Abstract
Transcriptional mechanisms mediated by the binding of transcription factors to cis-acting regulatory DNA elements play critical roles in directing gene expression. While transcription factors have been extensively studied, less effort has gone towards the identification and functional characterization of cis-regulatory elements and associated epigenetic modulation. However, due to methodological and analytical advances, more comprehensive studies of regulatory elements and mechanisms are now possible. Here we summarize recent progress in integrative analyses of these regulatory components in the development of the cerebral neocortex, the part of the brain involved in cognition and complex behavior. These studies are uncovering not only the underlying transcriptional regulatory networks, but also how these networks are altered across species and in neurological and psychiatric disorders.
Keywords: transcription factor, cis-regulatory element, chromatin, epigenetics, gene expression, neuron, disease
cis-regulatory elements are integral components of transcriptional regulatory networks
A fundamental issue in developmental biology is how the multitude of structures necessary for a functioning organism arise from the shared genomic heritage of a single zygote [1]. The transformation from a zygote to an adult organism is composed of a series of developmental events that each rely on the precise spatio-temporal regulation of gene expression [1–3]. These programs are, in large part, mediated by trans-acting factors [i.e., transcription factors (TFs)] that bind specific genomic sequences [4, 5] in cis-regulatory elements (CREs). According to their structural and functional features, TFs can be categorized into many distinct families [4]. Depending on their function, CREs can be divided into functionally different classes, with the proximal promoters, enhancers, insulators and silencers being the most widely studied [6–10]. While other regulatory elements operating in trans- or in cis-, including but not limited to chromatin remodelers, RNA-binding proteins, regulatory RNAs, and cis-regulatory RNA elements also exist and function at the transcriptional or post-transcriptional level [11], the complex sets of interactions among TFs and CREs form the central core of transcriptional regulatory networks (TRNs) (BOX 1) [1].
Box 1. Core classes of cis-regulatory elements in DNA.
Cis-regulatory elements (CREs) in genomic DNA can be divided into functional classes, with promoters, enhancers, insulators, and silencers being the core components that have historically been the most widely studied. Promoters may be proximal promoters, located near the transcription start site and occupied by the primary transcriptional machinery, or distal promoters, bound by TFs but having a weaker regulatory influence. In either case, the promoter serves as a scaffold where general TFs such as TAF1 bind to initiate transcription. Besides RNA polymerase II, active promoters may be associated with histone H3 trimethylated lysine 4 (H3K4me3) or other markers of promoter activity [114].
Enhancers, silencers, and insulators can influence target gene transcription more globally. Enhancers may facilitate gene expression by serving as scaffolds to recruit both TFs and proteins with histone acetyltransferase activity. This allows the decondensation of chromatin, permitting the general transcription machinery greater access to promoters. In contrast to enhancers, silencers and insulators act as negative regulators of gene expression. These regulatory elements may prevent the spread of heterochromatin or prevent the activation of promoters unrelated to nearby enhancers. The effects of these CREs are orientation independent and may be temporally or tissue specific.
As with promoters, there are a number of markers for active enhancers, silencers, and insulators. For example, active enhancers may be associated with H3.3 [115], H2A.Z [7], H3K27ac [115], H3K4me1, or other markers. Among the markers for active silencers and insulators are REST [116], SUZ12 [117] and CCCTC-binding factor (CTCF) [118].
Super enhancers consist of clusters of enhancers spanning up to 50 kb, are densely occupied by master TFs and Mediator complexes, and play crucial roles in defining cell identity in many differentiated cell types including mouse progenitor B cells, T helper cells, and macrophages [112, 119]. Super enhancers may also activate the expression of master TFs and other genes that play important roles in cell type specification, potentially making the expression of genes regulated by super enhancers sensitive to perturbations impacting master TFs or Mediator complex.
Numerous other regulatory components exist. Other cis-elements affecting gene expression, if not always transcription, are 3′ untranslated regions, splicing regulatory elements, and epigenetic factors including DNA methylation, histone modifications, chromosomal conformation, and chromatin state. Additional trans-regulatory elements include regulatory RNAs (i.e. miRNAs, lincRNAs and so on), RNA-binding proteins, splicing factors, and chromatin modifiers/remodelers.
Interactions between TFs and CREs are complex, dynamic, and modulated by various epigenetic processes [12]. While individual CREs may be regulated by the competitive or cooperative binding of many TFs, multiple CREs may also confer transcriptional control over the expression of any single gene [12]. Furthermore, TFs and CREs regulate transcription together with their cognate transcriptional co-factors, chromatin regulators, epigenetic modifications, RNA binding proteins, and non-coding RNAs [2, 13]. This combinatorial and multi-level transcriptional regulation greatly increases the complexity of gene expression regulation, facilitating the unique spatio-temporal patterns necessary for development and function [3].
Researchers have long recognized the importance of TFs in development, evolution, function and pathogenesis. Although the contributions of CREs have also been explicitly recognized, our understanding of their key biological roles and associated TRNs has been more elusive due to the difficulties associated with their systematic identification and functional characterization. Fortunately, thanks to a number of recent methodological and analytical advances, more comprehensive and functionally relevant studies of CREs are now possible in various complex tissues and physiological contexts (BOX 2).
Box 2. Advances in the study of transcription factors and cis-regulatory elements.
Improving global scientific infrastructure has facilitated the attainment, preservation, and dispersal of organisms and tissues, including human and non-human primate brains, previously not easily available to the scientific community. This has allowed comparative genomics to flourish, leading to the identification of potential CREs through the recognition of sequence conservation across species [120, 121]. Similarly, improvements in tissue and cell processing methods have allowed the isolation of cells and discrete morphological areas on a level previously impossible [105, 122–126]. This has aided the creation of high-quality databases profiling the genome, transcriptome, epigenome, and regulome of multiple neural cell types, tissues, developmental periods, species, and diseases [10, 96, 125, 127] [97, 105] (see www.genome.gov/encode/; www.1000genomes.org; www.roadmapepigenomics.org; www.gtexportal.org/; www.brain-map.org; www.genepaint.org; enhancer.lbl.gov; www.brainspan.org and www.commonmind.org).
