Abstract
The ability of the cell to sense environmental conditions and alter gene expression in response to them is critical to its survival. In Saccharomyces cerevisiae, the Tor1/2 serine/threonine kinases are global regulators situated at the top of a signal cascade reported to receive and transmit nutritional signals associated with the nitrogen supply of the cell. At the other end of that cascade is Gln3, one of two transcriptional activators responsible for most nitrogen catabolic gene expression. When nitrogen is in excess, Tor1/2 are active, and Gln3 is phosphorylated and localizes to the cytoplasm. If Tor1/2 are inhibited by rapamycin or mutation, Gln3 becomes dephosphorylated, accumulates in the nucleus, and mediates nitrogen catabolite repression (NCR)-sensitive transcription. The observations that Gln3 also accumulates in the nuclei of cells provided with poor nitrogen sources or during nitrogen starvation has led to the conclusion that Tor1/2 control intracellular Gln3 localization and NCR-sensitive transcription by regulating Gln3 phosphorylation/dephosphorylation. To test this model, we compared Gln3 phosphorylation states and intracellular localizations under a variety of physiological conditions known to elicit different levels of NCR-sensitive transcription. Our data indicate that: (i) observable Gln3 phosphorylation levels do not correlate in a consistent way with the quality or quantity of the nitrogen source provided, the intracellular localization of Gln3, or the capacity to support NCR-sensitive transcription. (ii) Gln3-Myc13 is hyperphosphorylated during nitrogen and carbon starvation, but this uniform response does not correlate with Gln3 intracellular localization. (iii) Gln3-Myc13 dephosphorylation and nuclear localization correlate with one another at early but not late times after rapamycin treatment. These data suggest that rapamycin treatment and growth with poor nitrogen sources bring about nuclear accumulation of Gln3 but likely do so by different mechanisms or by a common mechanism involving molecules other than Gln3 and/or other than the levels of Gln3-Myc13 phosphorylation thus far detected by others and ourselves.
The ability to sense and respond to a constantly changing and sometimes hostile environment is critical to the survival of free living organisms. To cope with these demands, cells have evolved sophisticated signal transduction pathways that sense important components of their environment, such as nitrogen and carbon supplies. These transduction pathways culminate in the activation of transcription factors that adjust gene expression to maximize the potential for survival, growth, and reproduction. In Saccharomyces cerevisiae, Tor1 and Tor2 are thought to be central components of the transduction pathway responsible for collecting, integrating, and responding to nutritional signals. Tor1/2 are serine/threonine kinases that also possess regions of homology with phosphatidylinositol 3-lipid kinases (1–9). They are inhibited by the immunosuppressant and antineoplastic drug rapamycin complexed with peptidylprolylisomerase FKBP12/Rbp1 (1–9). Inactivation of the Tor proteins either by mutation or with rapamycin elicits broad changes in cellular activities including decreased translational initiation, eIF-4G instability, inhibition of cell cycle progression, aberrant actin cytoskeleton reorganization, decreased polymerase I and III activities, increased autophagy, increased protein degradation, and increased expression of retrograde and nitrogen catabolic genes (1–9).
The present study centers on Tor1/2 regulation of nitrogen catabolic gene expression. S. cerevisiae are able to utilize good nitrogen sources (e.g. glutamine and in some strains ammonia) in preference to poor ones (e.g. proline, urea). This capability derives from a regulatory process designated nitrogen catabolite repression (NCR).1 In the presence of good nitrogen sources, GATA family transcriptional activators Gln3 and Gat1 are restricted to the cytoplasm (10–14). As a result, NCR-sensitive transcription of the genes required for transport and degradation of poor nitrogen sources does not occur (8, 15). In the presence of a poor nitrogen source, Gln3 and Gat1 accumulate in the nucleus and activate NCR-sensitive transcription (8, 15).
Tor1 and Tor2 are thought to control Gln3 intracellular localization by regulating its phosphorylation/dephosphorylation (10, 13). The prevailing model posits that active Tor1/2 phosphorylate Tap42 and Tip41 which, in a way not fully understood, results in inactivation of Sit4, one of two phosphoprotein phosphatases suggested to be responsible for Gln3 dephosphorylation (10, 13, 17, 18). Tor1/2 are also suggested by some investigators to be the kinases responsible for Gln3 phosphorylation itself (13, 16). Ure2 complexes with phosphorylated Gln3 and prevents its nuclear entry either because (i) the phosphorylated form of Gln3, stabilized in the complex, is unable to enter the nucleus (13) or (ii) Gln3 cannot enter the nucleus when complexed with Ure2 (10). Inactivation of Tor1/2 by rapamycin prevents Tor1/2-mediated phosphorylation reactions and thereby generates the opposite outcomes, i.e. Gln3 dephosphorylation, dissociation of the Gln3·Ure2 complex, nuclear localization of Gln3, and derepressed NCR-sensitive gene expression.
The above model is based on several correlations. (i) Inactivation of Tor1/2 by rapamycin treatment results in derepressed expression of many NCR-sensitive genes, nuclear localization of Gln3, dissociation of Gln3 from its negative regulator Ure2, and dephosphorylation of Gln3 (10–14, 19). (ii) Many of the wide ranging cellular activities altered markedly by inactivation of Tor1/2 are similarly impacted by nutrient starvation (1–9).
Although the model convincingly explains the above correlations, data have begun to accumulate which are not as easily explained as those above (8, 9, 20–22). The most difficult observations to rectify were from studies of Gln3 localization in cells transferred from minimal glutamine to minimal proline medium or after the addition of rapamycin to ammonia- or glutamine-grown cultures. Both the time courses of and extents to which Gln3 accumulated in nuclei after these transfers was dependent upon the nitrogen source provided (20). This was unexpected because rapamycin specifically inhibits Tor1/2 that are posited to be situated downstream of the nutrient supply signal (10–13, 19). More recently, Gln3-Myc13 accumulation in the nuclei of cells transferred from glucose-glutamine to glucose-proline medium was shown to require an intact actin cytoskeleton, whereas similar nuclear accumulation of Gln3-Myc13 in response to rapamycin treatment does not exhibit this requirement.2 These observations prompted us to revisit the published correlative data and query whether we could find additional information that would resolve this paradox. Although we could confirm all of the previously reported correlations we tested, two critical correlations required by the current Tor1/2 model have not been tested, i.e. the electrophoretic mobility of Gln3 as a function of good versus poor nitrogen sources and quantitative comparison of Gln3 nuclear accumulation and electrophoretic mobility after changes in the nutritional environment of a cell.
Results of experiments designed to provide this missing information indicate that the detectable extent of Gln3 phosphorylation/ dephosphorylation does not correlate with its intracellular localization in good versus poor nitrogen sources. Further, the only condition for which Gln3 dephosphorylation and nuclear accumulation of Gln3 do correlate in a consistent way is at early, but not late, times after rapamycin treatment. These data suggest that rapamycin inhibition of Tor1/2 generates the same outcomes with respect to intracellular Gln3 localization as growth in a poor nitrogen source but do not support the contention that this intracellular localization is the result of changes in Gln3 phosphorylation which have thus far been detected by others and ourselves.
MATERIALS AND METHODS
Strains and Media
S. cerevisiae strain TB123 (MATa leu2–3,112, ura3–52, rme1, trp1, his4, GAL+, HMLa, GLN3-Myc13[KanMX]) (10) was grown at 30 °C to mid-log phase (A600 nm = 0.4–0.5) in YNB (without ammonium sulfate or amino acids), containing 2% glucose, 120 µg/ml leucine, 20 µg/ml uracil, 20 µg/ml histidine, 20 µg/ml tryptophan, and the nitrogen source (0.1% final concentration) noted in the figure legend. Rapamycin (dissolved in 10% Tween 20 + 90% ethanol) was added directly to the medium in which cells had grown overnight (final concentration of 0.2 µg/ml). Experiments, in which yeast were transferred into carbon or nitrogen starvation media, were performed as described previously (20). Media contained the required auxotrophic supplements as noted above. Samples were collected at the indicated times for immunofluorescence assays or protein or RNA isolation.
