Abstract
Regulated intracellular localization of Gln3, the transcriptional activator responsible for nitrogen catabolite repression (NCR)-sensitive transcription, permits Saccharomyces cerevisiae to utilize good nitrogen sources (e.g. glutamine and ammonia) in preference to poor ones (e.g. proline). During nitrogen starvation or growth in medium containing a poor nitrogen source, Gln3 is nuclear and NCR-sensitive transcription is high. However, when cells are grown in excess nitrogen, Gln3 is localized to the cytoplasm with a concomitant decrease in gene expression. Treating cells with the Tor protein inhibitor, rapamycin, mimics nitrogen starvation. Recently, carbon starvation has been reported to cause nuclear localization of Gln3 and increased NCR-sensitive transcription. Here we show that nuclear localization of Gln3 during carbon starvation derives from its indirect effects on nitrogen metabolism, i.e. Gln3 does not move into the nucleus of carbon-starved cells if glutamine rather than ammonia is provided as the nitrogen source. In addition, these studies have clearly shown Gln3 is not uniformly distributed in the cytoplasm, but rather localizes to punctate or tubular structures. Analysis of these images by deconvolution microscopy suggests that Gln3 is concentrated in or associated with a highly structured system in the cytosol, one that is possibly vesicular in nature. This finding may impact significantly on how we view (i) the mechanism by which Tor regulates the intracellular localization of Gln3 and (ii) how proteins move into and out of the nucleus.
The budding yeast Saccharomyces cerevisiae is often used as a model with which to elucidate the functions of important mammalian proteins. Few such proteins have generated greater excitement than Tor1/2 and their mammalian counterpart mTor, which are inhibited by the immunosuppressant and antineoplastic drug rapamycin (1–5). Tor1 and the essential Tor2 proteins are reported to be situated at the top of the Tor signal transduction cascade and through it to regulate an exceedingly large number of diverse cellular processes, including: translational initiation, G1-phase progression, autophagy, RNA polymerase I/III function, actin cytoskeleton organization, membrane protein stability, nitrogen catabolite repression (NCR)1-sensitive and retrograde gene expression (1–9). In this work, we focus on the last two of these processes.
NCR is the physiological process by which S. cerevisiae selectively uses good nitrogen sources in its environment in preference to poor ones (5, 10–12). In the presence of good nitrogen sources (glutamine, asparagine, and in some strains, ammonia), expression of genes encoding the permeases and enzymes needed for utilization of poor nitrogen sources is held at low levels, i.e. expression is repressed. When only poor (proline) or limiting amounts of good nitrogen sources are available, these genes are then expressed at the higher levels needed to scavenge whatever is available in the environment (5, 10–12). Two GATA family transcriptional activators (Gln3 and Gat1/Nil1) are responsible for NCR-sensitive transcription and Ure2 (a yeast prion precursor) inhibits their ability to carry out this function (5, 10–12). In glucose-proline medium, Gln3 and Gat1 are bound to the GATA sequences of the NCR-sensitive CAN1 gene, whereas in glucose-glutamine medium or in a gln3Δgat1Δ strain, these GATAs are unoccupied and available to serve as surrogate TATA elements (13). Correlating with these observations, Gln3 and Gat1 are localized to the nucleus in the former medium and to the cytoplasm in the latter (13).
Work in four laboratories, using the immunosuppressant drug, rapamycin, has provided insights into the mechanism regulating intracellular localization of Gln3 (14–17). They found that inhibiting Tor1/2 with rapamycin resulted in dephosphorylation of Gln3 and, in some laboratories, Ure2, and its entry into the nucleus (14, 16, 17). Loss of NCR-sensitive gene expression in rna1 and srp1 mutants (19, 20), led Carvalho et al. (20) to conclude that these proteins are required for Gln3 entry into the nucleus and that Crm1 mediates its exit. Tap42, a kinase that itself is phosphorylated by Tor (21, 22), Tap41, type 2 phosphatases Sit4 and Pph3, Mks1 and Ure2 are the remaining members of the Tor signal transduction cascade reported to be situated between Tor and Gln3/Gat1 (1–3, 6–9, 14–18, 23, 24). Recent work (24), however, demonstrated that Mks1 functions only indirectly in the regulation of Gln3/Gat1.
A second set of metabolic genes, those associated with the retrograde response have also been reported to be regulated by the Tor signal transduction cascade (6 –9, 24–26). The retrograde genes are those whose expression is increased in cells with damaged mitochondria (27–30). These genes, including CIT2, ACO1, IDH1/2, and DLD3, are thought to be responsible for synthesizing α-ketoglutarate, which is needed for glutamate and its biosynthetic products when: (i) the normal tricarboxylic acid cycle is shut down, (ii) cells are fermenting high concentrations of glucose, or (iii) mitochondria are otherwise unable to function. Consistent with this physiological function, retrograde gene expression is low when cells are provided with glutamate as nitrogen source. Retrograde gene expression is mediated by the transcription activators Rtg1/3 (24–30), whose differential phosphorylation levels and intracellular localization are regulated in a remarkably analogous way to those of Gln3 and Gat1. Like Gln3 and Gat1, nuclear localization of Rtg1/3 occurs when cells are treated with rapamycin (9), which also results in changes in Rtg3 phosphorylation levels (9). Although consensus remains to be established on whether nuclear localization of Rtg3 occurs with the hyperphosphorylated form, hypophosphorylated form, or both, correlations between differences in transcription factor phosphorylation and retrograde gene expression are convincing (9, 26, 28, 33). Rtg2 is required for Rtg1/3 to mediate retrograde gene expression, and Mks1, originally thought also to be required for Rtg1/3-mediated transcription (6, 8, 9), is now shown to be a strong negative regulator of Rtg1/3-mediated transcription (24, 26, 33).