Technological advances have also facilitated the discovery and characterization of CREs. While the increasingly long lists of TFs annotated for their involvement in any given development process has provided progressively more “bait” for chromatin immunoprecipitation (ChIP) assays [23, 128], the ability to couple ChIP with next generation sequencing, as in ChIP-seq, has allowed global assessments of CRE activity. The use of ChIP-seq using more general markers associated with specific CREs, such as EP300 (also known as p300), RNAp II (Pol II), and histone H3 acetyl Lys27 (H3K27ac), has also allowed these global searches for CREs to be performed independently of knowledge of (or antibodies against) TFs [10, 12, 96, 129] [130]. Recently developed techniques for mapping chromosomal interactions over long distances, such as ChIA-PET (chromatin-interaction analysis with paired-end tag sequencing), couple ChIP-seq with assays that identify both the distal and proximal gene regulatory elements of promoters by tethering conformationally related chromosomal regions [131–133]. Taken together, these advances allow both the identification of hundreds of thousands of CREs and novel opportunities to correlate promoter and enhancer occupancy with gene expression.
With the identification of so many regulatory elements has come an increased ability to validate and functionally characterize those elements. The Massively Parallel Reporter Assay (MPRA) and other similar approaches allow the rapid validation of hundreds of putative CREs in a single experiment [134]. Once validated, CREs can be functionally characterized through the use of BAC transgenesis [69], site-directed mutagenesis by CRISPR/Cas technology [135, 136], or other techniques drawn from an expanding molecular toolkit.
Among the most complex biological tissues, with a myriad of cell types and a very protracted development, is the cerebral neocortex. The neocortex is a portion of the brain unique to mammals, expanded in humans, and critical for cognition, perception and complex behavior [14–16]. The development of the neocortex, and consequently its function, depends on the highly orchestrated regulation of gene expression. Examples of the contributions of TFs to the developing neocortex, and to neurological and psychiatric disorders, are abundant [17–20], but, as is true for many complex biological systems, knowledge concerning CREs and their integration with TFs and other regulatory components into TRNs has lagged far behind.
Here we summarize recent progress in the study of selected key TFs, CREs, and associated regulatory processes in the control of gene expression underlying the development, evolution, and pathogenesis of the neocortex (BOX 3).
Box 3. Epochs of neocortical development.
The vast majority of cells occupying the adult neocortex originate either from radial glia (RG) in the ventricular zone (VZ), intermediate progenitor cells (IPCs) in the subventricular zone (SVZ), or progenitor pools located within the ganglionic eminences. In these proliferative niches, multiple TFs act to maintain progenitor cells in a proliferative state. Other TFs promote the production of differentiated neurons, glia, and astrocytes. The mechanisms mediating the choices between proliferation and differentiation and neurogenesis and gliogenesis are tightly regulated.
As nascent neurons exit mitosis, they migrate from these proliferative niches to their ultimate destinations. Interneurons arising from the ventrally located ganglionic eminences tend to migrate tangentially along cortical layers. Cells arising in the VZ or SVZ of the dorsal telencephalon migrate radially along a radial glial scaffold and deposit themselves in a time-dependent manner. Preplate neurons, including Cajal-Retzius cells, are born first. These cells secrete Reelin, an instructive guidance molecule for the formation of neocortical layers. Newly born neurons migrate radially, first splitting the preplate and forming the cortical plate between the marginal zone and the subplate. Subsequently born neurons migrate into the cortical plate in an inside-out manner as progressively younger neurons migrate past their older siblings. At the end, later-born, superficial-layer neurons reside in layers 2 through 4 and earlier-born deep-layer neurons in L5–6. A schematic depicting this process can be seen in Figure 1.
Cellular diversification and differentiation generates the wide diversity of neuronal types present in the adult neocortex. While strong evidence for the prepatterning of cells within the ventricular zone exists, many of the TFs implicated in the development of specific cell fates only begin to be expressed as cells complete their terminal mitosis. At this time cells may express TFs essential for the attainment of multiple distinct cell fates. However, through a series of feedback mechanisms including mutual cross-repression, cells ultimately settle on a final identity. An example of a transcriptional regulatory network involved in cell type specification is shown in Figure 2.
Additionally the assembly and function of neuronal circuits is essential for normal function. Like differentiation, the genetic and molecular components essential for both synaptogenesis and connectivity may be expressed during or be influenced by earlier developmental epochs. Other factors, including extracellular stimuli, can influence the expression of TFs, the activity of CREs, and the function of TRNs.
Transcriptional regulation of neocortical neurogenesis and gliogenesis
The neocortex is mainly composed of a myriad of neuronal and glial cell types. Neurons are broadly classified into two major groups, glutamatergic excitatory projection neurons (also known as pyramidal neurons) and GABAergic inhibitory local circuit interneurons, which follow substantially different developmental programs [17–23]. Projection neurons, which account for approximately 80% of neocortical neurons, originate from stem/progenitor cells within the neocortical wall in the dorsal forebrain (Fig. 1), whereas interneurons arise primarily from progenitor cells in the ventral forebrain and migrate tangentially into the cortex. Glial cells, classified mainly as astrocytes, ependymal cells, and oligodendrocytes, also originate from the same lineage of neural progenitors [24].
Figure 1. Schematic illustration of neurogenesis in the mammalian neocortex.