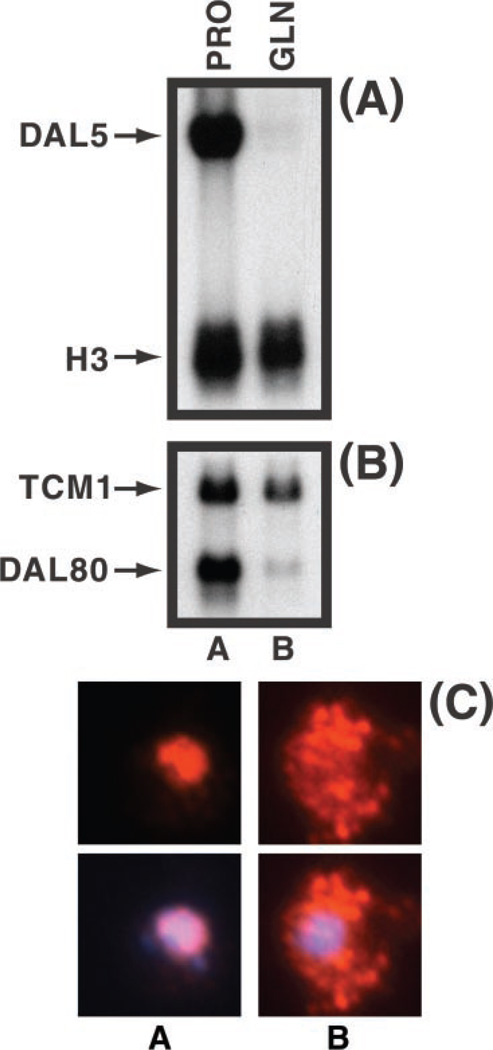
Strain differences have significantly influenced results from studies of the Tor1/2 regulatory pathway and complicated their interpretation (22). To avoid such strain differences and complications here, we performed the present analyses with the same strain initially used to demonstrate Tor1/2 regulation of intracellular Gln3 localization (10) and confirmed (in that strain) that Gln3 accumulates in the nuclei of cells in which NCR-sensitive transcription (DAL5 and DAL80, Fig. 1, A and B) is high and localizes to the cytoplasm of cells where transcription is low (Fig. 1C); we used growth with proline and glutamine as sole nitrogen source, respectively, to achieve these results (Fig. 1). A parallel correlation is observed when cells growing in rich medium are treated with rapamycin, i.e. Gln3 localizes to the cytoplasm of untreated cells but accumulates in the nuclei of most treated cells (data not shown) (1–9).
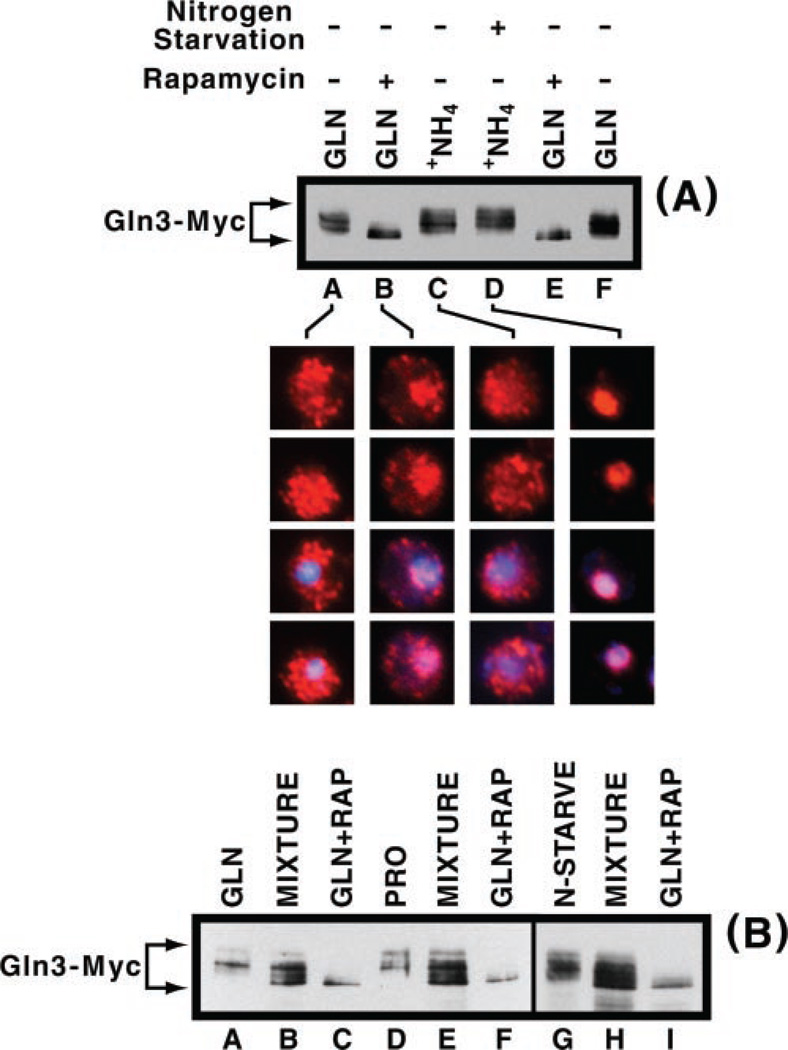
Fig. 1. Effect of nitrogen source on NCR-sensitive genes and Gln3-Myc13 localization in TB123.
S. cerevisiae cultures (TB123) were grown to mid-log phase in 2% glucose, YNB (without ammonium sulfate and amino acids), and either 0.1% proline (PRO) or 0.1% glutamine (GLN) as the nitrogen source. RNA blots were hybridized with 32P-labeled DNA probes against DAL5 (allantoate permease, A) or DAL80 (GATA family transcription factor, B) mRNA. Hht-1 histone3 (H3, A) and a ribosomal protein (TCM1, B) were used as controls. Intracellular localization of Gln3-Myc13 (red; DAPI, blue) was monitored in cells grown as described in A and B and processed as described under “Materials and Methods.” The bottom row of merged images (Gln3-Myc13 + DAPI) images are of the same cells as the top row.
Northern Blot Analysis
RNA was isolated by the method of Schmitt et al. (24). Northern blots were performed as described earlier (20).
Western Analysis
Cells were harvested by centrifugation (5 min at 3,000 rpm and 4 °C), resuspended in ice-cold extraction buffer (120 mm NaCl, 50 mm Tris, pH 7.5, 2 mm EDTA, 2 mm phenylmethylsulfonyl fluoride, 10 mm NaF, 10 mm p-nitrophenyl phosphate, 10 mm sodium pyrophosphate, 10 mm β-glycerophosphate, and a commercial mixture of protease inhibitors (Minicomplete, Roche Applied Science) and lysed with glass beads by vortex mixing (four times for 20 s each with 30-s intervening cooling intervals on ice). Unbroken cells and glass beads were removed by centrifugation for 1 min at 9,000 rpm, 4 °C, and the supernatant was clarified further by centrifugation for 15 min at 9,000 rpm, 4 °C. Protein concentrations were determined using the Bradford (Bio-Rad) assay. Samples for electrophoresis were diluted with sample buffer and incubated at 37 °C for 15 min. They were then resolved by SDS-PAGE (6% polyacrylamide) and transferred to a nitrocellulose membrane in transfer buffer without SDS (25). The membrane was incubated with a 1:1,000 dilution of 9E10(c-myc) (Covance MMS-150P) monoclonal antibody. The membrane was then washed and incubated with a 1:20,000 dilution of goat anti-mouse IgG (H-L)-horseradish peroxidase conjugate (Bio-Rad). After washing, immunoreactive species were detected by enhanced chemiluminescence (SuperSignal West Pico chemiluminescence substrate, Bio-Rad). We observed too much variation in the intensity of staining in duplicate samples to permit quantitative lane-to-lane comparisons of the amounts of Gln3-Myc13 protein that were present.
