Abstract
NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2), encoding a subunit of mitochondrial complex I, is a candidate gene for several neuronal diseases; schizophrenia, bipolar disorder and Parkinson disease (PD). We screened the entire coding region of NDUFV2 in 33 familial PD patients of North African Arab-Berber ethnicity in which all known genetic forms of PD had been excluded. We detected one novel substitution p.K209R (c.626A>G) in one PD. Segregation analysis within the family is inconclusive due to small sample size, but consistent with autosomal dominant mode of inheritance. Subsequent screening of this mutation in ethnically-matched sporadic PD patients (n=238) and controls (n=371) identified p.K209R in one additional patient. The clinical features of the mutation carriers revealed a mild form of parkinsonism with a prognosis similar to idiopathic PD. Our findings suggest further studies addressing the role of NDUFV2 variation in PD may be warranted.
Keywords: Parkinson disease, NDUFV2, mutation, genetics
Introduction
Parkinson disease (PD) is the second most common neurodegenerative disorder in elderly populations. Studies to date have identified several genetic causes of PD and early-onset parkinsonism including; alpha-synuclein (SNCA), leucine-rich repeat kinase 2 (LRRK2), parkin (PRKN), PTEN-induced kinase 1 (PINK1), and oncogene DJ1 [1]. We have previously reported a high frequency of LRRK2 c.6055G>A (p.G2019S) (30%) and PINK1 homozygote mutation carriers (15%) within PD patients of Arab-Berber ethnicity in North Africa [2, 3]. In addition a small percentage (4%) of families with PD were explained by homozygote and compound heterozygote mutations in PRKN [4]. However, 33 familial PD probands (51%) did not present mutations in known PD genes. The NADH dehydrogenase ubiquinone flavoprotein 2 (NDUFV2; NIM 600532) located on chromosome 18p11.31-p11.2 has been nominated as a causative gene in PD, as well as other neurological diseases including schizophrenia, and bipolar disorder [5–7]. In addition, association studies with common variants in NDUFV2 have shown positive results in Asian populations, although they did not replicate in a Caucasian series [5, 8, 9]. NDUFV2 is also a good biological candidate as it encodes a 24-kDa subunit of mitochondrial complex I and mitochondrial dysfunction has been associated with several neurodegenerative disorders, including PD [10].
Herein, we describe the identification of a novel mutation Lysine 209 Arginine (p.K209R) c.626A>G in NDUFV2 in one familial PD proband and one sporadic PD patient. Segregation analysis supports pathogenicity, although further confirmation is needed.
Material and methods
Subjects
This study genetically characterized NDUFV2 in a total of 33 familial probands with PD from the Institut National de Neurologie, Tunis. This group presented a mean age at disease onset of 53.0 ± 15.2 years (mean ± S.D) and a 12:21 male to female ratio. A previously described ethnically matched case-control series consisting of 238 sporadic PD cases and 371 controls was genotyped for the novel variant herein described [2]. The Institut National de Neurologie, Tunis provides a specialized neurological service to the entire country of Tunisia [11]. The site obtained local ethics committee approval before beginning recruitment (06-004383). Informed written or proxy consent for the study was given by all subjects. Individuals were diagnosed as “affected” if they satisfied the United Kingdom PD Society Brain Bank (UKPDS) criteria [12].
Sequencing and genotyping
Genomic DNA was extracted from peripheral blood lymphocytes using standard protocols. Primer pairs for NDUFV2 were used to sequence all 8 exons and exon-intron boundaries. PCR products were purified using Agencourt bead technology (Beverly, MA) with Biomek FX automation (Beckman Coulter, Fullerton, CA). Genotyping of the p.K209R was performed on a Sequenom MassArray iPLEX platform (San Diego, CA, US). All primer sequences are available on request.