Neuroepithelial cells (NPCs) undergo symmetric cell division to produce an initial pool of cortical progenitors that later transform into apical radial glia cells (aRGCs). Neocortical aRGCs line the ventricular zone from where they extend a long radial process stretching to the basal surface. aRGCs begin asymmetric cell division to generate another aRGC and a nascent projection neuron. The neuron then migrates radially from the ventricular zone (VZ) along the basal process of a RGC into the cortical plate (CP). The earliest born neurons migrate to form the preplate. Later migrating neurons split the preplate into the marginal zone (MZ) and subplate (SP). The MZ also consists of Cajal-Retzius cells originating from multiple sites in the forebrain including the cortical hem. As neurogenesis proceeds, diverse subtypes of neurons are generated through the successive asymmetric division of RGCs. Early-born nascent projection neurons settle in the deep layers (Layers 5 and 6; red layers) and later-born projection neurons settle in towards mid neurogenesis stage. Additionally, some populations of RGC daughter cells become intermediate progenitor cells (IPC) or basal radial glial cells (bRGC) in the subventricular zone (SVZ). After the neurogenic stages, the radial scaffold detaches from the apical surface and aRGCs become gliogenic, generating astrocytes, or transform into ependymal cells. Tangential migration of interneurons is observed in the MZ, intermediate zone (IZ) and SVZ. Neocortical projection neurons mature into cortical projection neurons (CPNs), which show layer- and subtype–specific morphology and axonal projection patterns. Subcerebral projection neurons, which extend axons to the striatum, thalamus, brainstem, and spinal cord, are located in the deep layers. Upper layer projection neurons project axons within the telencephalon (i.e., cerebrum). Adapted from Kwan et al, 2012.
Mediating the transitions between proliferation and differentiation of neural progenitor/stem cells in the ventricular zone (VZ) and subventricular zone (SVZ) is essential for determining neocortical size and other aspects of normal development [14]. A number of TFs have been implicated in controlling the onset, progression, and termination of neocortical neurogenesis. Among these, EMX1, EMX2, FOXG1, HES1, HES5, LHX2, and PAX6 are highly expressed by cortical progenitor cells throughout neurogenesis and have been shown to play critical roles in promoting the maintenance of a progenitor state [25–27] [28, 29]. Other TFs, such as FEZF2 (FEZL or ZFP312), ID4, NGN1, NGN2, and NR2E1 (TLX) are enriched in early progenitor cells [30–32], whereas TFAP2C (AP2-gamma), CUX1, CUX2, POU3F3, POU3F2 (BRN1 and BRN2, respectively) and TBR2 are enriched in later progenitor cells [31, 33–36]. Each of these TFs may promote neurogenesis in some developmental contexts. For example, several TFs, such as INSM1, NGN2, TBR2, and TFAP2C, have been shown to promote the generation of basal progenitors in the SVZ from VZ progenitor cells [33, 36–38].
Knowledge of the CREs involved in these processes is essential for understanding how these suites of TFs are integrated to promote the appropriate developmental pathways. Unfortunately, the known repertoire of CREs active in different types of cortical progenitor cells during neurogenesis is largely incomplete and ones that have been functionally characterized in this context are rare. One well-studied example of such a CRE concerns the seemingly paradoxical ability of PAX6 to promote both the expression of genes for progenitor self-renewal as well as those for neuronal differentiation. These conflicting abilities are resolved in part through a low-affinity enhancer, E1, of NGN2 [39]. When concentrations of PAX6 are low, PAX6 is unable to bind E1 and does not induce the expression of the proneural gene Ngn2. Instead, PAX6 induces the expression of genes promoting self-renewal while HES1 directly represses several genes that promote differentiation [40]. However, when levels of PAX6 are higher, PAX6 is able to bind E1 and induce the expression of NGN2. Other TFs, including FEZF1 and FEZF2, are subsequently induced and directly repress Hes1 and the related Hes5 [41]. This shifting ratio between PAX6 and HES1 is directly tied to neurogenesis [40].
Other examples of regulatory elements mediating neurogenesis also exist. The BAF (mSWI/SNF) chromatin remodeling complex is involved in regulating the size and thickness of the neocortex [42] and the ability of PAX6 to bind to specific CREs and regulate downstream target genes is modulated by this complex during adult neurogenesis [43]. These studies suggest that epigenetic mechanisms such as chromatin remodeling [11] may play a critical role in the transcriptional regulation of neurogenesis throughout development. In addition, specific non-coding RNAs have been shown to regulate cortical neurogenesis by targeting multiple key TFs [44, 45], providing a regulatory feedback mechanism and another layer of complexity.
TFs and CREs also control the switch from neurogenesis to gliogenesis that generally occurs later in development. Here again, key examples highlight the importance of epigenetic regulation, such as DNA methylation, in controlling transcriptional regulatory interactions and gene expression. During neocortical neurogenesis, promoters of astrocyte-specific genes, such as glial fibrillary acidic protein (Gfap), are heavily methylated and silenced [46]. Astrocyte fate is impeded by NGN1, which promotes neuronal fate by competing with STAT3 for EP300-SMAD activator complexes [47]. However, at the end of neurogenesis, the Gfap promoter is demethylated to facilitate both astrocyte differentiation and the transformation of the radial scaffold of progenitor cells into glial progenitors, differentiated astrocytes, and ependymal cells [46]. This is possible because the suppression of NGN1 expression, coupled with the demethylation of promoters of astrocyte-specific genes begun by NFIA [48], allows the JAK-STAT3 and BMP pathways to initiate the expression of pro-astrocytic genes.
Another major class of glial cells, oligodendrocytes, develop from proliferating oligodendrocyte precursor cells that arise in proliferative zones in multiple regions of the ventral and dorsal forebrain, migrate throughout the developing white matter, and divide a limited number of times before they terminally differentiate. Among the TFs that promote oligodendrocyte specification and maturation are OLIG1, OLIG2, SOX10, YY1, MRF, and ZFP219 [49, 50]. Interestingly, DLX1 and DLX2, TFs necessary for interneuron fate specification, and OLIG1 and OLIG2 control neuronal versus oligodendroglial cell fate acquisition in the developing ventral forebrain [51]. DLX1/2 promote neurogenesis at the expense of the oligodendrocyte lineage through repression of Olig2 expression [51, 52]. By contrast, OLIG1 directly binds to the Dlx1/2 I12b intergenic enhancer to repress Dlx1/2 expression, leading to the acquisition of an oligodendrocyte fate [52].
Transcriptional regulation of neuronal migration
Following neurogenesis, nascent projection neurons move toward the dorsal (pial) surface of the developing brain to ultimately form the six-layered architecture of the mature cerebral neocortex. Several TFs implicated in other epochs of neuronal development, including SOX5, TBR2, NGN1, POU3F3, and POU3F2, are also involved in the radial migration of nascent projection neurons in the dorsal telencephalon [53–55]. Presumably, these TFs regulate the expression of genes involved in cytoskeletal dynamics and other aspects of cell movement, but until recently the details of this regulation have remained unknown. Recent work concerning a CRE of RND2 offers insight into these processes.