The retrograde gene expression experiments just described collectively demonstrate a relationship between nitrogen and carbon metabolism in S. cerevisiae. Bertram et al. (31) have recently reported a second major bridge linking carbon and nitrogen metabolism. They reported that starving cells for carbon, like starving them for nitrogen, results in Gln3 being localized to the nucleus and mediating transcription of the NCR-sensitive genes, GAP1, GDH2, and PUT1 (31). Carbon-starved nuclear localization of Gln3 also correlates with its hyperphosphorylation. Finally, Snf1 has been shown to be required for these carbon starvation-induced changes in Gln3 localization, in phosphorylation, and for the transcription it mediates (31).
Our interest was piqued by these carbon starvation experiments, because here Gln3 is nuclear during the nitrogen excess, which exists during carbon starvation. Investigating this phenomenon, we find that whether or not Gln3 is localized to the nucleus during carbon starvation is dictated by the nature of the nitrogen source, i.e. Gln3 nuclear localization occurs with ammonia but not glutamine as nitrogen source. The effects of carbon starvation with ammonia as the nitrogen source are indirect and most likely caused by the inability of ammonia to be assimilated into glutamate during carbon starvation due to the lack of α-ketoglutarate, the carbon skeleton of glutamate. More important, however, we also found that Gln3 is not uniformly distributed in the cytoplasm during growth in glutamine medium. It appears to be sequestered in or associated with globular/tubular structures. The implications of this distribution generate an alternative way of viewing how Tor influences Gln3 intracellular distribution and NCR-sensitive transcription.
MATERIALS AND METHODS
Strains and Culture Conditions
Strains used in this work were JK9-3da (MATa, leu2–3,112, ura3–52, rme1, trp1, his4, GAL+, HMLa) and TB123 (MATa, leu2–3,112, ura3–52, rme1, trp1, his4, GAL+, HMLa, GLN3-myc[kanMX4]). Cultures were grown overnight at 30 °C in Difco YNB (without ammonium sulfate or amino acids), 2% glucose, and the indicated nitrogen source. Appropriate auxotropic requirements were supplied when media other than synthetic complete (SC) (31) were used. Experiments in which yeast were transferred from one medium to another were performed as follows. A sample of the exponentially growing cells (A600 nm = 0.4–0.7) was harvested from the initial medium for immunofluorescence or RNA analysis. The remaining cells in the culture were collected, washed with the new medium, recollected, and resuspended in an equal volume of the new medium. Additional samples were then collected and processed for RNA isolation or immunofluorescence at the indicated times. Precisely the same format was also used for the transfer of cells from rapamycin-free (initial medium) to rapamycin-containing (new medium) media. Rapamycin (Sigma), from a concentrated stock solution in 90% ethanol/10% Tween 20, was used at a final concentration of 200 ng/ml where indicated.
RNA Isolation and Northern Blot Analysis
Total RNA was prepared from cultures of strain JK9-3da as previously described (24). Total yeast RNA (9 µg) was separated on denaturing gels, transferred to a nylon membrane, and hybridized with 32P-labeled DNA probes, and the blots were washed as previously described (24).
Indirect Immunofluorescence
Immunofluorescent staining of yeast was carried out using a modification of the methods of Schwartz et al. (32). Cultures of strain TB123 were fixed by the addition of 1/10 volume of 37% formaldehyde and incubated with shaking at 30 °C for 10 min. They were then collected by centrifugation at room temperature and incubated in 3.7% formaldehyde in potassium phosphate buffer (40 mm, pH 6.5, containing 0.5 mm MgCl2) for 1 h. After washing and Zymolyase digestion as previously described, cells were applied to poly-l-lysine-coated microscope slides. The slides were blocked overnight at 4 °C using 0.5% bovine serum albumin, 0.5% Tween 20 in phosphate-buffered saline (pH 7.4). All further antibody incubations and washes were performed in this buffer. 9E10(c-myc) (Covance MMS-150P) was used at a dilution of 1:1000 as the primary antibody and either Texas Red-conjugated goat anti-mouse IgG (Jackson ImmunoResearch) or Alexa Fluor 488 goat anti-mouse IgG (Molecular Probes) was used at a dilution of 1:200 as the secondary antibody. To visualize nuclei, 4′,6′-diamino-2-phenylindole (DAPI) was added to the mounting media at a final concentration of 50 µg/ml, and images were collected immediately after mounting.
Immunofluorescence Microscopy
Cells in Figs. 1 and 2 were imaged using a Zeiss Axiophot microscope with a 63× Plan-Apochromat 1.40 oil objective. Images were acquired using AxioVision 3.0 (Zeiss) software and a Zeiss Axio camera. Alternatively, cells shown in Figs. 7–9 (see below) were imaged using a Zeiss Axioplan 2 imaging microscope with a 100× Plan-Apochromat 1.40 oil objective. Images were acquired using a Zeiss Axio camera and deconvolved using AxioVision 3.0 (Zeiss) software using the constrained iterative algorithm. Three-dimensional views (see Figs. 8, E–G, and 9 below) were produced from 15 images in a two-dimensional Z-stack, spanning about 1.5 µm of the center of a yeast cell using AxioVision Inside 4D (Zeiss) software. Images were rendered using either the maximum projection in which only pixels of the highest intensity along the axis are displayed (Fig. 8, E–G) or the surface mode in which non-transparent surfaces are calculated from gray values (Fig. 9).