Phosphatase Treatment
Crude protein extracts were prepared as described above with the following extraction buffer: 50 mm Tris-HCl, pH 7.5, 120 mm NaCl, 2 mm EDTA, 2 mm phenylmethylsulfonyl fluoride, and a commercial mixture of protease inhibitors (Roche Applied Science). The crude extracts were incubated with 20 units of calf intestinal alkaline phosphatase (Roche Diagnostics) or 20 units of calf intestinal alkaline phosphatase plus phosphatase inhibitor, 40 mm sodium pyrophosphate, for 10 min at 30 °C. Samples were boiled 5 min prior to loading onto the gel.
Indirect Immunofluorescence
Immunofluorescence staining was carried out as described by Cox et al. (20). Gln3-Myc13 was visualized using 9E10(c-myc) (Covance MMS-150P) monoclonal antibody at a dilution of 1:1,000 and Alexa Fluor 594 goat anti-mouse IgG (Molecular Probes) at a dilution of 1:200. Nuclei were stained with DAPI as described previously (20). Cells were imaged using a Zeiss Axioplan 2 imaging microscope with a 100× Plan-Apochromat 1.40 oil objective. Images were acquired with a Zeiss Axio camera using AxioVision 3.0 (Zeiss) software. For quantitation of intracellular Gln3 localization, at least 100 cells from randomly selected fields were counted.
RESULTS
Gln3-Myc13 Phosphorylation Profiles Generated in Response to Nitrogen Limitation Differ from Those Generated by Rapamycin Treatment
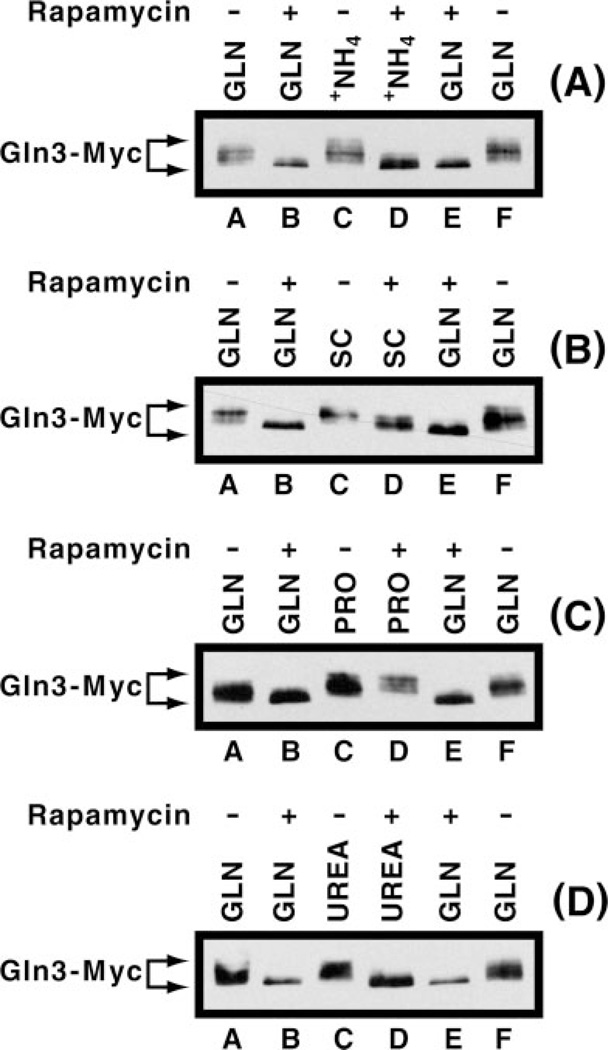
A critical correlation required by the current model describing Tor1/2 regulation of NCR-sensitive transcription is that Gln3 be dephosphorylated in cells provided with a poor nitrogen source and phosphorylated when cells are growing in rich medium. To test this correlation, we compared Gln3 phosphorylation (assayed by its electrophoretic mobility in Western blots) in rapamycin-treated cells versus cells provided with nitrogen sources ranging from good to poor. Ordered by the levels of NCR-sensitive transcription they support, the poor to good nitrogen sources we used were proline > urea > ammonia > synthetic complete (SC) > glutamine (26, data not shown). Gln3-Myc13 mobility is slower in cells provided with glutamine, a rich nitrogen source, than it is when glutamine-grown cells are treated with rapamycin (Fig. 2, all panels, lanes A versus B and F versus E), precisely the results expected if Gln3-Myc13 were phosphorylated in the former case and dephosphorylated in the latter (10, 13). Because these results are identical to those observed by multiple laboratories after rapamycin treatment of cells provided with rich nitrogen sources, we used these conditions in all subsequent experiments to establish positive and negative electrophoretic mobility standards. Gln3-Myc13 mobilities observed with rapamycin-treated and untreated ammonia-grown cells were similar to those seen with glutamine as nitrogen source (Fig. 2A, lanes C and D). This was the expected result because ammonia, like glutamine, is a good nitrogen source. With SC medium results were similar, but not identical, to those with glutamine and ammonia (Fig. 2B, lanes C and D). Gln3-Myc13 mobility increased upon rapamycin treatment but not to the same degree observed with glutamine or ammonia.
Fig. 2. Effect of rapamycin on the electrophoretic mobility of Gln3-Myc13 isolated from cells grown in various nitrogen sources.
Cultures of strain TB123 were grown to mid-log phase in YNB (without ammonium sulfate and amino acids) containing 2% glucose and either 0.1% ammonium sulfate (A), 0.1% proline (PRO, C), 0.1% urea (UREA, D), or 0.1% glutamine (GLN, A–D) as nitrogen source. Alternatively, cells were grown to mid-log phase in SC medium (B). Protein was isolated from these cells either before (lanes A, C, and F) or after a 30-min incubation in 0.2 µg/ml rapamycin (lanes B, D, and E). Gln3-Myc13 electrophoretic mobility was assayed by Western blot analysis. Protein isolated from glutamine-grown cells (lanes A and F) and rapamycin-treated, glutamine-grown cells (lanes B and E) is included in each panel as well as in subsequent figures as electrophoretic mobility references. We refer to Gln3-Myc13 as phosphorylated or dephosphorylated when the mobilities are those observed in untreated and rapamycin-treated glutamine-grown cells, respectively. We refer to Gln3-Myc13 as hyper- and hypophosphorylated when mobilities are less than observed in untreated glutamine-grown cells, and between those observed in untreated and rapamycin-treated glutamine-grown cells, respectively.
When this experiment was repeated with proline, a poor nitrogen source, we expected to see the more rapidly migrating species characteristic of dephosphorylated, nuclear localized Gln3-Myc13, both before and after rapamycin treatment. However, only the slower mobility species were observed irrespective of whether or not rapamycin was added to the proline-grown cultures (Fig. 2C, lanes C and D). In fact, if anything, Gln3-Myc13 migrated slightly slower in proline-grown, rapamycin-treated cells than in untreated cells. When cells provided with urea were analyzed, Gln3-Myc13 again unexpectedly migrated less rapidly in the absence of rapamycin treatment, but like ammonia, more rapidly when rapamycin was added to the medium (Fig. 2D, lanes C and D). In sum, Gln3-Myc13 electrophoretic mobility did not correlate with either the localization of Gln3-Myc13 or the levels of NCR-sensitive transcription supported by the various nitrogen sources.
