Abstract
GATA family activators (Gln3p and Gat1p) and repressors (Dal80p and Deh1p) regulate nitrogen catabolite repression (NCR)-sensitive transcription in Saccharomyces cerevisiae presumably via their competitive binding to the GATA sequences upstream of NCR-sensitive genes. Ure2p, which is not a GATA family member, inhibits Gln3p/Gat1p from functioning in the presence of good nitrogen sources. We show that NCR-sensitive DAL80 transcription can be influenced by the relative levels of GAT1 and URE2 expression. NCR, normally observed with ammonia or glutamine, is severely diminished when Gat1p is overproduced, and this inhibition is overcome by simultaneously increasing URE2 expression. Further, overproduction of Ure2p nearly eliminates NCR-sensitive transcription under derepressive growth conditions, i.e. with proline as the sole nitrogen source. Enhanced green fluorescent protein-Gat1p is nuclear when Gat1p-dependent transcription is high and cytoplasmic when it is inhibited by overproduction of Ure2p.
Two complementary modes of regulation are employed by Saccharomyces cerevisiae to achieve selective utilization of “good” nitrogen sources (ammonia, glutamine or asparagine) in preference to “poor” ones (proline), a process designated nitrogen catabolite repression (NCR)1 (1–3). Both depend upon NCR-sensitive transcription being mediated through UASNTR elements, containing GATAA sequences at their cores (4), and four members of the GATA-binding family of transcription factors, Gln3p, Gat1p/Nil1p, Dal80p, and Deh1p/Gzf3p/Nil2p (3, 5–12). The preeminent mode of regulation is that exerted by Ure2p, which inhibits the ability of the GATA activators, Gln3p and Gat1p, to function during times of nitrogen excess (3, 13, 14); ure2 mutants become insensitive to NCR (15, 16). The second mode, whose function we consider to be one of fine tuning, is proposed to depend upon the GATA repressors Dal80p and Deh1p competing with Gln3p and Gat1p for binding to their DNA targets (13, 17).
Success of the regulatory mechanism hypothesized above requires the relative amounts of Gln3p, Gat1p, Dal80-p, and Deh1p to be rigorously and finely controlled. Such fine control has been proposed to be achieved through autogenous and cross-regulation of GATA factor production (6, 11), a view supported by (i) the presence of multiple UASNTR elements situated upstream of the DAL80, DEH1, and GAT1 genes and (ii) effects of gln3Δ, gat1Δ, dal80∷hisG, and deh1Δ mutations on expression of the three genes (3, 11, 12). Although the model fits existing data, its central tenant has not been tested experimentally, i.e. that expression of one GATA factor, GAT1, influences expression of another, DAL80.
The purpose of this work was to test the above proposal by determining the effects of varying GAT1 expression on that of DAL80. The data obtained demonstrate that DAL80 expression is tightly linked to that of GAT1 and furthermore that NCR can be tightly linked to the relative production of Gat1p and Ure2p, i.e. that changing the relative levels of GAT1 and URE2 expression concomitantly changes the sensitivity of DAL80 expression to NCR. We also find that the intracellular localization of EGFP-Gat1p is influenced by the levels of GAT1 and URE2 expression. When Gat1p-dependent transcription is high, EGFP-Gat1p is predominantly nuclear, whereas when it is inhibited by overproduction of Ure2p, Gat1p is cytoplasmic.
MATERIALS AND METHODS
Strains and Culture Conditions
The strains used are: TCY1 (Matα lys2 ura3), TCY5 (Matα lys2 ura3 trp1∷hisG), TCY29 (Matα lys2 ura3 trp1∷hisGdal80∷hisG), TCY46 (Matα lys2 ura3 trp1∷hisG GAL1,10-GAT1), TCY48 (Matα lys2 ura3 trp1∷hisG dal80∷hisG GAL1,10-GAT1), TCY57 (Matα lys2 ura3 trp1∷hisG leu2∷hisG GAL1,10-URE2), TCY60 (Matα lys2 ura3 trp1∷hisG leu2∷hisG dal80∷hisG GAL1,10-GAT1 GAL1,10-URE2), RRJ715 (Matα, lys2, ura3, his3, gat1Δ∷hisG), RTC57 (Matα, lys2,trp1,his3,ura3,URE2-GAL1-10), and GYC86 (Mata, his3,leu2,trp1,ura3/Matα, his3,leu2,trp1,ura3). Minimal medium was 0.17% YNB without amino acids or (NH4)2SO4 (Difco) with indicated carbon and nitrogen sources and auxotrophic requirements.