Rnd2 is a gene encoding a rhoA-like GTPase involved in regulating cytoskeletal dynamics and implicated in neuronal migration [56]. The expression of RND2 is regulated by competitive binding of NGN2 and ZBTB18 (ZFP238 or RP58) to a CRE in the 3′ region of Rnd2. In the absence of NGN2 binding, RND2 is not expressed and the natively RND2-expressing neurons are incapable of migrating from the SVZ to the cortical plate [57]. In contrast to NGN2, ZBTB18 represses Rnd2 [58]. Neurons depleted of Zbtb18, which show an upregulation of Rnd2, also cannot migrate from the SVZ due to the failure of the multipolar-bipolar transition [59]. Furthermore, NGN2 activates Zbtb18 transcription by binding to its promoter [60], whereas ZBTB18 represses Ngn2 [59, 61]. NGN2 and ZBTB18 therefore form a feedback loop operating through a shared CRE to fine-tune the expression of Rnd2, thus controlling the radial migration of projection neurons.
Transcriptional regulation of neuronal diversification and differentiation
The organization of the neocortex into functionally distinct layers and areas is rooted in the diversification of developing projection neurons into distinct subtypes. The early progenitor pool gives rise to subplate (SP) and deep-layer (L5–6) neurons, whereas the late progenitor pool generates upper-layer (L2–4) neurons (Fig. 1). Further diversification into many subtypes occurs during the early postmitotic and postmigratory life of projection neurons (Fig. 1) [23, 54, 62–65].
Studies of laminar position and cortical projection neuron lineage specification illustrate both the importance of the spatial distribution of relevant TFs and the ability of CREs to integrate the effects of these TFs. These processes are also modulated by a number of other transcriptional and post-transcriptional regulatory processes. For example, FEZF2, which is enriched in L5 and, to a lesser extent, L6 projection neurons, is required for specifying subcerebral (corticofugal) projection neurons, including corticospinal neurons [66–70]. Acting along with FEZF2 in immature L5 corticospinal neurons are members of the SOX family of TFs (SOX4, SOX5, and SOX11) as well as BCL11B (CTIP2) (Fig. 3) [54, 69, 71, 72]. Similar suites of TFs exhibiting layer- and subtype- specific expression profiles are expressed by developing projection neurons in L6 (SOX5, TBR1, and NR2F1), L4 (NR2F1, LHX2, CUX1/2, and BHLHB5), and L2–4 (CUX1/2, BHLHB5, and SATB2).
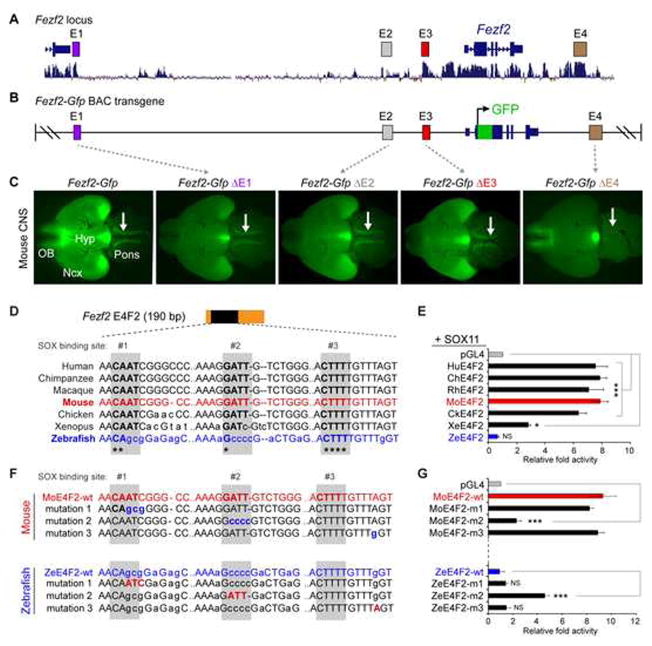
Figure 3. The identification and characterization of the E4 enhancer of Fezf2.
In this example of cis-regulatory element (CRE) discovery and functional characterization, transcriptome analysis and comparative genetics were coupled with BAC transgenesis to identify and characterize an enhancer of the Fezf2 gene. (A) Fezf2 is a transcription factor (TF) identified in a genome-wide search for genes enriched among corticospinal motor neurons and their predecessors as compared to other cortical projection neurons. Within the Fezf2 locus are four conserved non-exonic regions, labeled E1, E2, E3, and E4, with high levels of homology across species. The Fezf2 coding region is also indicated. (B) Schematic of a BAC reporter where a reporter gene, Gfp, is driven by the native regulatory environment governing Fezf2. (C) GFP expression, driven by the unmanipulated Fezf2-Gfp BAC, is present throughout the dorsal telencephalon and the corticospinal tract (arrow). This is also true when the putative enhancers E1, E2, and E3 are deleted (Δ). However, deletion of the E4 enhancer (right panel) results in a loss of GFP expression from the dorsal telencephalon and the apparent absence of the corticospinal tract. (D) Sequence-level homology of the E4 enhancer of Fezf2 in several species. Putative binding sites for SOX proteins are shaded. Note the generally high levels of homology except for the lack of homology in zebrafish at the second shaded SOX binding site. (E) SOX11 stimulates expression of a construct containing the E4 enhancer of Fezf2. The construct containing the zebrafish E4 sequence, which lacks the second consensus SOX binding site, is not induced by SOX11. (F) The putative SOX binding sites were mutagenized in mouse and zebrafish, effectively swapping mouse sequence into zebrafish and vice versa. (G) Swapping the sequences of the second SOX binding site between zebrafish and mouse reduces the ability of SOX11 to induce transcription from mouse E4 while increasing the ability of SOX11 to induce transcription from zebrafish E4. No change is observed when the sequences of the first and third putative SOX binding sites are swapped.