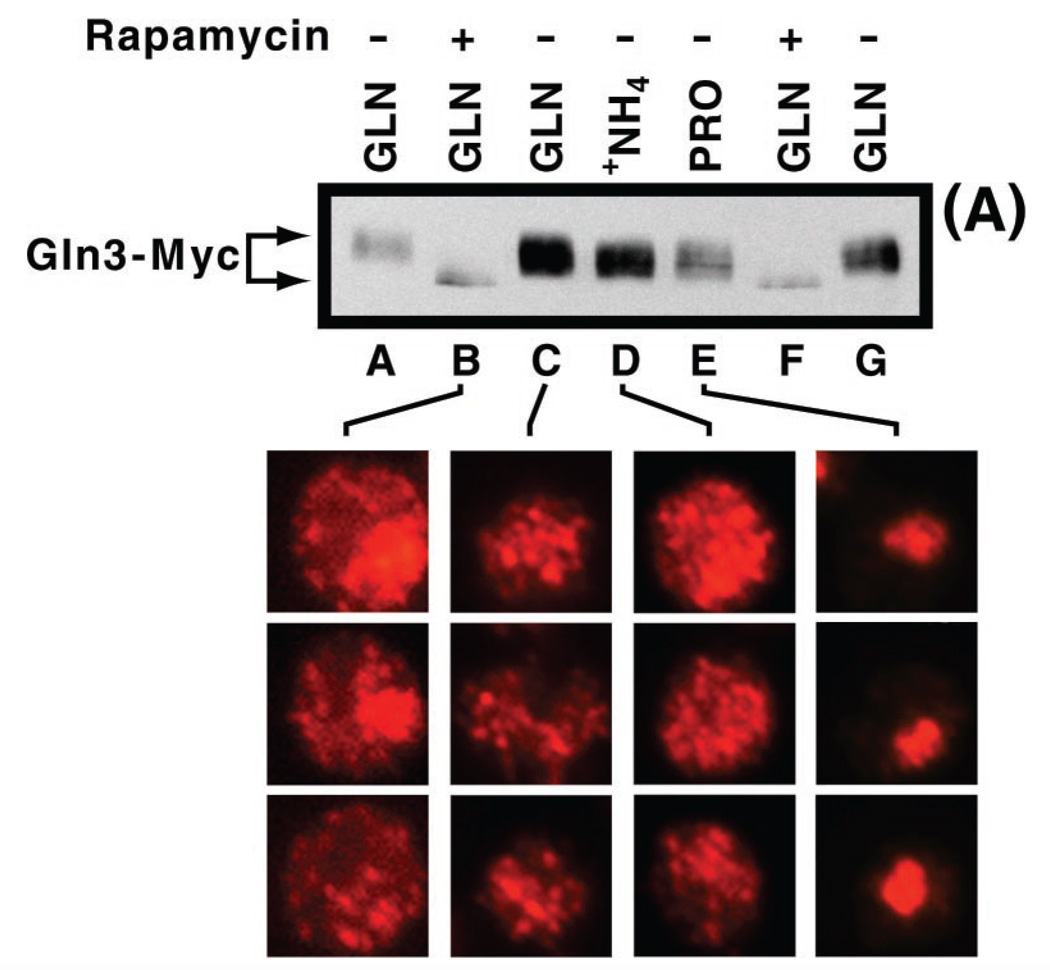
The above data suggested that Gln3-Myc13 electrophoretic mobility failed to correlate with intracellular localization and NCR-sensitive transcription predicted by the Tor1/2 regulatory model. Because these observations were not consistent with expectations, we were concerned that we might have missed small differences in the mobilities of Gln3 derived from cells provided with the nitrogen sources described in Fig. 2. To compare these Gln3-Myc13 mobilities more directly, we analyzed Gln3-Myc13 in extracts of glutamine-, ammonia-, and proline-grown cells in adjacent lanes of the gel. Gln3-Myc13 migrated identically regardless of the nitrogen source in the medium (Fig. 3, lanes C–E) and more slowly than occurred in rapamycin-treated, glutamine-grown cells (lanes B and F). This congruence of Gln3 mobilities occurred even though Gln3 was clearly cytoplasmic in glutamine- and ammonia-grown cells (Fig. 3, lanes C and D) and localized to the nuclei of cells provided with proline (Fig. 3, lane E). These observations contrast with those seen in rapamycin-treated and untreated cells growing with glutamine as the sole nitrogen source. Gln3-Myc13 mobility was greater in rapamycin-treated cells, where Gln3-Myc13 was largely nuclear, than in untreated cells, where it was cytoplasmic (Fig. 3, lanes B and C).
Fig. 3. Comparison of the effect of growth in various nitrogen sources on the intracellular localization and electrophoretic mobility of Gln3-Myc13.
Cells were grown to mid-log phase in YNB (without ammonium sulfate and amino acids) medium containing 2% glucose and either 0.1% glutamine (lanes A–C, F, and G), 0.1% ammonium sulfate (lane D), or 0.1% proline (lane E) as sole nitrogen source. Rapamycin was added to cells used for the preparations in lanes B and F. A, Western blot of isolated protein from cultures grown as described above. B, indirect immunofluorescence micrographs of three cells (grown as in the upper panel) showing Gln3-Myc13 intracellular localization. Gln3-Myc13 is cytoplasmic in one of the three cells shown in images derived from rapamycin-treated, glutamine-grown cells. This was done because only about half of the cells treated in this way contain nuclear Gln3-Myc13.