Northern Blot and β-Galactosidase Analyses
Yeast cultures for RNA analysis (see Figs. 2 and 4A) were grown in minimal proline galactose (2%) medium with glucose added to the final concentration indicated in the figure. For Fig. 4C, RRJ715, RRJ715/pRA27, and RTCY57/pRA27 were grown to A600 nm = 0.3–0.4 in Wickerham's minimal ammonia medium. RTCY57/pRA27 was washed and transferred to Wickerham's galactose medium for induction of URE2 (3 h). Total RNA was isolated (18, 19), resolved on formaldehyde-agarose gels, and transferred to Gene Screen Plus nylon 66 membranes (NEN Life Science Products) using 6× SSC as the buffer. Random priming was used to label PCR product or DNA fragment probes. Hybridization was in 50% formamide, 1 M NaCl, 1% SDS (42 °C, 17–20 h). Blots were washed twice each in 2× SSC at 25 °C, 2× SSC + 1% SDS at 65 °C and 0.1× SSC at 25 °C or as described (19). β-Galactosidase assays were based on 25 ml of culture. For GAL1,10-GAT1-lacZ or GAL1,10-URE2-lacZ expression, freshly inoculated cultures (glucose ammonia medium) were grown overnight to a cell density of A600 nm = 0.4–0.8.
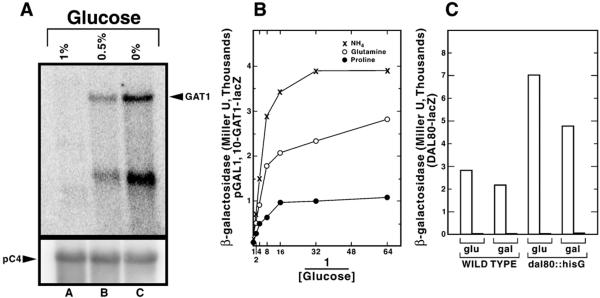
Fig. 2. GAL1,10-GAT1 expression profile measured by Northern blot analysis (A) and by β-galactosidase production supported by GAL1,10-GAT1-lacZ fusion plasmid (B).
Total RNA (20 μgm/lane) was prepared from GAL1,10-GAT1 strain TCY46 grown in minimal galactose proline medium plus the indicated amount of added glucose. [32P]dCTP-labeled probes were synthesized from the internal 1-kb SphI-StuI fragment of GAT1 and pC4 (33). β-Galactosidase production from pTSC666 (in strain TCY48) was measured following addition of various amounts of glucose (final concentration was 0, 0.016, 0.032, 0.063, 0.125, 0.25, 0.5 or 1%) to minimal galactose (2%) medium containing proline (●), glutamine (◯), or ammonium sulfate (×) as nitrogen source. C, DAL80-lacZ expression in wild type and dal80 strains grown in different carbon and nitrogen sources. β-Galactosidase activity supported by plasmid pTSC572 was assayed in wild type (TCY5) or dal80∷hisG (TCY29) strains. Open and filled bars denote proline and asparagine, respectively, as the nitrogen source. Glucose (glu) or galactose (gal) was used as the sole carbon source.
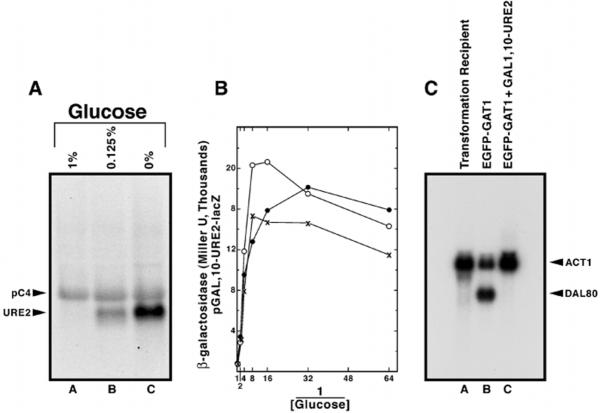
Fig. 4. GAL1,10-URE2 expression profile measured by Northern blot analysis (A) and by β-galactosidase production supported by GAL1,10-URE2-lacZ fusion (B).