Knowledge of the transcriptional interactions, CREs involved, and epigenetic machinery is necessary for understanding the cell-type specific expression of these TFs. SOX5 is required for L5/6 projection neuron migration and specification, but it also represses Fezf2 in L6 corticothalamic projection neurons through direct repression mediated by the E4 enhancer near Fezf2 [54, 71] (Fig. 3A,B). Conversely, SOX4 and SOX11 compete with SOX5 for the activation of the E4 enhancer [69] (Fig. 3A, B). The Fezf2 locus is also bound by TBR1, further promoting the repression of Fezf2 in L6 neurons [23, 73] (Fig. 3B). In turn, FEZF2, among other transcription factors including BCL11B, represses the Tbr1 locus (Fig. 3A). SATB2, another DNA-binding protein expressed by immature neurons across several cortical layers whose expression is indirectly regulated by FEZF2, activates the expression of TBR1 as well as suppresses the expression of BCL11B (Fig. 3C,D) [70, 74].
A problem inherent to the “closed” TRN described above, however, is that it must in some way be broken to allow the transition from the production of deep layer neurons to the production of upper layer neurons. This is at least partially mediated by the TF FOXG1. FOXG1 is expressed in neocortical progenitors and upper layer neurons and directly suppresses TBR1 expression by binding the 5′ region of the Tbr1 locus. This results in the upregulation of deep layer genes including Fezf2 and Bcl11b. The deep layer neurons then themselves produce a cell-extrinsic signal that, coupled with the repression of Tbr1 by FOXG1, makes progenitors competent to differentiate into upper layer neurons [75].
The cell-type-specific and sustained expression of TFs necessary for the specification, development, and later maintenance of neuronal cell types is mediated at least in part by epigenetic means [76]. This is also true for cortical projection neurons. For example, BCLL11B, which is expressed by the subcerebral projection neurons in L5/6, is a recently identified subunit of the BAF chromatin remodeling complexes [77]. As such, it likely cooperates with other TFs to target distinct chromatin loci containing CREs involved in the cell-type specific activation or repression of gene expression. A different example involves the interaction of AF9 with DOT1L histone H3 methyltransferase, a protein that mediates the methylation of histone H3 lysine 79 (H3K79) at the transcription start site of Tbr1 [78]. This epigenetic modification suppresses Tbr1 expression in upper layer neurons and promotes the fate maintenance of the intracerebral projection neurons in those layers. Similarly, SATB2 is itself a chromatin modifier and binds directly to matrix attachment regions. This allows SATB2 to recruit histone deacetylases and silence the expression from loci including Bcl11b [62, 63, 79].
OLIG1/2, discussed earlier as TFs necessary for gliogenesis, also play critical roles in oligodendrocyte differentiation. For example, OLIG2 transcriptionally pre-patterns the chromatin-remodeling factor SMARCA4 (BRG1) [80]. Other studies have also linked DNA methylation and chromatin modifications to glial differentiation and maintenance [81] and such findings will only increase in frequency as transcriptomes and epigenomes are compiled in parallel from diverse neuronal cell types.
Interneurons are subdivided into several types depending on their origin and molecular markers. Parvalbumin (PVALB)- and somatostatin (SOM)-positive interneurons are generated from the MGE and calretinin (CALB2 or CR)-positive interneurons arise from the caudal ganglionic eminence (CGE). A number of TFs have been implicated in controlling the specification and differentiation of these interneurons, including NKX2.1 (TITF1), ASCL1 (MASH1), LHX6, SOX6, NR2F1 and NR2F2 (COUP-TF1 and COUP-TF2, respectively), and members of the DLX family [21, 82, 83].
Whereas NKX2.1, LHX6, SOX6, NR2F1, and NR2F2 play discrete roles in the specification of different subtypes of interneurons, DLX1/2/5/6 and ASCL1 are important for the broad specification and differentiation of interneurons. For example, three CREs of DLX1, called URE2, I12b, and 156i, are associated with different subtypes of interneurons [84]. URE2 is active in PVALB/CALB2/NPY/NOS1-positive interneurons, whereas I12b and I56i are specifically active in SOM/VIP/CALB2- positive interneurons. This suggests distinct combinations of Dlx1 enhancers are used for the specification of interneurons. Taken together, these examples demonstrate that a combination of TFs, CREs, epigenetic modifications, and extrinsic signals function to first specify and then maintain diverse neural cell types.
Transcriptional regulation of the assembly and function of neural circuits
Developmentally regulated changes in TRNs continue long after neurogenesis has subsided and distinct cell fates have been determined. Sensory experience-driven synaptic activity during postnatal development initiates distinct transcriptional regulatory programs, allowing cellular functions and processes like synaptogenesis to be adapted to the environment [85, 86]. For example, BDNF is a member of the neurotrophin family and is implicated in neuronal survival, differentiation, neurite outgrowth, and synaptic plasticity. Some of the CREs of the Bdnf gene are bound and activated by CREB, ARNT2, NPAS4, CARF, and USF1, all proteins whose ability to induce gene expression is regulated by membrane activity [87].
Activity-dependent regulation is not unique to the BDNF gene. CREB-binding protein (CREBBP or CBP) is a DNA-binding transcriptional co-activator associated with enhancer activity that enables the general transcriptional machinery to access promoters, thus facilitating gene expression. Upon membrane depolarization, the number of CBP binding sites in the genome increases from approximately 1,000 to approximately 28,000 [88]. This includes, as assessed by the presence of the enhancer marker histone H3 lysine 4 monomethylation (H3K4me1), approximately 12,000 activity-regulated enhancers. Many of these enhancers are bound by FOS [85]. Greatly increased binding of CREB, NPAS4, and SRF to both enhancers and promoters is also observed. The activity of multiple TRNs of potentially thousands of genes may therefore be regulated by synaptic activity and, by extension, the cellular environment and experience. Additionally, here again knowledge of the CREs and regulatory mechanisms involved is necessary for understanding patterns of gene expression.