Fig. 1. Effect of nitrogen source on intracellular localization of Gln3 in carbon-starved cells grown in SC media.
Yeast cultures (TB123) were grown to mid-log in 2% glucose synthetic complete (SC) or 2% glucose synthetic complete with 0.1% glutamine replacing ammonia, the normal nitrogen source in SC (SC + Gln), and transferred to the same media without glucose. Aliquots were removed just prior to carbon starvation (0 min) or at times following transfer to glucose-free medium and processed for immunolocalization. Top panel, micrographs of cells grown in SC (A and B) or SC + glutamine medium (C and D) prior to (A and C) and 180 min after the onset of carbon starvation (B and D). Bottom panel, percentage of the total yeast cells in which Gln3 was localized to the nucleus at various times after transfer to carbon starvation medium.
Fig. 2. Effect of ammonia or glutamine on intracellular localization of Gln3 in carbon-starved cells grown in YNB media.
The experiment was performed as in Fig. 1 except for changes in the media. Top panel, micrographs of cells after 180 min of incubation in YNB-glucose-free ammonium sulfate (A) or YNB-glucose-free glutamine (B) media. Bottom panel, as in Fig. 1.
Fig. 7. Intracellular distribution of Gln3 under various growth conditions, visualized by conventional and deconvolution microscopy.
Yeast cultures (TB123) were grown to mid-log phase in YNB-glutamine medium and processed for immunolocalization (row 3). Alternatively, cultures were grown to mid-log phase in YNB-proline and transferred to YNB-glutamine medium. Aliquots were removed just prior to (row 1), and 1 min after (row 2) the transfer from proline to glutamine medium. Images are presented in pairs: Micrographs A, C, E, and G were imaged using a Zeiss Axioplan 2 imaging microscope. 0.1-µm sections were collected as a Z-stack, and one image from the center of the cell is shown. Micrographs B, D, F, and H show these same images deconvolved with AxioVision 3.0 (Zeiss) software using the constrained iterative algorithm.
Fig. 9. The three-dimensional image described in Fig. 8 was rendered using the surface mode in which non-transparent surfaces are calculated from gray values.
Fig. 8. TB123 cells were grown to mid-log phase in YNB-glutamine medium and processed for immunolocalization.
Micrographs A–D were imaged using a Zeiss Axioplan 2 imaging microscope. 0.1-µm sections were collected as a Z-stack, and one image from the center of the cell is shown. The images are shown either as raw data (A) or as deconvolved images (B–D) of Gln3p-mediated (A, B, and D) or of DAPI-mediated fluorescence (C and D). Three-dimensional views (E–G) were produced from 15 images in a two-dimensional Z-stack, spanning about 1.5 µm of the center of a yeast cell using AxioVision Inside 4D (Zeiss) software. Images were rendered using the maximum projection in which only pixels of the highest intensity along the axis are displayed. Three-dimensional images have been rotated on all three axes to provide different views of Gln3 intracellular localization.
RESULTS
Nitrogen Source Determines Intracellular Localization of Gln3 during Carbon Starvation
A recent report indicates that carbon starvation results in Snf1-dependent nuclear localization of Gln3 (31). These data, coupled with earlier reports that nitrogen starvation or limitation also results in nuclear localization of Gln3 and Gat1p (13–17), create a paradox. Cells grown in medium devoid of nitrogen are by definition growing in carbon excess. Conversely, cells grown in medium devoid of carbon are by definition growing in nitrogen excess. It was difficult to understand why Gln3 would react similarly to physiologically opposite conditions, i.e. nitrogen starvation and excess. Therefore, we investigated the phenomenon in greater detail, first by performing and then extending the critical published experiment (31). To acquire a crude estimate of the time course of Gln3 redistribution, following perturbation, we scored the intracellular localization of Gln3 in ~100 cells from random fields collected at increasing times following the onset of starvation. Multiple samples were independently scored in duplicate by two different individuals. Insignificant variations were observed in these duplicate counts. Following transfer of cells from SC to glucose-free SC medium, little change was observed at 30 min (Fig. 1B, green bars); representative microscopic images are shown in Fig. 1A. Nuclear-localized Gln3 rose from 7% of the cells at 1 h to over 40% at 2 h, and up to about 60% at 3 h (Fig. 1B). These results are similar to the published work, except that Gln3 response appeared to be slower in the strain we used. Gln3 appeared nuclear in the 30-min image of Bertram et al. (31).
A principal consequence of carbon starvation is the loss of carbon skeletons needed for biosynthetic reactions, such as those involving glutamate and glutamine. The loss of the ability to synthesize these amino acids would in turn generate serious consequences on nitrogen metabolism and NCR. Therefore, we repeated the carbon starvation experiment but accounted for this secondary consequence by performing it in SC + 0.1% glutamine rather than SC medium. Even though the SC + glutamine medium, like that used above, was devoid of glucose, Gln3 was nuclear in less than 10% of the cells at 2 h and decreased to near undetectable levels thereafter (Fig. 1B, red bars).