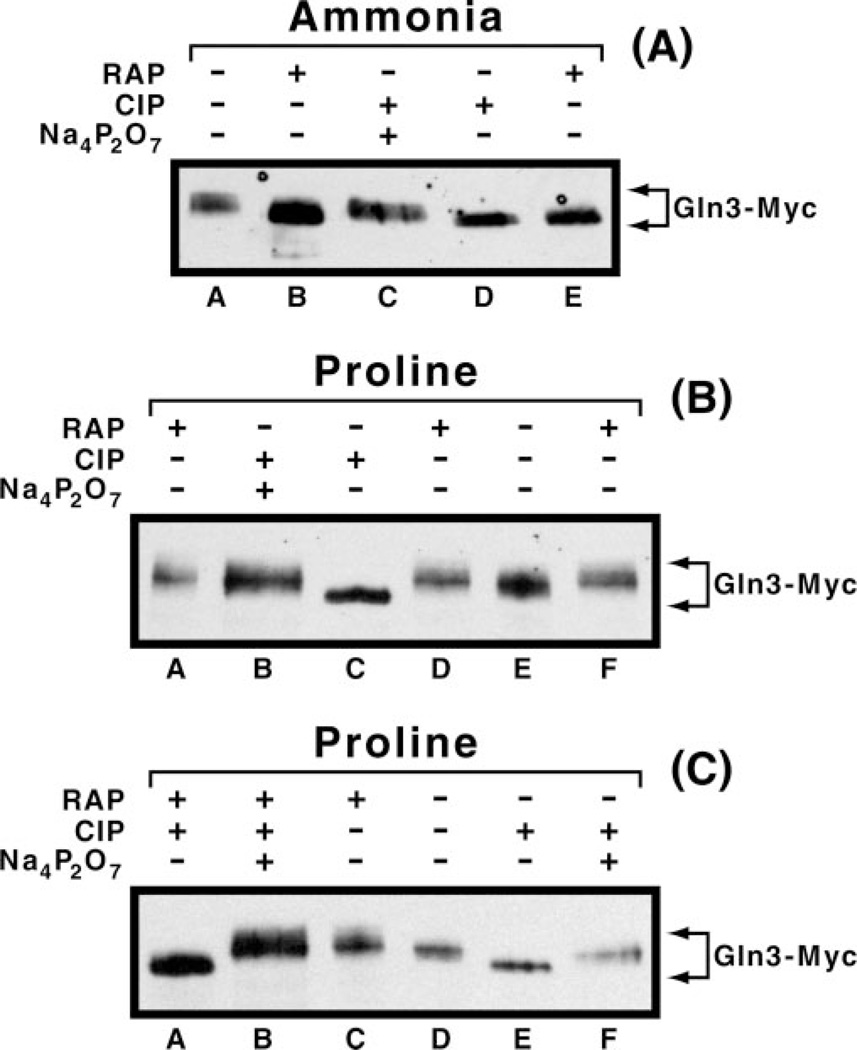
Inconsistency between Gln3-Myc13 electrophoretic mobilities observed with various nitrogen sources and those predicted by the Tor1/2 regulatory model required that we verify the changes in mobility did, in fact, derive from changes in phosphorylation. Treatment of ammonia-grown cells with rapamycin or incubation of untreated, ammonia-grown cell extracts with calf intestinal phosphatase resulted in the more rapidly migrating form of Gln3-Myc13 (Fig. 4A, lanes B, D, and E), the expected results if rapamycin treatment resulted in Gln3-Myc13 dephosphorylation (10, 13). Gln3-Myc13 from untreated cells or rapamycin-treated cell extracts incubated with both calf intestinal phosphatase and phosphatase inhibitor, sodium pyrophosphate, migrated more slowly, indicative of it being phosphorylated (Fig. 4A, lanes A and C). Similar results were obtained with proline-grown cells (Fig. 4B), except that rapamycin treatment had no effect on Gln3-Myc13 mobility (Fig. 4B, lanes A, D, and F). The fact that Gln3-Myc13 mobility did not increase in rapamycin-treated, proline-grown cells raised the possibility that a modification other than phosphorylation might be responsible for the observed lack of effect. To assess this, we incubated extracts from rapamycin-treated, proline-grown cells with calf intestinal phosphatase in the absence or presence of sodium pyrophosphate (Fig. 4C, lanes A and B). The results were consistent with those expected if slow migration of Gln3-Myc13 derived from increased phosphorylation. In addition, we also wished to verify that changes in Gln3-Myc13 phosphorylation did not occur artifactually after cells were broken. We prepared extracts from cultures of rapamycin-treated and untreated cells and a mixture of those cultures made just prior to cell breakage. As shown in Fig. 5B, the mobilities of Gln3-Myc13 from the mixed cultures remained the same as seen in the individual components of the mixture irrespective of nitrogen source or rapamycin treatment.
Fig. 4. Effect of alkaline phosphatase treatment on the electrophoretic mobility of Gln3-Myc13 isolated from cells cultured under various conditions.
Cultures of strain TB123 were grown to mid-log phase in YNB (without amino acids and ammonium sulfate) containing 2% glucose and 0.1% ammonium sulfate (A) or 0.1% proline (B and C). Protein extracts were prepared (as described under “Materials and Methods”) either before (A, lanes A, C, and D; B, lanes B, C, and E; and C, lanes D, E, and F) or after the addition of 0.2 µg/ml rapamycin (A, lanes B and E; B, lanes A, D, and F; and C, lanes A, B, and C). Calf intestine alkaline phosphatase (CIP) (Roche Applied Science) was added along with 10× phosphate buffer (Roche Applied Science, supplied with calf intestinal alkaline phosphatase) (A, lanes C and D; B, lanes B and C; and C, lanes A, B, E, and F). 40 mm Na4P2O7 was added as calf intestinal alkaline phosphatase inhibitor in A, lane C; B, lane B; and C, lanes B and F.
Fig. 5. Effect of nitrogen starvation on Gln3-Myc13 intracellular localization and electrophoretic mobility.
A, cells were grown to mid-log phase in YNB (without ammonium sulfate and amino acids) medium containing 2% glucose and 0.1% ammonium sulfate (upper panel A, lane C). Half of the culture was subsequently transferred to nitrogen-free YNB medium and incubated for 30 min (upper panel, lane D). Crude extracts of these cultures were prepared and analyzed by Western blot analysis as described under “Materials and Methods.” Protein isolated from glutamine-grown (upper panel, lanes A and F) and rapamycin-treated, glutamine-grown cells (upper panel, lanes B and E) was included in the Western blot analysis as electrophoretic mobility references. Lower panels, indirect immunofluorescence micrographs of two cells showing Gln3-Myc13 intracellular localization under the growth conditions described for the upper panel. The bottom two rows of images in the lower panel s how colocalization (magenta) of Gln3-Myc13 (red) and DNA (DAPI, blue). The bottom two rows of images are of the same cells as the top two rows. B, cultures were grown as described in Figs. 2, lanes A–F, and this figure, A, lanes G–I. Culture samples from lanes A and C, D and F, and G and I were mixed just prior to breaking the cells (these are the samples in lanes B, E, and H, respectively). All samples contained the same quantities of cells at the time of breakage. Thereafter, all of the samples were processed as in Fig. 2. 2–2.5 times more protein was loaded into lanes B, E, and H than into the other lanes because these lanes were expected to contain half as much of each species as the other lanes.
Gln3-Myc13 Phosphorylation Profiles Generated by Nitrogen Starvation and in Response to Rapamycin Treatment Differ from One Another
Nitrogen starvation has been concluded to affect Gln3 phosphorylation and intracellular localization in the same way as rapamycin treatment (23). However, this correlation has been tested directly for only a single nitrogen source (SC) and time of starvation. Although nitrogen starvation is the limit case of nitrogen limitation, there is a fundamental difference between growth on a poor nitrogen source, such as a nonlimiting concentration of proline, and nitrogen starvation. In the former case cells continue to divide indefinitely, albeit more slowly. In contrast, nitrogen starvation is a more stressful condition in which cell division ceases, and cells accumulate in G1. Therefore, it would not, a priori, be unreasonable to expect that changes in Gln3-Myc13 mobilities and intracellular localization generated by nitrogen starvation and rapamycin treatment would be similar to one another and yet could be different from those observed with good versus poor nitrogen sources. To evaluate these possibilities, we monitored Gln3-Myc13 mobility and intracellular localization in cells transferred from glucose-ammonia to glucose-nitrogen-free medium or as controls in cells growing in glucose-glutamine versus glucose-glutamine medium containing rapamycin. As shown in Fig. 5A, nitrogen starvation and rapamycin treatment resulted in nuclear accumulation of Gln3-Myc13 (lanes D and B). Similar observations have been made by others as well (23). In contrast, Gln3-Myc13 was localized to the cytoplasm of untreated or unstarved cells (lanes A and C). There was, however, no correlation between Gln3-Myc13 mobility and its intracellular localization, i.e. Gln3-Myc13 mobilities in nitrogen-starved and unstarved cells were similar even though Gln3-Myc13 was nuclear in the former case and cytoplasmic in the latter (Fig. 5A, lanes C and D). A similar lack of correlation was observed between results with nitrogen starvation and rapamycin treatment (Fig. 5A, lanes B and D).