Results
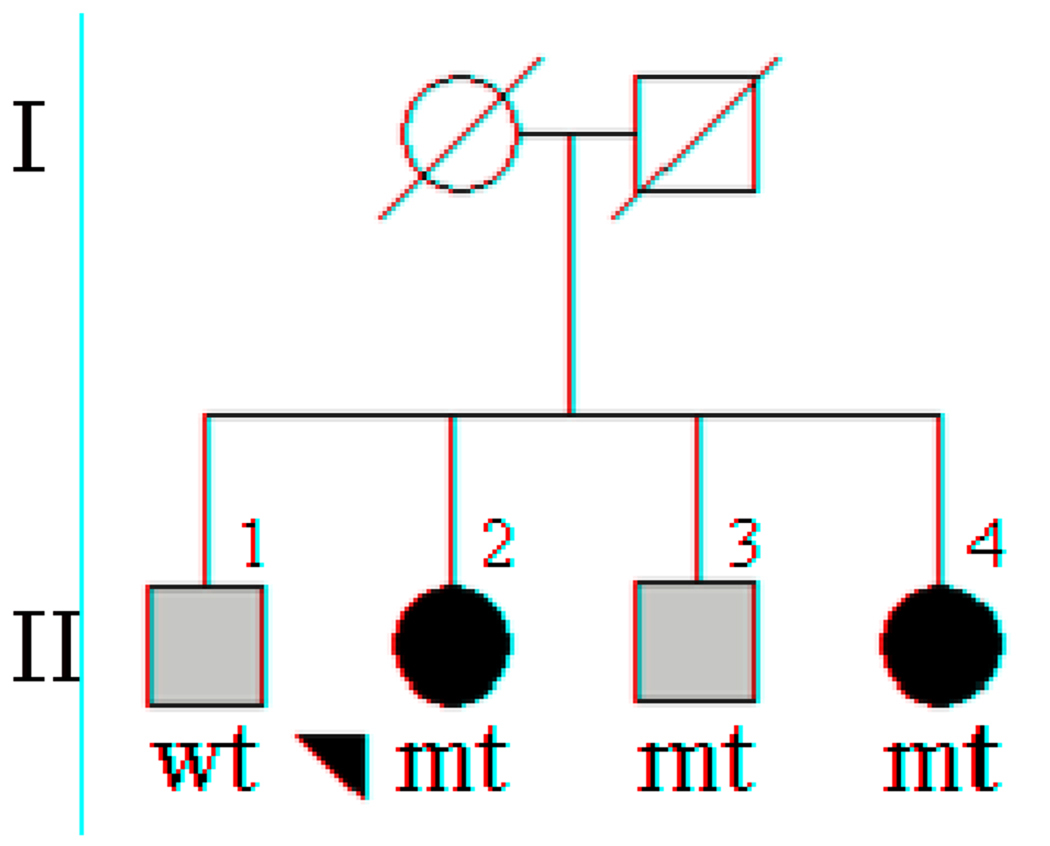
Sequencing analysis of the NDUFV2 in familial probands identified one novel missense mutation leading to a lysine to arginine substitution at aminoacid position 209 (p.K209R, c.626A>G). Genotyping of additional family members from this pedigree revealed a total of three mutation carriers, two diagnosed with PD and one with essential tremor (ET). One additional family member not carrying the mutation had also been diagnosed with ET (Figure 1). The clinical features of patient II-2 consisted of an age-at-onset of 57 years, with a disease duration of 13 years, Hoehn and Yahr stage I and MMSE 20/30. Patient II-4 presented disease with and age-at-onset of 66 years, and 17 years of disease duration, Hoehn and Yahr stage I and MMSE 22/30. Both presented with a mild form of parkinsonism and a typical clinical course. The last patient harboring this mutation (II-3) was diagnosed with ET and did not present any signs of parkinsonism at 75 years of age. Subsequent screening of 238 sporadic PD cases and 371 controls from the same ethnicity and geographical location identified one additional male patient diagnosed with PD at 60 years of age, harboring the p.K209R mutation; none of the controls were positive for this mutation.
Figure 1. Segregation analysis of NDUFV2 p.K209R.
A) Males are represented by squares, female by circles, and the probands is arrowed. Individuals diagnosed with Parkinson disease are indicated with black filled symbols, grey filled symbols indicate patients diagnosed with essential tremor. NDUFV2 p.K209R mutation carriers are indicated with mt, non carriers with wt.
Discussion
Association studies of NDUFV2 gene have suggested it may be a genetic factor in a number of disorders including PD, bipolar disorder or schizophrenia. Hattori et al. first reported an association with the polymorphism p.A29V and PD in the Japanese population [5], however they did not identify any pathogenic mutations. In this study we evaluated the presence of novel mutations in 33 PD probands of Arab-Berber ethnicity. Sequencing the entire coding region of NDUFV2 identified one novel lysine to arginine substitution (p.K209R), later identified in one additional sporadic patient with PD. Segregation analysis of p.K209R is consistent with pathogenicity, but inconclusive due to the small size of the pedigree. The existence of a mutation carrier (II-3) diagnosed with ET rather than PD, suggests that if pathogenic, the p.K209R could result in different disease phenotypes. Although this amino acid position appears to be highly conserved in many species, the mutant amino acid is the wild-type residue in rat (Figure 2). Although bioinformatics analysis using PolyPhen, and the presence of arginine in the wild-type protein sequence of rat may be taken as indicative of non-pathogenicity; equivalent data exists for the confirmed pathogenic α-synuclein p.A53T mutation [13].
Figure 2. Conservation of NDUFV2 p.K209R.
Protein homologues were aligned using ClustalW; p.K209R position is highlighted in black, non-conserved amino acid positions are highlighted in grey. GeneBank accession numbers: Homo sapiens, NP_066552; Pan troglodytes, NP_001065254; Macaca mulatta, XP_001099724; Mus musculus, NP_082664; Rattus norvegicus, NP_112326; Canis familiaris, XP_537328; Bos Taurus, NP_776990; Gallus gallus, XP_001232141; Ornithorhynchus anatinus, XP_001507932.