Details of the Northern blot analysis were as in Fig. 2 using strain TCY57 was grown in galactose-proline minimal medium with the indicated amount of glucose added. 32P-Labeled probes were synthesized from the 0.45-kb BsrGI-ApaI fragment from URE2 and yeast pC4 β-Galactosidase production from pTSC668 (in strain TCY1) was measured as in Fig. 2. C, Northern blot analysis of total RNA (9 μg/lane) from strain RRJ715, untransformed (lane A) or transformed pRA27 (lane B), and strain RTCY57 transformed with pRA27 (lane C). Strains were grown in minimal glucose ammonia medium, and RTCY57 was additionally transferred to galactose ammonia medium and induced for 3 h prior to assay. Full-length DAL80 and ACT1 probes were used.
GAL1,10-GAT1, and GAL1,10-URE2 Strains
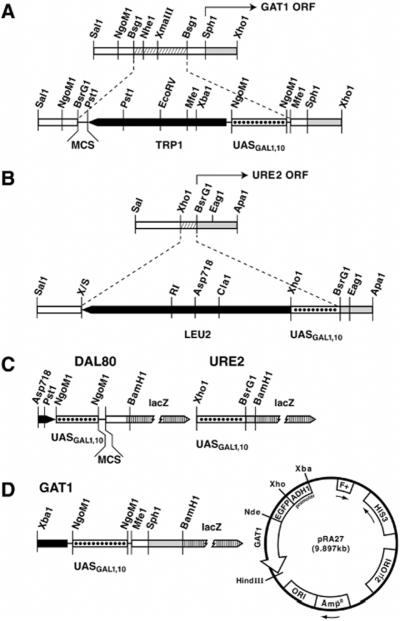
GAL1,10-GAT1 and GAL1,10-URE2 fusion fragments were used to replace genomic GAT1 and URE2. For GAT1, the 0.8-kb SalI-XhoI promoter fragment was cloned into pBSKS(+) (Stratagene) yielding pTSC613. The 0.8-kb ApaI-SalI GAT1 fragment of pTSC613 was cloned into pLitmus38 yielding pTSC614. pTSC614 was digested with BsgI, and the backbone was isolated, deleting the 0.3-kb GAT1 BsgI fragment containing clustered GATAs. A blunt-ended polylinker (BsrGI to MfeI) from pLitmus38 was cloned into the BsgI site of the plasmid backbone, following Klenow treatment of the linearized DNA, yielding pTSC620. A Klenow-treated, EcoRI TRP1 fragment from YRp7 was digested with PstI and cloned into pTSC620 digested with HindIII, treated with Klenow, and then digested with PstI, yielding pTSC627. The GAL1,10 promoter fragment was generated by Pwo PCR with oligonucleotide primers, 5′-GATCGCCGGCCCTTCTCTTTGGAACTTTCAGTAAT-3′ and 5′-GATCGCCGGCTCGCTGATTAATTACCCAGAA-3′, and pEG(KG) (20) as template. The PCR product was digested with NgoMI and cloned into pTSC627 partially digested with NgoMI. Restriction site analysis and DNA sequencing (GAL1,10-GAT1 junction) confirmed the resulting construct, pTSC645. The SalI-XhoI fragment of pTSC645 was purified and used to transform relevant host strains.
The 1.1-kb SalI-XhoI URE2 fragment from pRD17 (21) was cloned into pLitmus38, yielding pTSC638. The BsrG1-XhoI fragment of pTSC638 (containing 170 bp of the URE2 promoter) was deleted and replaced by a PCR-generated GAL1,10 promoter fragment (primers: 5′-CCGCTCGAGCCGAAGGAAGACTCTCCTCCG-3′ and 5′-CGCTCTACAGACGTTAAAGTATAGAGGTAT-3′, template, pEG(KG) (20)). Because the URE2 promoter lacks a TATA box, the above PCR product included the GAL1,10 TATA box and transcription start site(s) (22). The BsrGI-XhoI GAL1,10 promoter fragment was ligated into the backbone of pTSC638, yielding pTSC650. A SalI-XhoI LEU2 fragment (2) was then cloned into the unique XhoI site of pTSC650 yielding pTSC657. The SalI-XhoI and SalI-ApaI fragments of pTSC645 and pTSC657, respectively, were purified and used to transform the relevant strains. Integrants were verified by Southern blot analysis using multiple restriction endonucleases (data not shown).