To ensure that the complex nitrogen mixture contained in SC medium was not misleading us, we performed the experiment in YNB to which was added a single nitrogen source, ammonia or glutamine. In these less complex media, the time course profiles were even clearer. Gln3 localized to the nuclei of a gradually increasing number of cells cultured in carbon-free ammonia medium (Fig. 2B, green bars), whereas with carbon-free glutamine medium, Gln3 remained cytoplasmic throughout starvation. However, in contrast to SC medium in which 1 h was required for Gln3 to become nuclear in 7% of the cells, in YNB-ammonia medium Gln3 was nuclear in 5% of the cells at the outset. By 1 h, this value increased to 45% and reached over 80% by the end of the experiment (Fig. 2B, green bars). With glutamine as sole nitrogen source, we were unable to detect nuclear localization of Gln3 (Fig. 2B, red bars). The data collectively argue that nuclear localization of Gln3 is a function not of carbon starvation but of the nitrogen source provided in the medium in which starvation is carried out. Carbon starvation is inconsequential to the cellular localization of Gln3 if sufficient glutamine is present, i.e. NCR is operating.
The Expression of Only Some NCR-sensitive Genes Correlates with the Nuclear Localization of Gln3
To determine the strength of correlation between the intracellular Gln3 localization and NCR-sensitive gene expression, we measured mRNA levels for several well-studied nitrogen-related genes. Rather than use all of the conditions we studied microscopically, the three most likely to be informative were chosen: YNB-ammonia, YNB-glutamine, and SC media, with samples harvested just prior to and 10, 30, and 120 min following the onset of carbon starvation. The first thing we noticed was that genes routinely used to assess loading and RNA transfer were not useful when performing carbon starvation experiments. H3, ACT1, and TCM1 mRNA levels all decreased following the onset of carbon starvation regardless of the medium used, arguing that carbon starvation rather than nitrogen source was responsible for the effects (Fig. 3, B and C). The only probe we found usable as a standard was one (designated pC4) used by investigators for studying the onset of meiosis and sporulation. This standard also followed bulk RNA in its quantitation.
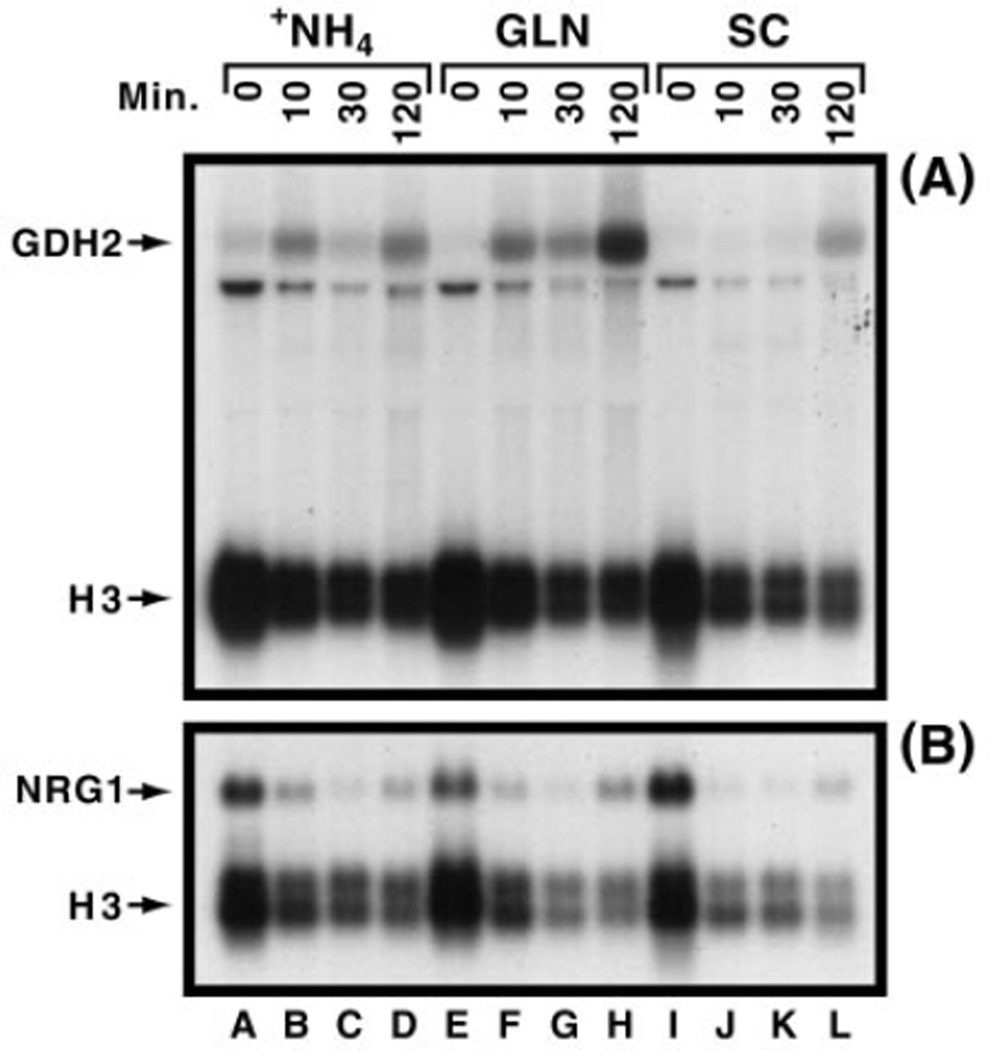
Fig. 3. Effect of nitrogen source on the transcription of selected “control” and NCR-sensitive (DAL5, DAL80) genes.