Evolution of neocortical cis-regulatory elements
Although the neocortex is present in all mammals, there are substantial variations in its development and organization that may underlie some species-specific aspects of cognition and behavior. Changes in coding sequences undoubtedly contribute to the phenotypes unique to any given species [89–91], but a significant portion of that uniqueness may be due to changes in TRNs including CREs and other regulatory components [92–94]. Moreover, changes in CRE sequence and activity have been linked with evolution across mammals [69, 92, 93, 95]. For example, only 58% of loci occupied by the enhancer-associated proteins EP300/CBP in mid-fetal human neocortex were similarly enriched for ChIP-seq reads at the orthologous site in the mouse genome [96].
While the functional consequences of such changes in CRE activity are largely unknown, these data suggest massive changes in the regulatory architecture governing the emergence and development of the neocortex. Recent studies demonstrate the reliance of this architecture on CREs, as the main regional domains of the forebrain can be further divided into several subdomains demarcated by enhancer activities [96, 97]. Additionally, these results suggest how the activities of a relatively small number of TFs with broad expression gradients may be integrated to produce comparatively fine distinctions within a tissue. CREs are therefore potentially powerful substrates for evolution to act upon in the creation of new subdomains and cell types within the telencephalon and elsewhere.
Specific examples of critical new insights into the functional relevance of species differences in the sequence and activity of CRE sequences on neocortical development and evolution have started to emerge. For example, the acquisition by mammals (and possibly birds) of the E4 enhancer sequence of Fezf2 may have contributed to the emergence of the corticospinal projection system, thus enabling the highly skilled motor control typical of these species [69] (Fig. 2C, D). Such examples are not limited to TF binding sites, as DNA sequence changes in other regulatory sites, including sites bound by RNA-binding proteins in neocortically expressed transcript have also undergone evolutionary changes. One example of this is the human NOS1 mRNA and FMRP [98].
Figure 2. Transcriptional regulatory networks controlling specification and differentiation of cortical projection neurons.
(A) Key transcription factors (TFs) involved in neurogenesis and cell type specification. Solid lines indicate direct activation or repression of a downstream target by the upstream TF indicated. Dashed arrows indicate regulation by unknown mechanisms. Orange arrows indicate indirect results of the effects of the TFs indicated. (B–D) Models for the regulation of the transcription of Fezf2, Bcl11b (aka Ctip2), and Tbr1 during cell type specification. (B) Regulation of Fezf2 transcription. SOX4 and SOX11 activate Fezf2 transcription while SOX5 suppresses this expression by competing for the E4 enhancer. The physical interaction was tested by EMSA and ChIP [69]. TBR1 suppresses Fezf2 expression by binding to another CRE. This physical interaction was tested by ChIP [23]. (C) SATB2 represses Bcl11b expression by binding to MAR (matrix attachment regions) sequences (orange boxes) [62]. (D) SATB2 induces the expression of Tbr1 by binding to three MAR sequences [70].
Neocortical cis-regulatory elements in neurological and psychiatric disorders
Alterations to TRNs have been linked to many disorders, including neurological and psychiatric disorders. Such alterations can arise from trans-regulatory changes that modify the expression or function of TFs or from cis-regulatory changes that affect the binding of TFs, RNA-binding proteins, and non-coding RNAs. For example, many copy number variants or mutations in TFs previously implicated in neocortical circuitry development, including ARX2, FEZF2, TBR1, SATB2, and SOX5 [65, 99–104], have been linked to neurological and psychiatric disorders.
A growing number of studies, in particular genome-wide association studies, have also identified genetic polymorphisms or expression quantitative trait loci (eQTL) in putative CREs and regulatory non-coding RNAs that can lead to variation in gene expression levels between neural cell types, brain regions, timepoints or individuals [105–107]. Other brain disease-associated SNPs occur in non-coding regions that overlap with putative CREs [12, 108] [109–111]. Some of these, such as SNPs associated with Alzheimer’s disease, have also been reported to be enriched in super enhancers, which mediate the establishment and maintenance of cell type identity, as compared to more typical enhancers [112]. However, the respective functional contributions of these cis- and trans-regulatory changes, as well as the vast majority of eQTLs, to molecular and cellular processes have remained largely unexplored.
Importantly, CREs allow the data from studies on evolution and brain disorders to synergize. A comparison of the distribution of H3K4me3, a promoter-specific histone modification associated with active transcription, in the prefrontal cortices of human, chimpanzee, and macaque brains found 410 regions enriched for H3K4me3 that were unique, and 61 regions that were uniquely lost, in humans [113]. Among the genes potentially affected were DPP10, CNTN4, CHL, and other genes previously linked to brain disorders. Analyses of TRNs involving these genes may provide insights into how the human brain may have become increasingly and possibly uniquely susceptible to certain neurological and psychiatric disorders as compared to non-human primates.
Future directions
Our focus here on TFs and the CREs that mediate, either directly or indirectly through changes in chromatin structure and epigenetic modifications, the binding of TFs has led us to generally exclude from discussion many other regulatory modalities, including regulatory RNAs and chromatin remodelers, utilized by TRNs. Furthermore, we limited the scope to examples where molecular interactions between TFs and CREs were well-defined, and many studies that have assessed CREs, including eQTL, on a global scale were therefore not discussed. Despite these omissions, recent advances make clear that CREs do not function merely as passive conduits carrying developmental cues linearly along from TFs. Rather, they integrate diverse sets of developmental programs promoted by the cellular environment, external stimuli, and TFs themselves. This is seen in the ability of CREs to mediate competition or synergism between multiple TFs [54, 69, 71, 96] and the ability of CREs to make large swaths of the genome accessible, as in the new availability of the thousands of CBP binding sites present following membrane depolarization [85, 88]. Such integration and regulation is bi-directional; super enhancers may recruit some master TFs and regulate the expression of others while at the same time DNA-binding proteins modify CREs to regulate the activity of super enhancers [112].