GAL1,10-GAT1-lacZ, GAL1,10-URE2-lacZ, and GAL1,10-DAL80-lacZ Plasmids
A BamHI oligonucleotide (10-mer) was ligated into GAT1 pTSC645 digested with XhoI and treated with Klenow polymerase. The 0.9-kb XbaI-BamHI fragment containing the 5′ end of TRP1, GAL1,10 UAS and the 5′ end of GAT1 (to position +193) was isolated and ligated into plasmid pHP41 (23) yielding pTSC666 (Fig. 1). For GAL1,10-URE2, a BamHI oligonucleotide (10-mer) was ligated onto EagI-digested, Klenow-treated pTSC657. The 0.7-kb XhoI-BamHI fragment (containing the GAL1,10 UAS and the 5′ end of URE2 to position +103) was ligated into pHP41 treated with SalI and BamHI, yielding pTSC668 (Fig. 1). For GAL1,10-DAL80, the 0.9-kb Asp718-PvuII fragment from pTSC674 was cloned into Litmus 28 to yield pTSC678. A BamHI oligonucleotide (10-mer) was ligated onto the blunt ends of pTSC678 digested with PvuII. The 0.9-kb XbaI-BamHI fragment from this plasmid (containing the 3′ end of HIS3, GAL1,10 UAS and DAL80 5′ end to position +46) was isolated and ligated into pHP41, yielding pTSC679 (Fig. 1).
Fig. 1. Fusion plasmids and genes used in this work.
The native GAT1 and URE2 genomic regions are shown before and after promoter substitutions have occurred. Open bars, native sequences; hatched bars, deleted sequences; filled bars, selected gene; dotted bars, GAL1,10 regulatory sequences; shaded bars, open reading frames; vertically striped bars, lacZ. GAT1 and URE2 open reading frames begin at the SphI and near the BsrG1 sites, respectively. X/S is a SalI-XhoI junction.
EGFP-GAT1 pRA27 Was Constructed by Cloning a 1.7-kb NdeI
HindIII GAT1 fragment from pRR351 into pRMS2 (pADH1-EGFP, a 2 μ plasmid carrying a HIS3 marker) allowing constitutive overexpression of EGFP-GAT1) (24). All fusion junctions were sequenced to ensure they were in frame.
Fluorescence Microscopy
Cytological experiments were performed as described elsewhere (25). The distribution of fluorescent material was the same whether cells were grown in Wickerham's or YNB minimal medium.
RESULTS
GAT1 Expression under Control of the GAL1,10 Promoter
Dissecting the regulatory relationships of the GATA transcription factors is hampered by their extensive autogenous- and cross-regulation (11, 12). To overcome this, we replaced the genomic GAT1 promoter with a GAL1,10 fragment (UASGAL), thereby removing GAT1 expression from nitrogen and GATA factor control and placing it under galactose regulation. Because our experiments depended upon the transcription profile of the GAL1,10-GAT1 construct, we measured steady-state levels of GAT1-specific mRNA following three different glucose additions to the medium. GAT1 mRNA increased as glucose decreased (Fig. 2A). Next, we substituted the lacZ open reading frame for the GAT1 open reading frame in CEN-based GAL1,10-GAT1 pTSC666, measured β-galactosidase production under conditions similar to those in Fig. 2A, and found β-galactosidase activity decreased as the amount of glucose added to the medium increased (Fig. 2B). The inverse of the glucose addition was plotted on the abscissa throughout this work to facilitate visualization of the effects of GAL1,10 driven GAT1 or URE2 expression. As expected, GAL1,10-GAT1 expression is NCR-insensitive (Fig. 2B). In fact, GAL1,10-GAT1-lacZ expression is 2- and 4-fold higher, respectively, with glutamine and ammonia compared with proline. This effect probably derives from ammonia and glutamine being significantly better nitrogen sources than proline and hence supporting higher synthetic capacities (26).
DAL80 Expression Is Strongly Influenced by the Level of GAT1 Expression
A predicted consequence of the cross-regulation model of GATA factor control (11) is that the level of GAT1 expression influences DAL80 expression. We tested this prediction using a DAL80-lacZ reporter. We recognized from the outset that interpretation of our results could be compromised by: (i) down-regulation of DAL80-lacZ expression by Dal80p derived from the genomic DAL80 gene responding to increased Gat1p (6) and (ii) secondary effects of growing cells with galactose as carbon source. We eliminated the first complication by performing our experiments in dal80∷hisG disruption strain TCY48. To evaluate the second potential complication, we compared β-galactosidase production in wild type (TCY5) and dal80∷hisG disruption (TCY29) strains transformed using DAL80-lacZ pTSC572. The only difference in the DAL80-lacZ expression profile was a 20–30% decrease with galactose as carbon source (Fig. 2C), a relatively modest effect relative to that of NCR.