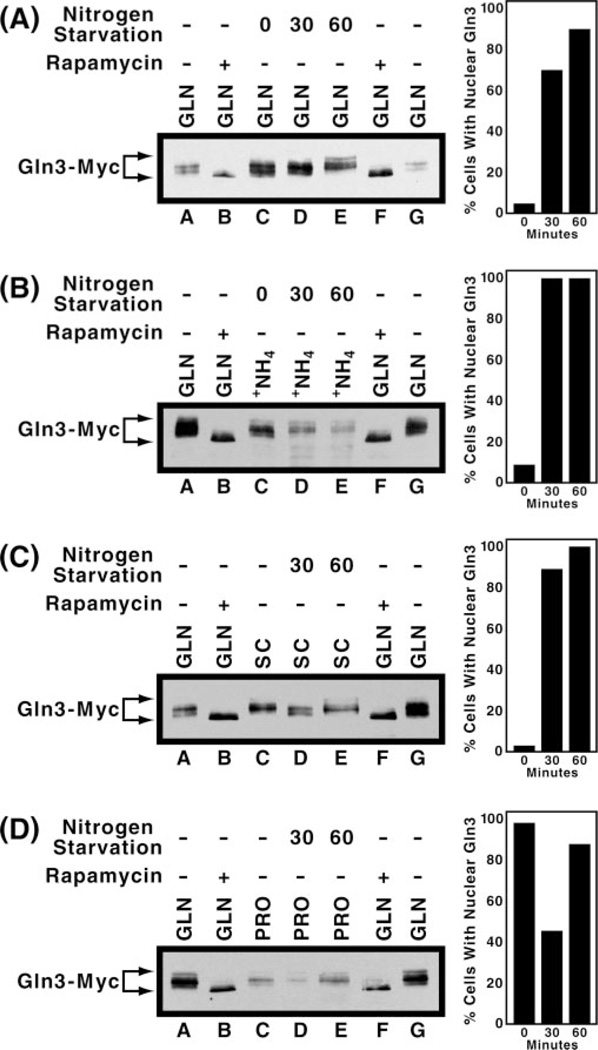
Because these results led to conclusions distinctly different from those in the literature, we queried whether subtle differences in our experimental conditions might account for them; the strain used was the same as in earlier studies (10) so this was not a variable. However, we had previously observed nitrogen source-dependent differences in the timing of nuclear Gln3-Myc13 accumulation (20), thus raising nitrogen source and length of starvation as potential variables. To account for these variables, we provided cells with one of four different nitrogen sources prior to starvation and monitored Gln3-Myc13 mobilities and intracellular localization 30 and 60 min after the onset of starvation. 30 min is the time used by other investigators (23), whereas 60 min takes into account the possibility that the onset of starvation is slower than expected from the literature. After 30 min of starvation, Gln3-Myc13 localized to the nuclei of 70 and 100% of cells pregrown with glutamine or ammonia, respectively. These values increased to 90 and 100% after 60 min of starvation (Fig. 6, A and B, right panels). Gln3-Myc13 mobilities decreased slightly at 30 min and further at 60 min in contrast with increased mobility of Gln3-Myc13 from rapamycin-treated cells (Fig. 6A and B, left panels, lanes D and E versus B and F). We confirmed that the slower migration of Gln3-Myc13, from nitrogen-starved cells pregrown with glutamine, derived from increased phosphorylation. This was done using phosphatase treatment as in Fig. 4 (data not shown). We also determined that differences in Gln3-Myc13 mobilities did not derive artifactually after cells were broken, using mixed cultures as described in Fig. 5B, lanes A–F. Gln3-Myc13 mobilities in the mixed culture were the sum of those seen in the component cultures alone (Fig. 5B, lanes G–I).
Fig. 6. Comparison of the effects of nitrogen starvation on the intracellular localization and electrophoretic mobility of Gln3-Myc13 isolated from cultures pregrown in various nitrogen sources.
Cells were grown to mid-log phase in YNB (without ammonium sulfate and amino acids) medium containing 2% glucose and 0.1% glutamine (A), 0.1% ammonium sulfate (B), or 0.1% proline (D). Alternatively, cells were grown to mid-log phase in SC medium (C). After a sample of the culture was removed (lane C), the remainder was washed once and transferred to nitrogen-free medium and incubated for 30 (lane D) or 60 min (lane E). Protein was isolated and analyzed by Western blotting. Protein isolated from glutamine-grown cells (lanes A and G) and rapamycin-treated, glutamine-grown cells (lanes B and F) was included as electrophoretic mobility references. Graphs in the right panels depict the intracellular localization of Gln3-Myc13, assayed by indirect immunofluorescence, in cells grown under conditions identical to those in lanes C–E of the left panels.
When SC medium was provided prior to starvation, Gln3-Myc13 intracellular localization behaved similarly to that with ammonia- or glutamine-grown cells (Fig. 6C, right panel). In this case, Gln3-Myc13 mobility increased at 30 min as reported by Bertram and co-workers (23), and then decreased to the level observed in unstarved cells at 60 min. The increase in Gln3-Myc13 mobility at 30 min was less than observed in extracts from rapamycin-treated cells (Fig. 6C, lanes B–D). In the case of proline, Gln3-Myc13 was almost totally nuclear prior to starvation. The percentage of cells containing nuclear localized Gln3-Myc13 decreased by about half at 30 min poststarvation and by 60 min increased to levels similar to those observed prior to the onset of starvation (Fig. 6D, right panel). The transient decrease in nuclear localization of Gln3-Myc13 when proline-grown cells are transferred to fresh medium has been observed before.2 Gln3-Myc13 mobility was the same prior to and 30 min after the onset of starvation and decreased slightly at 60 min. We also observed an instance in which Gln3-Myc13 mobility from cells growing in minimal proline medium prior to starvation increased slightly at 30 min after starvation and then decreased in the same way as observed with SC medium (data not shown). The source of this transient variability in mobility is not known. In no case did Gln3-Myc13 mobility achieve that observed with extracts from rapamycin-treated cells even though Gln3-Myc13 is nuclear in both rapamycin-treated and nitrogen-starved cells. Although subtle differences exist in Gln3-Myc13 mobilities observed after nitrogen starvation of cells pregrown in various nitrogen sources, overall Gln3-Myc13 mobilities did not correlate with rapamycin treatment or intracellular localization.