Yeast cultures (JK9-3da) were grown to mid-log in 2% glucose, YNB (without ammonium sulfate and amino acids) and either 0.1% ammonium sulfate (+NH4) or 0.1% glutamine (GLN) as nitrogen source. Alternatively, cells were grown in SC medium. Cultures were transferred to the same medium but devoid of glucose, and samples were removed prior to (0 min) or at times following transfer to glucose-free media. RNA blots were hybridized with 32P-radiolabeled DAL5 (allantoate permease) (panel A), H3 (Hht1-histone3) (panels A and C), ACT1 (actin) (panel B), DAL80 (GATA-family transcription factor) (panel B), and TCM1/RPL3 (a ribosomal protein) probes. PC4 (a control used for sporulation assays whose expression mirrors ribosomal RNA concentration) was used as a control.
Next we analyzed two highly NCR-sensitive genes, DAL5 and DAL80. In both cases, expression reached detectable levels only after 2 h of carbon starvation with ammonia as the nitrogen source. DAL5 or DAL80 expression was not detected in SC or YNB-glutamine grown cells (Fig. 3, A and B). The GDH2 expression profile was quite different. In YNB-ammonia medium GDH2 expression was low at the outset of starvation, increased at 10 min, fell at 30 min, and increased again at 2 h (Fig. 4A). In SC medium GDH2 expression was detectable only after 2 h of carbon starvation. Still different were the results with YNB-glutamine. Here, GDH2 expression began low, increased at 10 min, decreased marginally if at all at 30 min, and increased to very high levels after 2 h of starvation. On the other hand, expression of NRG1, encoding a carbon-regulated gene, was high at the outset of starvation and for the most part decreased thereafter regardless of the nitrogen source employed (Fig. 4B).
Fig. 4. Effect of nitrogen source on the transcription of the GDH2 and NRG1 genes.
The experiment was performed as in Fig. 3.
The most striking characteristic of these data is the lack of a consistent pattern of expression. At a more discriminating level, however, there are visible patterns. If one considers a gene, known to be regulated by carbon, NRG1, the pattern is reasonably consistent and largely independent of the nitrogen source. Similarly, with the gene whose expression has to date been demonstrated to be regulated by nothing other than NCR (DAL5 or DAL80), the pattern is again consistent and behaves as predicted from the level of NCR generated by each nitrogen source. The difficulty arises with genes like GDH2, assayed both in our work and that of Betram et al. (31). GDH2 expression fluctuates in both laboratories, but not congruently. However, when one takes into consideration that GDH2 expression is reported to be regulated in four distinctly different ways, the variation is perhaps not so difficult to envision. As carbon starvation progresses, intracellular levels of carbon and nitrogen compounds are constantly changing. Because the level of GDH2 expression is the sum of four different types of regulation, such fluctuation is expected rather than surprising. Therefore, unless the gene chosen is subject to a single type of regulation whose response to all of the experimental perturbations can be reliably predicted, data like that seen with GDH2 are to be expected. Instances of complex profiles can be traced to equally complex and differing modes of regulation to which many nitrogen related genes are subjected, in addition to NCR. For example, the CAR1 promoter has 14 distinct promoter elements and 9 trans-acting factors that function during expression (25).
Time Course of Gln3 Nuclear Import and Export
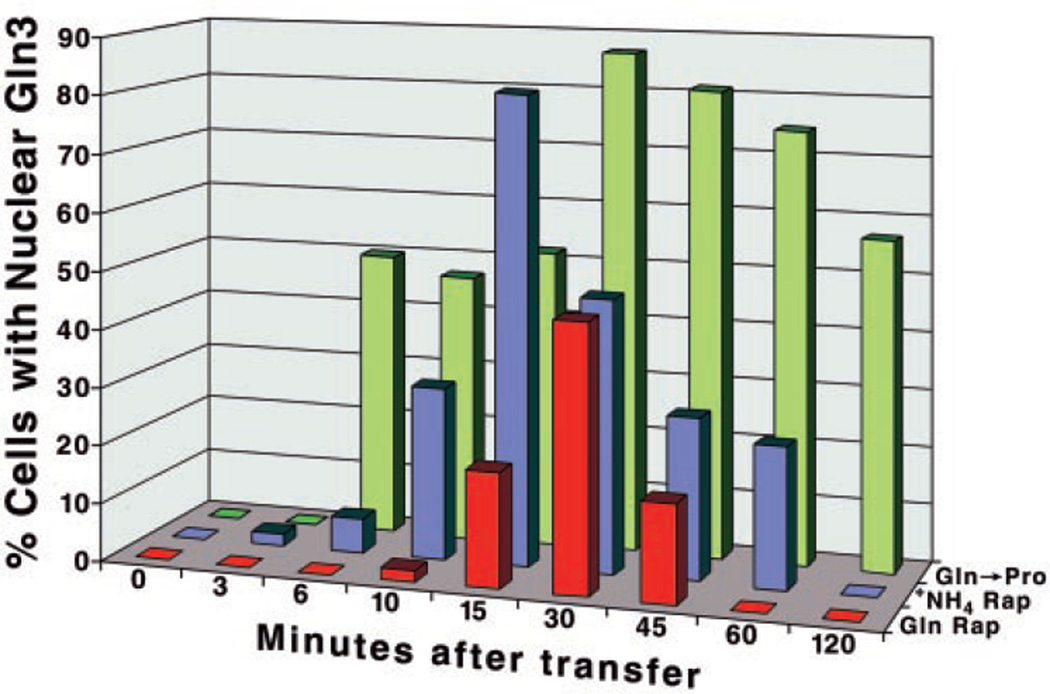
Studies of the mechanism by which NCR is accomplished have depended heavily upon the ability of rapamycin-treatment to mimic nitrogen starvation (14–17). The correlation of observations generated by nitrogen starvation and rapamycin-treatment are indeed among the most important reasons for concluding that Gln3 intracellular localization, and thereby NCR, are regulated by the Tor1/2 signal cascade (6, 14–17). This prompted us to compare the time course of events triggered by metabolic and rapamycin-mediated perturbation. We first compared Gln3 distribution following the onset of carbon starvation or after transfer of cells from glucose-glutamine to glucose-proline media. Ammonia was used as a nitrogen source for the carbon starvation experiment, because, with glutamine, Gln3 isn’t detected in the nucleus. As shown in Fig. 5, the two perturbations generate time-course profiles that differ in two important respects. Gln3 movement into the nucleus becomes detectable between 3 and 6 min after transferring glutamine-grown cells to proline and localizes there in nearly 90% of the cells by 30 min (Fig. 5, green bars). In glucose starvation the onset of Gln3 movement is slower, being first detected at 30 min, and is far less complete, attaining localization in about half of the cells at 120 min (Fig. 5). We suggest the different profiles derive from the differences in the times required for the metabolite that controls NCR to decrease sufficiently for NCR to be relieved thereby permitting Gln3 to enter the nucleus. That more time is required for glucose-generated α-ketoglutarate to become limiting in carbon starvation than glutamine in a glutamine to proline shift makes reasonable physiological sense. We noted an additional correlation with the glutamine to proline transition. The fraction of cells in which Gln3 is nuclear increases sharply between 3 and 6 min, plateaus between 6 and 15 min, and again rises sharply between 15 and 30 min. The cytoplasm of cells from the 30-min sample exhibited much less fluorescence than at 15 min, arguing that nuclear localization of Gln3 became more pronounced after 15 min (data not shown).
