186/1471-2148-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Fu Q, Aximu-Petri A, Glocke I, Nickel B, Arsuaga JL, Martínez I, Gracia A, de Castro JMB, Carbonell E, Pääbo S. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 2013;505:403–406. doi: 10.1038/nature12788. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Shen J, Tang Z, Bacanu SA, Fraser D, Warren L, Aponte J, Zawistowski M, Liu X, Zhang H, Zhang Y, Li J, Li Y, Li L, Woollard P, Topp S, Hall MD, Nangle K, Wang J, Abecasis G, Cardon LR, Zollner S, Whittaker JC, Chissoe SL, Novembre J, Mooser V. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin AG, Newton JR, Potekhina ID. Mitochondrial haplogroup C in ancient mitochondrial DNA from Ukraine extends the presence of East Eurasian genetic lineages in Neolithic Central and Eastern Europe. J Hum Genet. 2012;57:610–612. doi: 10.1038/jhg.2012.69. [DOI] [PubMed] [Google Scholar]
- Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantinga TS, Alonso S, Izagirre N, Hervella M, Fregel R, van der Meer JW, Netea MG, de la Rúa C. Low prevalence of lactase persistence in Neolithic South-West Europe. European J Hum Genet. 2012;20:778–782. doi: 10.1038/ejhg.2011.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TD. Europe’s First Farmers. Cambridge University Press; New York: 2000. [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, Li H, Mallick S, Dannemann M, Fu Q, Kircher M, Kuhlwilm M, Lachmann M, Meyer M, Ongyerth M, Siebauer M, Theunert C, Tandon A, Moorjani P, Pickrell J, Mullikin JC, Vohr SH, Green RE, Hellmann I, Johnson PLF, Blanche H, Cann H, Kitzman JO, Shendure J, Eichler EE, Lein ES, Bakken TE, Golovanova LV, Doronichev VB, Shunkov MV, Derevianko AP, Viola B, Slatkin M, Reich D, Kelso J, Pääbo S. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puit I, Sajantila A, Simanainen J, Georgiev O, Schaffner W, Pääbo S. Mitochondrial DNA sequences from Switzerland reveal striking homogeneity of European populations. Biol Chem Hoppe-Seyler. 1994;375:837–840. [PubMed] [Google Scholar]
- Rannala B, Reeve JP. High-resolution multipoint linkage-disequilibrium mapping in the context of a human genome sequence. Am J Hum Genet. 2001;69:159–178. doi: 10.1086/321279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, Sellitto D, Cruciani F, Kivisild T, Villems R, Thomas M, Rychkov S, Rychkov O, Rychkov Y, lge MG, Dimitrov D, Hill E, Bradley D, Romano V, Calı F, Vona G, Demaine A, Papiha S, Triantaphyllidis C, Stefanescu G, Hatina J, Belledi M, Di Rienzo A, Novelletto A, Oppenheim A, Nørby S, Al-Zaheri N, Santachiara-Benerecetti S, Scozzari R, Torroni A, Bandelt HJR. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. 2000;67:1251. [PMC free article] [PubMed] [Google Scholar]
- Risch N, de Leon D, Ozelius L, Kramer P, Almasyz L, Singer B, Fahn S, Breakefield X, Bressman S. Genetic analysis of idiopathic torsion dystonia in Ashkenazi Jews and their recent descent from a small founder population. Nat Genet. 1995;9:152–159. doi: 10.1038/ng0295-152. [DOI] [PubMed] [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim AN, Amos W, Armenteros M, Arroyo E, Barbujani G, Beckman G, Beckman L, Bertranpetit J, Bosch E, Bradley DG, Brede G, Cooper G, Corte-Real HBSM, de Knijff P, Decorte R, Dubrova YE, Evgrafov O, Gilissen A, Glisic S, lge MG, Hill EW, Jeziorowska A, Kalaydjieva L, Kayser M, Kivisild T, Kravchenko SA, Krumina A, Kucinskas V, Lavinha JO, Livshits LA, Malaspina P, Maria S, McElreavey K, Meitinger TA, Mikelsaar AV, Mitchell RJ, Nafa K, Nicholson J, Nørby S, Pandya A, Parik JR, Patsalis PC, Pereira LI, Peterlin B, Pielberg G, Prata MJO, Previdere C, Roewer L, Rootsi S, Rubinsztein DC, Saillard J, Santos FR, Stefanescu G, Sykes BC, Tolun A, Villems R, Tyler-Smith C, Jobling MA. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet. 2000;67:1526. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sams A, Hawks J. Patterns of population differentiation and natural selection on the celiac disease background risk network. PLoS One. 2013;8:e70564. doi: 10.1371/journal.pone.0070564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Quinto F, Schroeder H, Ramirez O, Avila-Arcos MC, Pybus M, Olalde I, Velazquez A, Marcos MEP, Encinas JMV, Bertranpetit J. Genomic affinities of two 7,000-year-old Iberian hunter-gatherers. Curr Biol. 2012;22:R631–R633. doi: 10.1016/j.cub.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Serre JL, Simon-Bouy B, Mornet E, Jaume-Roig B, Balassopoulou A, Schwartz M, Taillandier A, Boue J, Boue A. Studies of RFLP closely linked to the cystic fibrosis locus throughout Europe lead to new considerations in populations genetics. Hum Genet. 1990;84:449–454. doi: 10.1007/BF00195818. [DOI] [PubMed] [Google Scholar]
- Simoni L, Calafell F, Pettener D, Bertranpetit J, Barbujani G. Geographic patterns of mtDNA diversity in Europe. Am J Hum Genet. 2000;66:262–278. doi: 10.1086/302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Malmström H, Raghavan M, Storå J, Hall P, Willerslev E, Gilbert MTP, Götherström A, Jakobsson M. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336:466–469. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Rannala B. Estimating allele age. A Rev Genom Hum Genet. 2000;1:225–249. doi: 10.1146/annurev.genom.1.1.225. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Menozzi P. Spatial autocorrelations of HLA frequencies in Europe support demic diffusion of early farmers. Am Nat. 1982;119:1–17. [Google Scholar]
- Sokal RR, Oden NL, Wilson C. Genetic evidence for the spread of agriculture in Europe by demic diffusion. Nature. 1991;351:143–145. doi: 10.1038/351143a0. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO NHLBI Exome Sequencing Project. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2006;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvelebil M, Dolukhanov P. The transition to farming in Eastern and Northern Europe. J World Prehist. 1991;5:233–278. [Google Scholar]