With the experimental system established, we determined the effect of varying GAT1 expression on DAL80-lacZ expression in TCY48. As the glucose concentration decreased (GAT1 expression increased), DAL80-lacZ expression increased (Fig. 3). In addition to the DAL80 expression expected when proline was provided as nitrogen source, increasing GAT1 expression unexpectedly increased DAL80-lacZ expression with ammonia and glutamine as nitrogen sources as well (Fig. 3). To place this result in perspective, DAL80-lacZ expression with ammonia as nitrogen source in Fig. 3 (3,969 units) is about 300-fold greater than normal (glutamine, 12 units) (11). In short, increasing GAT1 expression overcomes the repressive effects of preferred nitrogen sources on DAL80-lacZ expression.
Fig. 3. DAL80-lacZ expression observed at various levels of GAT1 expression.
β-Galactosidase production, supported by DAL80-lacZ pTSC572, was measured in a transformant of GAL1,10-GAT1 strain TCY48 growing in minimal medium containing proline (◯), ammonium sulfate (×), or glutamine (▵) as sole nitrogen source. GAT1 EXP. indicates GAT1 expression.
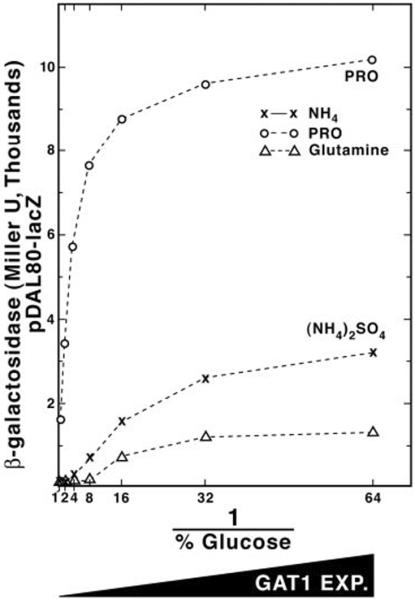
Increased URE2 Expression Suppresses the Effects of High Level GAT1 Expression
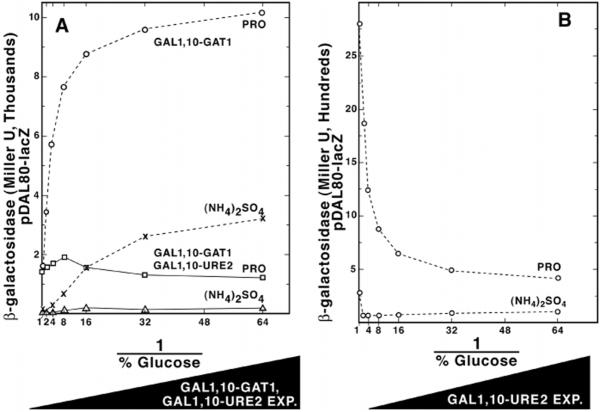
The current model of NCR proposes that the binding of Ure2p to Gln3p inhibits its ability to function as a transcriptional activator (27). Because Ure2p also regulates Gat1p-dependent NCR-sensitive transcription (9), we determined whether increasing URE2 and GAT1 expression simultaneously would inhibit the ability of GAT1 to suppress the NCR sensitivity of DAL80-lacZ expression. Because the GAL1,10-URE2 construct could not be made in exactly the same way as GAL1,10-GAT1, we first determined whether the overall GAL1,10-URE2 transcription profile was similar to that of GAL1,10-GAT1 (Fig. 2). Fig. 4 (A and B) shows that it was, except for a modest decrease in β-galactosidase production at the highest levels of URE2 expression; the reason for this decrease is unknown. We found that increasing URE2 and GAT1 expression together (Fig. 5A, solid lines) dramatically suppressed the ability of GAT1 overexpression (Fig. 5A, dotted lines) to support high level DAL80-lacZ expression with proline or ammonia as nitrogen source. Moreover, DAL80-lacZ expression with GAT1 and URE2 both overexpressed was fully NCR-sensitive. These data show that high level URE2 expression neutralizes the effects of overexpressing GAT1. Furthermore, increasing GAL1,10-URE2 expression (in TCY57) abrogated the need of an excess nitrogen “signal,” i.e. as URE2 expression increased in a culture provided with a nonrepressive nitrogen source (proline), β-galactosidase production (DAL80-lacZ expression) correspondingly decreased (Fig. 5B).
Fig. 5.
A, expression of the DAL80-lacZ gene in strains expressing GAT1 or GAT1 1 URE2 at various levels. Strain TCY48 was grown with either proline (◯) or ammonium sulfate (×) as sole nitrogen source in galactose minimal medium, to which was added the indicated amounts of glucose. Strain TCY60, containing plasmid pTSC572, was grown with either proline (◻) or ammonium sulfate (▵) as sole nitrogen source. B, DAL80-lacZ expression (from pTSC572) in strain TCY57 expressing URE2 at various levels.