Fig. 5. Comparison of the time courses of Gln3 movement into nuclei of cells transferred from glutamine to proline medium versus YNB-ammonia to glucose-free YNB-ammonia medium.
The format of the experiment was as described in Fig. 1.
Equally pertinent comparisons are those made with rapamycin. Both rapamycin and glutamine or the physiologically significant signal are situated upstream of Tor1/2 in the signal transduction cascade hypothesized to regulate the cellular distribution of Gln3 (6, 14–17). Therefore, rapamycin should affect Gln3 distribution at least as quickly or perhaps more so than a shift from glutamine to proline. Three conditions were assayed to test this expectation: rapamycin addition to cells provided with (i) ammonia or (ii) glutamine as nitrogen source and (iii) transfer of cells from glutamine to proline. Contrary to expectation, marked differences were again observed (Fig. 6). Gln3 becomes nuclear more quickly when cells are shifted from glutamine to proline (50% at 6 min) than with rapamycin treatment irrespective of the nitrogen source (6% at 6 min with ammonia and 20% at 15 min with glutamine) (Fig. 6). Two things are exceptional in these data: (i) the fact that rapamycin acts more slowly than shifting glutamine-grown cells to proline, and (ii) the speed with which rapamycin acts is dependent upon the nitrogen source. The latter observation is also consistent with the fact that we find rapamycin “induces” retrograde and NCR-sensitive gene expression less well with glutamine than ammonia as the nitrogen source (24). This would not be expected a priori, because rapamycin is thought to act downstream of the signal metabolite.
Fig. 6. Comparison of the time courses of Gln3 movement into nuclei of cells transferred from YNB-glutamine to YNB-proline medium versus YNB-ammonia to YNB-ammonia + rapamycin versus YNB-glutamine to YNB-glutamine + rapamycin media.
The format of the experiment was as described for Fig. 1, including those media containing rapamycin.
The other striking difference is that transfer from glutamine to proline medium generates a more sustained response than rapamycin, with duration of the rapamycin effect again depending on the nitrogen source used. Two hours after shifting cells from glutamine to proline, Gln3 was still localized to the nucleus in over 50% of the cells (Fig. 6, green bars). In contrast, nuclear localization of Gln3 peaked at 15 and 30 min with ammonia and glutamine, respectively (Fig. 6, blue and red bars). Furthermore, Gln3 localization to the nuclei of rapamycin-treated, ammonia-grown cells peaks earlier, is higher, and is sustained longer than when glutamine is used (Fig. 6). With rapamycin-treatment, as with carbon starvation, the nitrogen source makes a noticeable difference in the time course of Gln3 entry and exit from the nucleus even though rapamycin is supposedly downstream of the sensed physiological signal.
Cytoplasmic Compartmentation of Gln3 during Nitrogen Catabolite Repression
During this work, we noticed that, when Gln3 was localized to the cytoplasm, more often than not, the distribution of fluorescence was not uniform. Gln3-derived fluorescence appeared to be concentrated in “clumps” or more defined punctate structures; several examples of this are shown in Fig. 7, row 3, images A, C, E, and G). This “clumping” was very reminiscent of the bright, punctate foci situated around the periphery of the cell or vacuole that we reported earlier to occur with both EGFP-Gln3 and EGFP-Ure2 (see Figs. 9 and 10 of Ref. 13).