Gln3-Myc13 Phosphorylation Profiles That Result from Carbon Starvation Differ from Those Generated in Response to Rapamycin Treatment
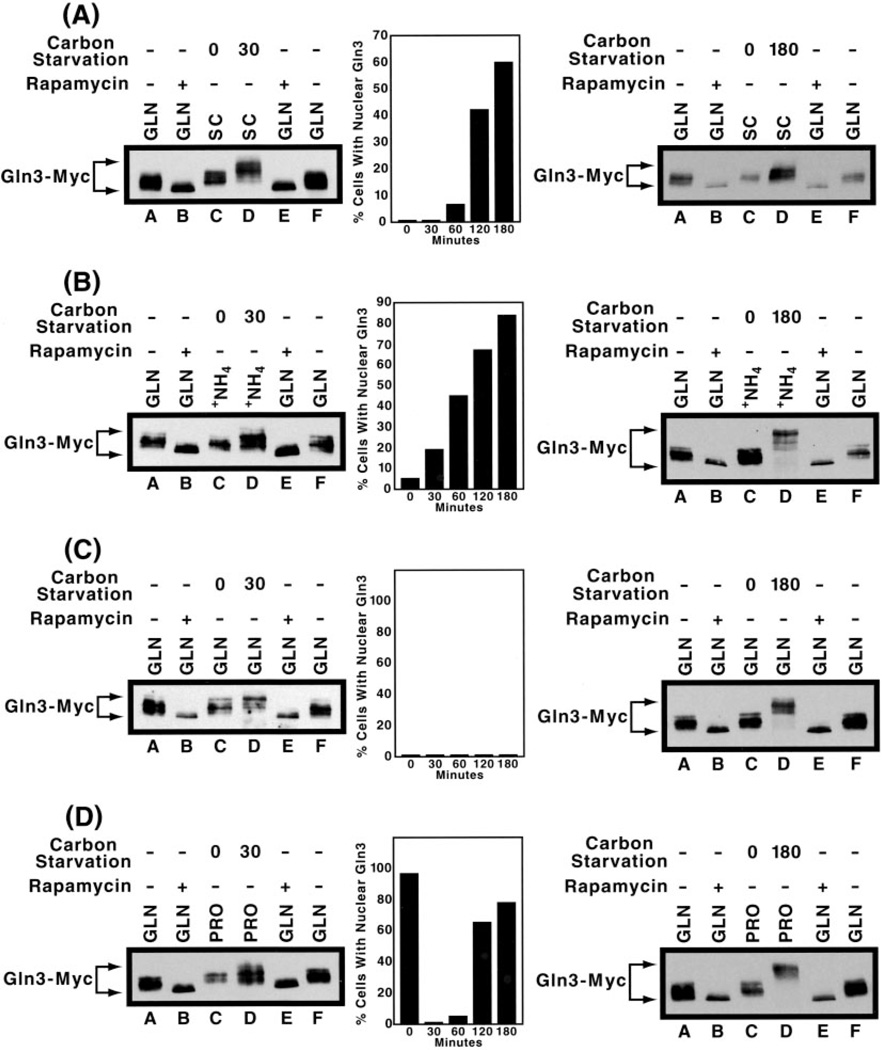
Nitrogen limitation, nitrogen starvation, and carbon starvation are the three nutrient conditions for which Tor1/2 has been reported to regulate Gln3 phosphorylation, intracellular localization, and function (1–9, 20, 23). Failure to observe expected correlations between Gln3-Myc13 mobility and intracellular localization for the first two conditions motivated us to evaluate carbon starvation, as well. We performed measurements at two different times after carbon starvation of cells pregrown in one of the four nitrogen sources used in the nitrogen starvation experiments above. 30 min was the time used by other investigators in the field (23), whereas 180 min was the one at which we previously observed Gln3-Myc13 to be nuclear in the largest percentage of cells (20). As shown in Fig. 7A, left panel, and by other investigators (23), Gln3-Myc13 mobility decreased at 30 min after the onset of carbon starvation in cells pregrown with SC medium; similar Gln3-Myc13 mobilities were observed at 180 min (right panel). In agreement with data from the Zheng laboratory (23), Gln3-Myc13 accumulated in the nuclei of the carbon-starved cells (Fig. 7A, center panel), although it was not, in our hands, clearly measurable until after 60 min of starvation. Recent work has shown that nuclear accumulation of Gln3-Myc13 after carbon starvation of cells pregrown in SC medium derives indirectly as a result of nitrogen starvation brought about by loss of α-ketoglutarate production during starvation (20). When cells were pregrown in minimal ammonia medium prior to being starved for carbon (Fig. 7B), the data were qualitatively similar to those in Fig. 7A except that nuclear accumulation occurred more quickly (compare Fig. 7, A and B, center panels), and the decrease in Gln3-Myc13 mobility was somewhat greater at 180 min after the onset of starvation (compare Fig. 7, A and B, right panels).
Fig. 7. Comparison of the effects of carbon starvation on the intracellular localization and electrophoretic mobility of Gln3-Myc13 isolated from cultures pregrown in various nitrogen sources.
Cells were grown to mid-log phase in SC medium (A). Alternatively, cells were grown to mid-log phase in YNB (without ammonium sulfate and amino acids) medium containing 2% glucose, and 0.1% ammonium sulfate (B), 0.1% glutamine (C), or 0.1% proline (D). Samples of the cells were removed (lane C), and the remainder of the cultures were washed once and transferred to carbon-free medium and incubated for 30 (left panels, lane D) or 180 min (right panels, lane D). Protein was isolated and analyzed by Western blotting. Protein samples isolated from untreated (lanes A and F) and rapamycin-treated (lanes B and E), glutamine-grown cells were included as electrophoretic mobility references. Bar graphs in the center panels depict the intracellular localization of Gln3-Myc13 in starvation cultures prepared identically to those assayed by Western blot analysis.
In contrast to data in Fig. 7, A and B, Gln3-Myc13 did not accumulate in the nuclei of carbon-starved cells pregrown with glutamine as nitrogen source (Fig. 7C, center panel) even though Gln3-Myc13 mobility was similar to that seen with SC and minimal ammonia media in which nuclear accumulation was extensive (Fig. 7C, left and right panels). When proline was provided as the nitrogen source prior to carbon starvation, Gln3-Myc13 was completely nuclear at the outset of the experiment but became localized to the cytoplasm of nearly all of the cells after 30 min of starvation (Fig. 7D, center panel). Thereafter, Gln3-Myc13 slowly became nuclear once again as observed with the other nitrogen sources. The drastic changes in intracellular Gln3-Myc13 localization were not matched by similar changes in its electrophoretic mobility, i.e. it was hyper-phosphorylated at 30 min after the onset of starvation and even more so after 180 min (Fig. 7D, left and right panels). In sum, Gln3-Myc13 was hyperphosphorylated in all cases of carbon starvation irrespective of whether Gln3-Myc13 localized to the cytoplasm or nuclei of starved cells.
Gln3-Myc13 Phosphorylation Profiles Generated in Response to Rapamycin Treatment Do Not Always Correlate with Gln3-Myc13 Intracellular Localization
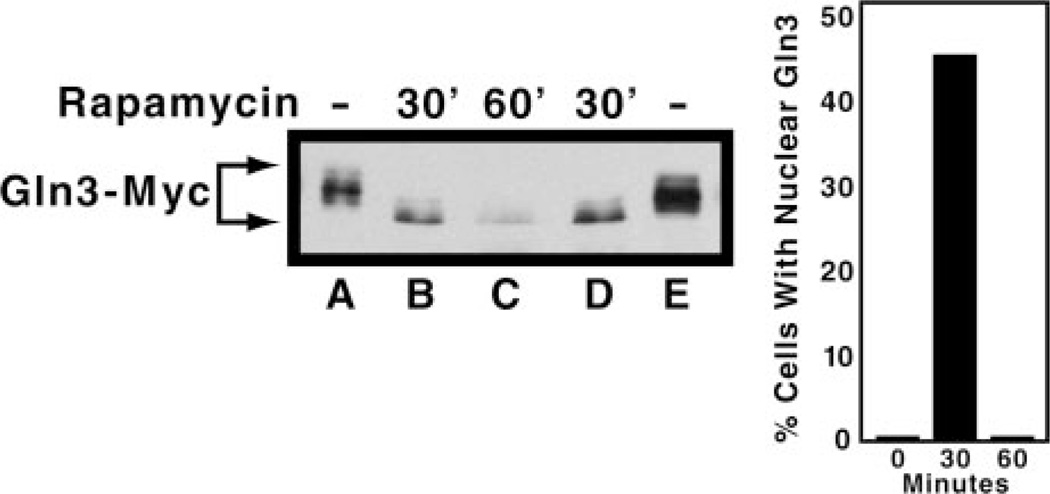
Rapamycin treatment affects Gln3 localization and NCR-sensitive transcription only transiently, in most cases reaching a maximum 30 min after treatment begins (11, 12, 20). This effect is shown quantitatively for Gln3-Myc13 intracellular localization in the right panel of Fig. 8. Gln3-Myc13 is totally cytoplasmic in glutamine-grown cells. Upon treating cells with rapamycin, Gln3-Myc13 accumulates in the nuclei of nearly half of the cells at 30 min but is totally cytoplasmic again at 60 min. According to the correlations presented in this work, and earlier by several others (11, 12), Gln3-Myc13 would be expected to exhibit a slower mobility prior to rapamycin treatment (0 min) and at 60 min after the onset of treatment rather than exhibiting the same mobility observed at 30 min postonset. As shown in Fig. 8 (left panel), Gln3-Myc13 mobility was the same 30 and 60 min after the addition of rapamycin and greater than before treatment. Therefore, even with rapamycin treatment, Gln3-Myc13 mobility does not correlate with its intracellular localization in a consistent way.
Fig. 8. Comparison of the time-dependent effects of rapamycin on the intracellular localization and electrophoretic mobility of Gln3-Myc13 in glutamine-grown cells.