Gat1p Appears in the Nucleus Only When There Is Gat1p-dependent Transcription
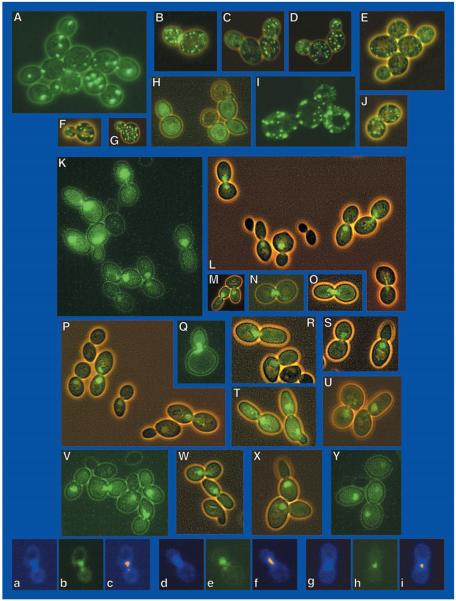
We previously showed a correlation between intracellular distribution of a GATA factor, Gln3p, and NCR-sensitive gene expression.2 Therefore, we wanted to determine whether DAL80 expression correlated with the localization of Gat1p. We constructed ADH1-EGFP-GAT1 pRA27 and used it to transform gat1Δ (RRJ715) cells. In this strain Ure2p is produced at wild type levels, and GAT1 is constitutively overexpressed. DAL80 was highly expressed in the transformants (Fig. 4C, lane B). When the experiment was repeated in strain RTCY57, which overproduces Ure2p, no DAL80 expression was detected (Fig. 4C), confirming the observations in Fig. 5A. When similarly prepared cultures were viewed microscopically, EGFP-Gat1p fluorescence was predominantly nuclear when URE2 was expressed at wild type levels (Fig. 6, K–Y) and co-localized with DAPI-stained material (panels a–i). On the other hand, no nuclear localization was observed when URE2 was overexpressed and DAL80 expression was inhibited (Fig. 4C). Fluorescence rather appeared in the cytoplasm as punctate spots (Fig. 6, A–J) similar to those observed when cultures were transformed with GFP-URE2 pNVS22 (data not shown, but it appears as in Fig. 9 of Cox et al.2 and as reported earlier by Wickner's laboratory (28)).
Fig. 6.
A–J, ADH1-EGFP-GAT1 expressed in RTCY57 that overexpresses GAL1,10-URE2. K—Y, ADH1-EGFP-GAT1 expressed in wild type GYC86 Panels a, d, and g were stained with 4,6-diamidino-2-phenylindole; EGFP-GAT1p was visualized in panels b, e, and h. In panels c, f, and i, the 4,6-diamidino-2-phenylindole-positive material in panels a, d, and g was pseudocolored red, and the images were superimposed on those in panels b, e, and h.
DISCUSSION
The cross-regulation model (11) of GATA factor regulation in S. cerevisiae posits that GAT1 expression directly regulates the level of DAL80 expression and hence Dal80p production. To test this prediction we circumvented the complex normal cross-regulation observed among GATA factor genes and their products by substituting GAL1,10 for the GAT1 and/or URE2 promoters and demonstrated that we could obtain cultures containing differing amounts of GAT1 and URE2 expression and mRNA, which implies they also produced differing amounts of Gat1p and Ure2p. Using this experimental system, we demonstrated that DAL80-lacZ expression (i) depended upon GAT1 expression, (ii) became significantly insensitive to NCR when GAT1 was overexpressed, and (iii) regained NCR sensitivity when URE2 was simultaneously overexpressed with GAT1.