It was conceivable the punctate appearance observed in those earlier studies derived from prion formation, because Ure2 was highly overproduced (13). In contrast, the images in Figs. 1, 2, and 7 derive from cells in which neither Ure2p nor Gln3 are overproduced, and so it is unlikely that punctate staining derives from prion formation. These data raised the possibility that Gln3 might not be uniformly distributed throughout the cytosol during growth in good nitrogen sources. To examine this more carefully, cultures were grown to mid-log phase in either 2% glucose YNB medium containing 0.1% proline or glutamine as nitrogen source. Cells in the proline-grown culture were then transferred to medium containing glutamine as the nitrogen source. At various times after the transfer, samples of the culture were taken and analyzed by immunofluorescence microscopy. The glucose-glutamine culture was not treated further before cell samples were taken. Fluorescence is sometimes distributed as dots around the periphery of the cell or ringing the vacuole (Fig. 7, row 2, images B, D, F, and H; row 3, images B, F, and H) and at others as tubes or networks of tubes (row 2, images B, D, and F; row 3, images D, F, and H). As expected, cells grown in glucose-proline medium exhibited nuclear localization of Gln3 (row 1). However, after only 1 min of transfer to glucose-glutamine medium, about 50% of the cells showed complete cytoplasmic localization of Gln3. In those cells that still retained some nuclear concentration of Gln3, fluorescence can be seen in projections emanating from the nucleus (row 2, images B, D, and H). Note that the images in Fig. 7 all appear as pairs, with fluorescent images (panels A, C, E, and G) followed by deconvolved views (panels B, D, F, and H).
To ascertain whether the fluorescent dots represented end views of tubular structures, we analyzed three-dimensional reconstructions of multiple images; an example is shown in Fig. 8. In the normal fluorescent image, clearly defined dots appear to circle the cell (Fig. 8, image A). They are much more apparent in the deconvolved reconstruction (image B, arrows). The nucleus was visualized with DAPI staining (image C) and appears along with Gln3 fluorescence in image D. As the plane of view is rotated about its axes, fluorescent dots (arrows in image B) turn into elongated tubes (Fig. 8, images E–G, arrows).
Finally, the three-dimensional image from Fig. 8 was rendered using a surface mode in which solid surfaces are constructed from the gray values of the pixels (Fig. 9). In this view, the structures appear similar to what would be expected if Gln3 was contained within or was attached to one of the vesicular systems of the cell cytoplasm. The nucleus (blue image, stained with DAPI) appears to come in close contact with several of the protrusions from this system. Although identity of the globular/tubular system responsible for the observed Gln3 distribution in the cytoplasm is not yet clear, the images are consistent with the suggestion that a significant fraction of Gln3 may not be not free in the cytoplasm but rather localizes to a subcellular network during NCR.
DISCUSSION
We initiated this work to resolve an apparent paradox, i.e. Gln3 is localized to the nucleus in both carbon and nitrogen starvation (31). During carbon starvation, nitrogen is in excess. That Gln3 is nuclear under conditions of nitrogen excess prompted us to ask why Gln3 was cytoplasmic under some conditions of nitrogen excess but not others.
We demonstrate that it’s not carbon starvation per se that results in Gln3 being localized to the nucleus but the indirect effects of carbon starvation on nitrogen metabolism and hence on NCR. The principal finding supporting this conclusion is that nuclear localization of Gln3 observed upon transferring cells from SC to carbon-free SC medium can be completely prevented if glutamine is added to the latter medium. This result was confirmed using more defined media containing single sources of nitrogen.
The time course of Gln3 localization points to a possible explanation of the paradox. Gln3 moves more quickly to the nucleus and in a greater percentage of the cells when cells are transferred from glutamine to proline compared with transfer from glucose-ammonia to glucose-free-ammonia medium. For ammonia to be assimilated into nitrogenous compounds and thereby exert NCR on GATA-mediated transcription, it must first be converted to glutamate and glutamine. The carbon skeletons for these amino acids derive from glucose. As glucose starvation worsens, carbon-containing compounds (including α-ketoglutarate) become increasingly limiting, thereby slowing ammonia assimilation with concomitant loss of NCR and movement of Gln3 into the nucleus.
This is not the first instance in which nutrient starvation or rapamycin-induced alteration of metabolite pools has generated indirect effects on nitrogen catabolic gene expression. Tate et al. (24) recently showed that Mks1, rather than being an important component of the Tor1/2 signal transduction pathway that positively regulates nuclear localization of Gln3 and Rtg1/3, is a negative regulator of Rtg1/3 function that only indirectly influences NCR-sensitive transcription.
The time course of Gln3 movement from cytoplasm to nucleus is also revealing from the standpoint of rapamycin-“induced” GATA-factor-mediated gene expression. It is currently accepted that Tor1/2, whose functions are inhibited by rapamycin, are situated downstream of the metabolite effector of NCR in the nutrient limitation signal transduction cascade (1–6, 14–17). Yet Gln3 movement into the nucleus following transfer of cells from glutamine to proline medium is faster, more complete, and longer lasting than in cells treated with rapamycin. Furthermore, the time course profiles seen with ammonia and glutamine following rapamycin addition are not the same. With glutamine, Gln3 movement into the nucleus occurs more slowly, in fewer cells, and remains nuclear for a shorter time than with ammonia. In other words, the effect of rapamycin on Gln3 localization, as with carbon starvation, depends on the nitrogen source. If Tor1/2 are indeed situated downstream of the nitrogen signal, the onset of Gln3 movement into the nucleus should occur equally or more quickly with rapamycin treatment and be independent of the nitrogen source. An important caveat, we can’t rigorously evaluate for lack of radioactive rapamycin, is the possibility that rapamycin uptake is strongly influenced by the nitrogen source. This, however, is unlikely in view of the speed with which rapamycin is reported to work and the fact that both ammonia and glutamine are good nitrogen sources in the strains we used (14). Therefore, incongruities exist between expected and observed time courses of rapamycin response when predictions are predicated on current models of Tor1/2 signal transduction.