As the necessity for incorporating CREs into TRNs is becoming all the more clear, so too is the need for the systematic discovery and characterization of CREs active in different neural cells, regions, timepoints, species, individuals and disease states. Combining these data with transcriptomic and other data sets, including interactomes and epigenomes, will further aid not only the identification of target genes but also the development of new insights into how changes in TRNs contribute to changes in gene expression patterns. Importantly, developing in parallel with new techniques for the identification of putative CREs are new methods for the large-scale validation of these CREs. The reconstruction of TRNs, in particular through the rapid introduction or manipulation of CREs in the mouse genome by CRISPR/CAS editing, further allows functional assessments of CREs and their roles in TRNs. Taken together, these advances will allow scientists to characterize TRNs in neocortical development, evolution, function and dysfunction as never before.
Highlights.
Recent advances have facilitated the study of neural cis-regulatory elements
This new work is reshaping our understanding of transcriptional regulatory networks
Brain-specific transcriptional regulatory networks are altered in evolution and disease
Acknowledgments
The authors acknowledge financial support from the National Institute of Health, the Simons Foundation, the James S. McDonnell Foundation, the Kavli Foundation, and the Foster-Davis Foundation (NARSAD). We apologize to all colleagues whose important work was not cited because of the large scope of this review and space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci U S A. 2005;102(14):4936–42. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152(6):1237–51. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spitz F, Duboule D. Global control regions and regulatory landscapes in vertebrate development and evolution. Adv Genet. 2008;61:175–205. doi: 10.1016/S0065-2660(07)00006-5. [DOI] [PubMed] [Google Scholar]
- 4.Vaquerizas JM, et al. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 5.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613–26. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 6.Pennacchio LA, et al. Enhancers: five essential questions. Nat Rev Genet. 2013;14(4):288–95. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet. 2012;13(7):469–83. doi: 10.1038/nrg3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2012;13(1):59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- 10.Nord AS, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155(7):1521–31. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Son EY, Crabtree GR. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am J Med Genet C Semin Med Genet. 2014;166C(3):333–49. doi: 10.1002/ajmg.c.31416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babu MM, et al. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol. 2004;14(3):283–91. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141(11):2182–94. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 15.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146(1):18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill RS, Walsh CA. Molecular insights into human brain evolution. Nature. 2005;437(7055):64–7. doi: 10.1038/nature04103. [DOI] [PubMed] [Google Scholar]
- 17.Rubenstein JL. Annual Research Review: Development of the cerebral cortex: implications for neurodevelopmental disorders. J Child Psychol Psychiatry. 2011;52(4):339–55. doi: 10.1111/j.1469-7610.2010.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139(9):1535–46. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greig LC, et al. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14(11):755–69. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leone DP, et al. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18(1):28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7(9):687–96. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- 22.Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505(7483):318–26. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han W, et al. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci U S A. 2011;108(7):3041–6. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallo V, Deneen B. Glial development: the crossroads of regeneration and repair in the CNS. Neuron. 2014;83(2):283–308. doi: 10.1016/j.neuron.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida M, et al. Emx1 and Emx2 functions in development of dorsal telencephalon. Development. 1997;124(1):101–11. doi: 10.1242/dev.124.1.101. [DOI] [PubMed] [Google Scholar]
- 26.Hanashima C, et al. Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J Neurosci. 2002;22(15):6526–36. doi: 10.1523/JNEUROSCI.22-15-06526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou SJ, O’Leary DD. Role for Lhx2 in corticogenesis through regulation of progenitor differentiation. Mol Cell Neurosci. 2013;56:1–9. doi: 10.1016/j.mcn.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imayoshi I, et al. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30(9):3489–98. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinn JC, et al. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev Biol. 2007;302(1):50–65. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yun K, et al. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004;131(21):5441–8. doi: 10.1242/dev.01430. [DOI] [PubMed] [Google Scholar]
- 31.Schuurmans C, et al. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. Embo J. 2004;23(14):2892–902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo C, et al. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron. 2013;80(5):1167–74. doi: 10.1016/j.neuron.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinto L, et al. AP2gamma regulates basal progenitor fate in a region- and layer-specific manner in the developing cortex. Nat Neurosci. 2009;12(10):1229–37. doi: 10.1038/nn.2399. [DOI] [PubMed] [Google Scholar]
- 34.Sugitani Y, et al. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16(14):1760–5. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cubelos B, et al. Cux-1 and Cux-2 control the development of Reelin expressing cortical interneurons. Dev Neurobiol. 2008;68(7):917–25. doi: 10.1002/dneu.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sessa A, et al. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60(1):56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farkas LM, et al. Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron. 2008;60(1):40–55. doi: 10.1016/j.neuron.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 38.Britz O, et al. A role for proneural genes in the maturation of cortical progenitor cells. Cereb Cortex. 2006;16(Suppl 1):i138–51. doi: 10.1093/cercor/bhj168. [DOI] [PubMed] [Google Scholar]
- 39.Scardigli R, et al. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development. 2003;130(14):3269–81. doi: 10.1242/dev.00539. [DOI] [PubMed] [Google Scholar]
- 40.Sansom SN, et al. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5(6):e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu T, et al. Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development. 2010;137(11):1875–85. doi: 10.1242/dev.047167. [DOI] [PubMed] [Google Scholar]
- 42.Tuoc TC, et al. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell. 2013;25(3):256–69. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Ninkovic J, et al. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13(4):403–18. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowakowski TJ, et al. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proc Natl Acad Sci U S A. 2013;110(17):7056–61. doi: 10.1073/pnas.1219385110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata M, et al. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31(9):3407–22. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takizawa T, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1(6):749–58. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 47.Sun Y, et al. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104(3):365–76. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- 48.Namihira M, et al. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell. 2009;16(2):245–55. doi: 10.1016/j.devcel.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 49.Li H, et al. Two-tier transcriptional control of oligodendrocyte differentiation. Curr Opin Neurobiol. 2009;19(5):479–85. doi: 10.1016/j.conb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468(7321):214–22. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 51.Petryniak MA, et al. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55(3):417–33. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silbereis JC, et al. Olig1 function is required to repress dlx1/2 and interneuron production in Mammalian brain. Neuron. 2014;81(3):574–87. doi: 10.1016/j.neuron.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hand R, et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48(1):45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 54.Kwan KY, et al. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A. 2008;105(41):16021–6. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McEvilly RJ, et al. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295(5559):1528–32. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- 56.Nakamura K, et al. In vivo function of Rnd2 in the development of neocortical pyramidal neurons. Neurosci Res. 2006;54(2):149–53. doi: 10.1016/j.neures.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 57.Heng JI, et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455(7209):114–8. doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