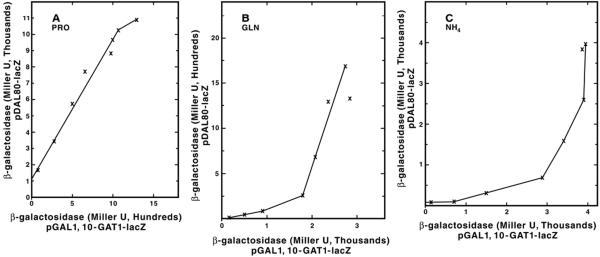
To further evaluate the relationship between GAT1 and DAL80 expression, we plotted GAL1,10-GAT1-lacZ expression data from Fig. 2B as a function of DAL80-lacZ expression (Fig. 3). A linear relationship exists at all but the highest levels of GAT1 expression with proline as nitrogen source (Fig. 7). This linear relationship is not observed with glutamine as nitrogen source until GAL1,10-GAT1-lacZ expression was much higher (Fig. 7B). A similar relationship was also observed with ammonia (Fig. 7C). These data are expected of a situation in which functional Gat1p is “titrating” an inhibitor with ammonia or glutamine as nitrogen sources, and increased DAL80-lacZ expression is not observed until Gat1p excedes a critical level, thereafter overcoming inhibition. Data in Fig. 5 suggest that the molecule Gat1p is likely titrating is Ure2p, because the effects of increased GAT1 expression are suppressed when URE2 expression is simultaneously increased. These results are consistent with the possibility that Gat1p forms a complex with Ure2p as has been suggested for Gln3p (27).
Fig. 7. DAL80-lacZ expression plotted as a function of GAT1-lacZ expression in medium containing proline (A), glutamine (B), or ammonia (C) as sole nitrogen source.
Microscopic evidence extended the relationship between Gat1p and Ure2p by demonstrating that Gat1p is localizes predominantly to the nucleus when Gat1p-dependent transcription is occurring and is cytoplasmic when this transcription is inhibited by overproduction of Ure2p. In other words, Ure2p prevents Gat1p from reaching its physiological target, the GATA elements in NCR-sensitive gene promoters. A similar conclusion was reached for control of Gln3p operation from analysis of CAN1 expression.2
Taken together, the data lead us to suggest that expression of GATA factor-regulated genes depends upon binding of the transcriptional activators Gln3p and Gat1p to their promoter targets and that once bound the activators are able to mediate transcriptional activation and gene expression. Although a negative observation, the finding that Gln3p tethered to DNA by LexAp mediates reporter gene transcription that exhibits little if any NCR-sensitive control (29) is consistent with this proposal. From this perspective, GATA factor regulation is achieved by regulating its binding to its promoter targets. At a coarse level this most likely occurs by regulating Gln3p and Gat1p access to the nucleus that is regulated by Ure2p. At a fine level, once the transcriptional activators have gained access to the nucleus, their overall level of operation is regulated by competition with the transcriptional repressors Dal80p and Deh1p for DNA binding. The fluidity and fine nature of the overall regulation is then achieved by the fact that production of three of the four GATA factors is autogenously and cross-regulated.
Data in Fig. 5B highlight an important characteristic of Ure2p participation in the NCR regulatory cascade. Increasing the intracellular concentration of Ure2p appears to eliminate the need for the physiological signal that intracellular nitrogen is in excess. This observation is consistent with the ideas that: (i) Ure2p exists as an inactive form that is activated in response to a signal indicating nitrogen excess and (ii) Ure2p exists in an active form that is inactivated in response to a signal indicating nitrogen limitation. By this reasoning, signal(s) generated in response to intracellular nitrogen excess or limitation can be circumvented by increasing the concentration of Ure2p or neutralizing negative control exerted by Ure2p by increasing the concentration of at least one of the GATA activators, in this case Gat1p. The simplest basis for such concentration-dependent control is protein-protein complex formation among the constituents of the regulatory circuit.
As this manuscript was being written, four reports simultaneously appeared reaching conclusions to which ours are both similar and complementary (30–33). Although there is not full agreement on the mechanistic details, all propose that Ure2p complexes with one form or another of Gln3p, thereby preventing its entry into the nucleus when cells are grown in rich medium, and one of them demonstrated the proposed Gln3p nuclear and cytoplasmic localization (30). A similar model was proposed for Gat1p, although the investigators were unable to show any interaction between Gat1p and Ure2p. Our work points to such an interaction and contributes to filling this missing link in the proposed models.
Acknowledgments
We thank Dr. R. Rai for pRR351 (T7–7-GAT1), Tim Higgins for preparing the artwork, and the UT Yeast Group for suggested improvements to the manuscript.
Footnotes
This work was supported by National Institutes of Health Grant GM-35642. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: NCR, nitrogen catabolite repression; PCR, polymerase chain reaction; kb, kilobase(s); EGFP, enhanced green fluorescent protein.
Cox, K. H., Rai, R., Distler, M., Daugherty, J. R., Coffman, J., and Cooper, T. G. (2000) J. Biol. Chem., in press.