The finding that Gln3 localization may not be uniformly distributed in the cytoplasm suggests an alternative view of the Tor1/2 signal transduction cascade. We suggest that movement of Gln3 and Gat1 into and out of the nucleus, like that of Rtg3, another phosphorylated transcription factor reported to be regulated by the Tor1/2 signal transduction pathway, involves the participation of a general protein trafficking pathway. By our hypothesis, Tor1/2 may not be situated at the top of the signal transduction pathway regulating NCR-sensitive and retrograde gene expression but are participants in or regulators of the general protein trafficking pathway through which Gln3/Gat1, Rtg1/3, and a growing list of other unrelated transcription factors enter and exit the nucleus. Just as Gln3 uses a generic nuclear import (Srp1) and export (Crm1) proteins, which are also used by other unrelated transcription factors, so too Gln3 may use a general protein trafficking system whose components or regulators include Tor1/2. We suggest the non-uniform distribution of Gln3 in the cytoplasm of glutamine-grown cells may derive from its sequestration in or localization to components of that general pathway.
Our challenge now is to determine whether the explanations suggested above will stand the test of experimental investigation, and if they do, to determine how they account for the large number of cellular processes that are influenced, directly or indirectly, by the Tor1/2 proteins and their inhibition by rapamycin. Furthermore, if verified experimentally, this interpretation of our data also provides a novel way of viewing the nucleocytoplasmic shuttling of proteins into and out of the nucleus.
Acknowledgments
We thank Dr. Michael Hall for very generously providing strains, Dr. Ted Powers for sharing a preprint of his work, Tim Higgins for preparing the artwork, and the University of Tennessee Yeast Group for suggestions to improve the manuscript.
Footnotes
This work was supported by National Institutes of Health Grant GM-35642.
The abbreviations used are: NCR, nitrogen catabolite repression; DAPI, 4′,6-diamidino-2-phenylindole.
REFERENCES
- 1.Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. Mol. Biol. Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dennis PB, Fumagalli S, Thomas G. Cur. Opin. Genet. Develop. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- 3.Schmelzle T, Hall MN. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 4.Rohde J, Heitman J, Cardenas ME. J. Biol. Chem. 2001;276:9583–9586. doi: 10.1074/jbc.R000034200. [DOI] [PubMed] [Google Scholar]
- 5.Cooper TG. FEMS Microbiol. Rev. 2002;737:1–16. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shamji AF, Kuruvilla FG, Schreiber SL. Cur. Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- 7.Edskes HK, Hanover JA, Wickner RB. Genetics. 1999;153:585–594. doi: 10.1093/genetics/153.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pierce MM, Maddelein ML, Roberts BT, Wickner RB. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13213–13218. doi: 10.1073/pnas.181486098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komeili A, Wedaman KP, O’Shea EK, Powers T. J. Cell Biol. 2000;151:863–878. doi: 10.1083/jcb.151.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman-Bang J. Mol. Biotechnol. 1999;12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- 11.ter Schure EG, van Riel NA, Verrips CT. FEMS Microbiol. Rev. 2000;24:67–83. doi: 10.1111/j.1574-6976.2000.tb00533.x. [DOI] [PubMed] [Google Scholar]
- 12.Cooper TG. In: Mycota III. Marzluf G, Bambrl R, editors. Berlin, Heidelberg: Springer Verlag; 1996. pp. 139–169. [Google Scholar]
- 13.Cox KH, Rai R, Distler M, Daugherty JR, Coffman JA, Cooper TG. J. Biol. Chem. 2000;275:17611–17618. doi: 10.1074/jbc.M001648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck T, Hall MN. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 15.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwick JS, Kuruvilla FG, Tong JF, Shamji AF, Schreiber SL. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan T-F, Zheng XFS. J. Biol. Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- 18.Kunz J, Schneider U, Howald I, Schmidt A, Hall MN. J. Biol. Chem. 2000;275:37011–37020. doi: 10.1074/jbc.M007296200. [DOI] [PubMed] [Google Scholar]
- 19.Bossinger J, Cooper TG. J. Bacteriol. 1976;126:198–204. doi: 10.1128/jb.126.1.198-204.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvalho J, Bertram PG, Wente SR, Zheng XFS. J. Biol. Chem. 2001;276:25359–25365. doi: 10.1074/jbc.M103050200. [DOI] [PubMed] [Google Scholar]
- 21.Di Como CJ, Arndt KT. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y, Broach JR. EMBO. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. Mol. Cell. 2001;8:1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- 24.Tate JJ, Cox KH, Rai R, Cooper TG. J. Biol. Chem. 2002;277:20477–20482. doi: 10.1074/jbc.M200962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smart WC, Coffman JA, Cooper TG. Mol. Cell. Biol. 1996;16:5876–5887. doi: 10.1128/mcb.16.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekito T, Liu Z, Thornton J, Butow RA. Mol. Biol. Cell. 2002;13:795–804. doi: 10.1091/mbc.01-09-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epstein CB, Waddle JA, Hale W, 4th, Dave V, Thornton J, Macatee TL, Garner HR, Butow RA. Mol. Biol. Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekito T, Thornton J, Butow RA. Mol. Biol. Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Butow RA. Mol. Cell. Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao X, Butow RA. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- 31.Bertram PG, Choi JH, Carvalho J, Chan TF, Ai W, Zheng XF. Mol. Cell. Biol. 2002;22:1246–1252. doi: 10.1128/MCB.22.4.1246-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz K, Richards K, Botstein D. Mol. Biol. Cell. 1997;8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dilova I, Chen C-Y, Powers T. Curr. Biol. 2002;12:389–395. doi: 10.1016/s0960-9822(02)00677-2. [DOI] [PubMed] [Google Scholar]