NDUFV2 is one of the many components of the mitochondrial oxidative phosphorylation pathway, variation in the gene leading to altered energy production and mitochondrial function has been postulated as a factor to alter the risk of developing PD [5]. In this study we genetically characterized NDUFV2 in an ethnically distinct population which led to the identification of a novel mutation in one kindred and one sporadic PD patient. Further studies characterizing NDUFV2 in larger series and ethnically distinct populations are necessary to evaluate the role of this gene in PD, and discern whether p.K209R is a rare polymorphism in the Tunisian population or a global risk factor for PD.
Acknowledgement
The authors wish to thank the patients and families who participated in the study. This work was supported by the Morris K. Udall Center at Mayo Clinic Jacksonville, National Institute of Neurological Disorders and Stroke P50 NS40256 and R21 NS64885 and the Michael J Fox Foundation for Parkinson’s Research. KN was supported by an Eli-Lilly scholarship and Herb Geist gift for Lewy body research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no financial or other conflict of interests.
References
Additional reference added at the request of the reviewers
- 1.Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nature reviews. 2006 Apr;7(4):306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 2.Hulihan MM, Ishihara-Paul L, Kachergus J, Warren L, Amouri R, Elango R, et al. LRRK2 Gly2019Ser penetrance in Arab-Berber patients from Tunisia: a case-control genetic study. Lancet neurology. 2008 Jul;7(7):591–594. doi: 10.1016/S1474-4422(08)70116-9. [DOI] [PubMed] [Google Scholar]
- 3.Ishihara-Paul L, Hulihan MM, Kachergus J, Upmanyu R, Warren L, Amouri R, et al. PINK1 mutations and parkinsonism. Neurology. 2008 Sep 16;71(12):896–902. doi: 10.1212/01.wnl.0000323812.40708.1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishioka K, Kefi M, Jasinska-Myga B, Wider C, Vilarino-Guell C, Ross OA, et al. A comparative study of LRRK2, PINK1 and genetically undefined familial Parkinson disease. Journal of neurology, neurosurgery, and psychiatry. 2010 Apr;81(4):391–395. doi: 10.1136/jnnp.2009.185231. [DOI] [PubMed] [Google Scholar]
- 5.Hattori N, Yoshino H, Tanaka M, Suzuki H, Mizuno Y. Genotype in the 24-kDa subunit gene (NDUFV2) of mitochondrial complex I and susceptibility to Parkinson disease. Genomics. 1998 Apr 1;49(1):52–58. doi: 10.1006/geno.1997.5192. [DOI] [PubMed] [Google Scholar]
- 6.Washizuka S, Kakiuchi C, Mori K, Kunugi H, Tajima O, Akiyama T, et al. Association of mitochondrial complex I subunit gene NDUFV2 at 18p11 with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2003 Jul 1;120B(1):72–78. doi: 10.1002/ajmg.b.20041. [DOI] [PubMed] [Google Scholar]
- 7.Washizuka S, Kametani M, Sasaki T, Tochigi M, Umekage T, Kohda K, et al. Association of mitochondrial complex I subunit gene NDUFV2 at 18p11 with schizophrenia in the Japanese population. Am J Med Genet B Neuropsychiatr Genet. 2006 Apr 5;141B(3):301–304. doi: 10.1002/ajmg.b.30285. [DOI] [PubMed] [Google Scholar]
- 8.Mizuta I, Tsunoda T, Satake W, Nakabayashi Y, Watanabe M, Takeda A, et al. Calbindin 1, fibroblast growth factor 20, and alpha-synuclein in sporadic Parkinson's disease. Human genetics. 2008 Aug;124(1):89–94. doi: 10.1007/s00439-008-0525-5. [DOI] [PubMed] [Google Scholar]
- 9.Swerdlow RH, Weaver B, Grawey A, Wenger C, Freed E, Worrall BB. Complex I polymorphisms, bigenomic heterogeneity, and family history in Virginians with Parkinson's disease. Journal of the neurological sciences. 2006 Sep 25;247(2):224–230. doi: 10.1016/j.jns.2006.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henchcliffe C, Beal MF. Mitochondrial biology and oxidative stress in Parkinson disease pathogenesis. Nature clinical practice. 2008 Nov;4(11):600–609. doi: 10.1038/ncpneuro0924. [DOI] [PubMed] [Google Scholar]
- 11.Ishihara L, Gibson RA, Warren L, Amouri R, Lyons K, Wielinski C, et al. Screening for Lrrk2 G2019S and clinical comparison of Tunisian and North American Caucasian Parkinson's disease families. Mov Disord. 2007 Jan;22(1):55–61. doi: 10.1002/mds.21180. [DOI] [PubMed] [Google Scholar]
- 12.Hughes AJ, Ben-Shlomo Y, Daniel SE, Lees AJ. What features improve the accuracy of clinical diagnosis in Parkinson's disease: a clinicopathologic study. Neurology. 1992 Jun;42(6):1142–1146. doi: 10.1212/wnl.42.6.1142. [DOI] [PubMed] [Google Scholar]
- 13.Polymeropoulos MH, Lavedan C, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997 Jun;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]