REFERENCES
- 1.Cooper TG. In: The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. Strathern JN, Jones EW, Broach J, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor; NY: 1982. pp. 39–99. [Google Scholar]
- 2.Wiame J-M, Grenson M, Arst H. Adv. Microb. Physiol. 1985;26:1–87. doi: 10.1016/s0065-2911(08)60394-x. [DOI] [PubMed] [Google Scholar]
- 3.ter Schure EG, van Riel NA, Verrips CT. FEMS Microbiol. Rev. 2000;24:67–83. doi: 10.1111/j.1574-6976.2000.tb00533.x. [DOI] [PubMed] [Google Scholar]
- 4.Bysani N, Daugherty JR, Cooper TG. J. Bacteriol. 1991;173:4977–4982. doi: 10.1128/jb.173.16.4977-4982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chisholm G, Cooper TG. Mol. Cell. Biol. 1982;2:1088–1095. doi: 10.1128/mcb.2.9.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham TS, Cooper TG. Mol. Cell. Biol. 1991;11:6205–6215. doi: 10.1128/mcb.11.12.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell AP, Magasanik B. Mol. Cell. Biol. 1984;4:2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minehart PL, Magasanik B. Mol. Cell. Biol. 1991;11:6216–6228. doi: 10.1128/mcb.11.12.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffman JA, Rai R, Cunningham T, Svetlov V, Cooper TG. Mol. Cell. Biol. 1996;16:847–858. doi: 10.1128/mcb.16.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanbrough M, Rowen DW, Magasanik B. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9450–9454. doi: 10.1073/pnas.92.21.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffman JA, Rai R, Loprete DM, Cunningham T, Svetlov V, Cooper TG. J. Bacteriol. 1997;179:3416–3429. doi: 10.1128/jb.179.11.3416-3429.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soussi-Boudekou S, Vissers S, Urrestarazu A, Jauniaux J-C, Andre B. Mol. Microbiol. 1997;23:1157–1168. doi: 10.1046/j.1365-2958.1997.3021665.x. [DOI] [PubMed] [Google Scholar]
- 13.Cooper TG. In: Mycota III. Marzluf G, Bambrl R, editors. Springer-Verlag; Berlin: 1994. pp. 139–169. [Google Scholar]
- 14.Courchesne WE, Magasanik B. J. Bacteriol. 1988;170:708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drillien R, Aigle M, Lacroute F. Biochem. Biophys. Res. Commun. 1973;53:367–372. doi: 10.1016/0006-291x(73)90671-2. [DOI] [PubMed] [Google Scholar]
- 16.Drillien R, Lacroute F. J. Bacteriol. 1972;109:203–208. doi: 10.1128/jb.109.1.203-208.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham TS, Dorrington RA, Cooper TG. J. Bacteriol. 1994;176:4718–4725. doi: 10.1128/jb.176.15.4718-4725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daugherty JR, Rai R, ElBerry HM, Cooper TG. J. Bacteriol. 1993;175:64–73. doi: 10.1128/jb.175.1.64-73.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox K, Pinchak AB, Cooper TG. Yeast. 1999;15:703–713. doi: 10.1002/(SICI)1097-0061(19990615)15:8<703::AID-YEA413>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell DA, Marshall TK, Deschenes RJ. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 21.Coffman JA, El Berry HM, Cooper TG. J. Bacteriol. 1994;176:7476–7483. doi: 10.1128/jb.176.24.7476-7483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston M, Davis R. Mol. Cell. Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park H-D, Luche RM, Cooper TG. Nucleic Acids Res. 1992;20:1909–1915. doi: 10.1093/nar/20.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Gene (Amst.) 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 25.Scott S, Dorrington R, Svetlov V, Beeser AE, Distler M, Cooper TG. J. Biol. Chem. 2000;275:7198–7204. doi: 10.1074/jbc.275.10.7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovari L, Sumrada R, Kovari I, Cooper TG. Cell Biol. 1990;10:5087–5097. doi: 10.1128/mcb.10.10.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blinder D, Coschigano P, Magasanik B. J. Bacteriol. 1996;178:4734–4736. doi: 10.1128/jb.178.15.4734-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edskes HK, Gray V, T., Wickner RB. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunningham TS, Svetlov V, Rai R, Cooper TG. J. Bacteriol. 1995;178:3470–3479. doi: 10.1128/jb.178.12.3470-3479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck T, Hall MN. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 31.Cardenas ME, Cutler NS, Lorenz M, Di Como CJ, Heitman J. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber S. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardwick JS, Tong JK, Schreiber SL. Nature Genetics. 1999;23:49. [Google Scholar]