Cells were grown to mid-log phase in YNB (without ammonium sulfate and amino acids) medium containing 2% glucose and 0.1% glutamine. Protein was isolated from these cells either before (lanes A and E) or after 30 (lanes B and D) or 60 min (lane C) of incubation in the presence of 0.2 µg/ml rapamycin. Gln3-Myc13 electrophoretic mobility was assayed by Western blotting. The bar graph in the right panel depicts the intracellular localization of Gln3-Myc13 (assayed by indirect immunofluorescence) in cells cultured under conditions identical to those assayed by Western blot analysis in lanes A–C.
DISCUSSION
An impressive body of work has led to development of a model positing that Tor1/2 regulates NCR-sensitive gene expression in response to nutrient limitation or starvation (3–9). Phosphorylation of the transcriptional activator Gln3 is the terminal and pivotal event contended to be directly (13) or indirectly (10) responsible for restricting it to the cytoplasm of cells growing in the presence of excess nitrogen. Dephosphorylation, on the other hand, is suggested to permit Gln3 to accumulate in the nuclei of cells provided with poor nitrogen sources or during starvation, thereby activating NCR-sensitive transcription. According to this model, we expected Gln3 electrophoretic mobility and intracellular localization to correlate with the quality and quantity of a the nutrient supply of a cell. Although our data confirm all of the previously reported Gln3-associated correlations we tested, we were unable to demonstrate the existence of this critical correlation. In contrast, we observed Gln3-Myc13 electrophoretic mobility to be consistent with suggesting that Gln3 is phosphorylated or hyperphosphorylated regardless of the nutrient conditions assayed or intracellular localization of Gln3 observed. The only condition generating dephosphorylated Gln3-Myc13 as suggested by the current Tor1/2 model was short term rapamycin treatment, an observation reported by others as well (11, 12). Beyond 30 min, however, rapamycin treatment caused Gln3-Myc13 dephosphorylation, but fluorescent signal was localized to the cytoplasm rather than nucleus.
It is important to emphasize several characteristics of the present data and prudent limitations that we recommend be placed upon their interpretation. (i) The experiments described here produced some of the same observations reported previously by others and from which the current model of Tor1/2 regulation was developed (10–13, 23). As such, it is unlikely that failure to observe correlations between Gln3-Myc13 electrophoretic mobility and nutrient availability predicted by the current Tor1/2 model derives from strain or experimental variations because the strains were the same, and we could reproduce the same results as others for short term rapamycin treatment. (ii) Data reported here do not at all rule out the possibility that Gln3 phosphorylation/dephosphorylation is the actual direct or indirect determinant of its intracellular localization. Rather they argue that data presented here and in the current literature are insufficient to conclude a direct cause-effect relationship between Gln3 phosphorylation and intracellular localization. (iii) Changes in Gln3-Myc13 electrophoretic mobility in response to environmental conditions are large. Mobility changes of this magnitude would most likely be associated with gross phosphorylation/dephosphorylation of multiple sites on the surface of the protein. A single phosphorylation/ dephosphorylation event might easily escape detection in all reported data from our laboratory and others and yet would be quite sufficient to regulate the intracellular localization of the molecule (10, 13, 23). Therefore, rather than ruling out such an event, our data argue that the question remains open. (iv) Most experimentation reported in the literature has focused on the phosphorylation/dephosphorylation of Gln3. It is equally likely that molecules other than Gln3 may be the physiologically significant targets of Tor1/2 regulation. For example, Gln3-Myc13 accumulation in the nuclei of cells transferred from glutamine to proline medium requires an intact actin cytoskeleton. Gln3-Myc13 nuclear localization in response to rapamycin treatment, on the other hand, does not exhibit this requirement.2
The absence of correlation among Gln3-Myc13 phosphorylation, intracellular localization, and physiologically significant changes in nutrient supply points to the possibility that inhibition of Tor1/2 activities by rapamycin and nutrient limitation generates the same cellular responses, but does so by distinctly different mechanistic pathways. This possibility may contribute to understanding other instances in which events predicted by the current Tor1/2 regulatory model are not congruent with those observed experimentally (8, 9). It also prompts the question of whether similar considerations are pertinent with respect to other systems reported to be regulated by Tor1/2 as well. In this regard, Tor1/2 control of retrograde gene expression has been recently reported to derive indirectly from alterations in nitrogen metabolism rather than direct Tor1/2 regulation (22). The new information presented here, and the questions it raises about Tor1/2-mediated regulation of nitrogen and other regulons, will open new avenues of investigation into the control of cellular activities by these fascinating proteins.
Acknowledgments
We thank Dr. Michael Hall for strain TB123, Dr. John Cox for helpful advice, Tim Higgins for preparing the artwork, and the University of Tennessee Yeast Group for suggestions to improve the manuscript.
Footnotes
This work was supported by National Institutes of Health Grant GM-35642.
The abbreviations used are: NCR, nitrogen catabolite repression; DAPI, 4,6-diamidino-2-phenylindole; SC medium, synthetic complete medium.
K. H. Cox, J. J. Tate, and T. G. Cooper, submitted for publication.
REFERENCES
- 1.Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dennis PB, Fumagalli S, Thomas G. Curr. Opin. Genet. Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- 3.Schmelzle T, Hall MN. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 4.Raught B, Gingras AC, Sonenberg N. Proc. Natl. Acad. Sci. U. S. A. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhode J, Heitman J, Cardenas ME. J. Biol. Chem. 2001;276:9583–9586. doi: 10.1074/jbc.R000034200. [DOI] [PubMed] [Google Scholar]
- 6.Cyert MS. J. Biol. Chem. 2001;276:20805–20808. doi: 10.1074/jbc.R100012200. [DOI] [PubMed] [Google Scholar]
- 7.Crespo JL, Hall MN. Microbiol. Mol. Biol. Rev. 2002;66:579–591. doi: 10.1128/MMBR.66.4.579-591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper TG. FEMS Microbiol. Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper TG. Curr. Gen. 2004 in press. [Google Scholar]
- 10.Beck T, Hall MN. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 11.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardwick JS, Kuruvilla FG, Tong JF, Shamji AF, Schreiber SL. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan T-F, Zheng XFS. J. Biol. Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- 14.Cox KH, Rai R, Distler M, Daugherty JR, Coffman JA, Cooper TG. J. Biol. Chem. 2000;275:17611–17618. doi: 10.1074/jbc.M001648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper TG. In: Mycota III. Marzluf G, Bambrl R, editors. Berlin: Springer Verlag; 1996. pp. 139–169. [Google Scholar]
- 16.Carvalho J, Bertram PG, Wente SR, Zheng XFS. J. Biol. Chem. 2001;276:25359–25365. doi: 10.1074/jbc.M103050200. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y, Broach JR. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. Mol. Cell. 2001;8:1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- 19.Shamji AF, Kuruvilla FG, Schreiber SL. Curr. Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- 20.Cox KH, Tate JJ, Cooper TG. J. Biol. Chem. 2002;277:37559–37566. doi: 10.1074/jbc.M204879200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tate JJ, Cox KH, Rai R, Cooper TG. J. Biol. Chem. 2002;277:20477–20482. doi: 10.1074/jbc.M200962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tate JJ, Cooper TG. J. Biol. Chem. 2003;278:36924–36933. doi: 10.1074/jbc.M301829200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertram PG, Choi JH, Carvalho J, Chan TF, Ai W, Zheng XF. Mol. Cell. Biol. 2002;22:1246–1252. doi: 10.1128/MCB.22.4.1246-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt ME, Brown TA, Trumpower BL. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Brooklyn, NY: Green Publishing Associates; 1991. [Google Scholar]
- 26.Cooper TG. In: Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. Strathern JN, Jones EW, Broach JR, editors. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1982. pp. 39–99. [Google Scholar]