- 58.Heng JI, et al. The Zinc Finger Transcription Factor RP58 Negatively Regulates Rnd2 for the Control of Neuronal Migration During Cerebral Cortical Development. Cereb Cortex. 2013 doi: 10.1093/cercor/bht277. [DOI] [PubMed] [Google Scholar]
- 59.Ohtaka-Maruyama C, et al. RP58 regulates the multipolar-bipolar transition of newborn neurons in the developing cerebral cortex. Cell Rep. 2013;3(2):458–71. doi: 10.1016/j.celrep.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 60.Ohtaka-Maruyama C, et al. The 5′-flanking region of the RP58 coding sequence shows prominent promoter activity in multipolar cells in the subventricular zone during corticogenesis. Neuroscience. 2012;201:67–84. doi: 10.1016/j.neuroscience.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 61.Xiang C, et al. RP58/ZNF238 directly modulates proneurogenic gene levels and is required for neuronal differentiation and brain expansion. Cell Death Differ. 2012;19(4):692–702. doi: 10.1038/cdd.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alcamo EA, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57(3):364–77. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 63.Britanova O, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57(3):378–92. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 64.Hevner RF, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29(2):353–66. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 65.Bedogni F, et al. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci U S A. 2010;107(29):13129–34. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Molyneaux BJ, et al. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47(6):817–31. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 67.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci U S A. 2005;102(47):17184–9. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen JG, et al. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci U S A. 2005;102(49):17792–7. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shim S, et al. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486(7401):74–9. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Srinivasan K, et al. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc Natl Acad Sci U S A. 2012;109(47):19071–8. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lai T, et al. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57(2):232–47. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 72.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45(2):207–21. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 73.McKenna WL, et al. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31(2):549–64. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tashiro K, et al. A mammalian conserved element derived from SINE displays enhancer properties recapitulating Satb2 expression in early-born callosal projection neurons. PLoS One. 2011;6(12):e28497. doi: 10.1371/journal.pone.0028497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toma K, Kumamoto T, Hanashima C. The timing of upper-layer neurogenesis is conferred by sequential derepression and negative feedback from deep-layer neurons. J Neurosci. 2014;34(39):13259–76. doi: 10.1523/JNEUROSCI.2334-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6(11):846–56. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 77.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45(6):592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buttner N, et al. Af9/Mllt3 interferes with Tbr1 expression through epigenetic modification of histone H3K79 during development of the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107(15):7042–7. doi: 10.1073/pnas.0912041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gyorgy AB, et al. SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur J Neurosci. 2008;27(4):865–73. doi: 10.1111/j.1460-9568.2008.06061.x. [DOI] [PubMed] [Google Scholar]
- 80.Yu Y, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152(1–2):248–61. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Namihira M, Nakashima K. Mechanisms of astrocytogenesis in the mammalian brain. Curr Opin Neurobiol. 2013;23(6):921–7. doi: 10.1016/j.conb.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 82.Azim E, et al. SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nat Neurosci. 2009;12(10):1238–47. doi: 10.1038/nn.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Batista-Brito R, et al. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron. 2009;63(4):466–81. doi: 10.1016/j.neuron.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ghanem N, et al. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J Neurosci. 2007;27(19):5012–22. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Malik AN, et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci. 2014 doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3(6) doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pruunsild P, et al. Identification of cis-elements and transcription factors regulating neuronal activity-dependent transcription of human BDNF gene. J Neurosci. 2011;31(9):3295–308. doi: 10.1523/JNEUROSCI.4540-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clark AG, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302(5652):1960–3. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- 90.Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3(6):e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437(7062):1153–7. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 92.McLean CY, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471(7337):216–9. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Prabhakar S, et al. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314(5800):786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 94.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188(4184):107–16. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 95.Haygood R, et al. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat Genet. 2007;39(9):1140–4. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- 96.Visel A, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152(4):895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pattabiraman K, et al. Transcriptional regulation of enhancers active in protodomains of the developing cerebral cortex. Neuron. 2014;82(5):989–1003. doi: 10.1016/j.neuron.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kwan KY, et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012;149(4):899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–21. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Talkowski ME, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149(3):525–37. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rosenfeld JA, et al. Small deletions of SATB2 cause some of the clinical features of the 2q33.1 microdeletion syndrome. PLoS One. 2009;4(8):e6568. doi: 10.1371/journal.pone.0006568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lamb AN, et al. Haploinsufficiency of SOX5 at 12p12.1 is associated with developmental delays with prominent language delay, behavior problems, and mild dysmorphic features. Hum Mutat. 2012;33(4):728–40. doi: 10.1002/humu.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155(5):997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–9. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Colantuoni C, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–23. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ramasamy A, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–28. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schizophrenia Psychiatric Genome-Wide Association Study, C. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43(10):969–76. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vernot B, et al. Personal and population genomics of human regulatory variation. Genome Res. 2012;22(9):1689–97. doi: 10.1101/gr.134890.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Psychiatric, G.C.B.D.W.G. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43(10):977–83. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wellcome Trust Case Control, C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shulha HP, et al. Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol. 2012;10(11):e1001427. doi: 10.1371/journal.pbio.1001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 115.Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol Cell Biol. 2008;28(1):227–36. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Johnson DS, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 117.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451(7180):796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 119.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Consortium EP, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–64. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cheung I, et al. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci U S A. 2010;107(19):8824–9. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lovatt D, et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods. 2014;11(2):190–6. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508(7495):199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Johnson MB, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62(4):494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pletikos M, et al. Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron. 2014;81(2):321–32. doi: 10.1016/j.neuron.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wenger AM, et al. The enhancer landscape during early neocortical development reveals patterns of dense regulation and co-option. PLoS Genet. 2013;9(8):e1003728. doi: 10.1371/journal.pgen.1003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vermunt MW, et al. Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep. 2014;9(2):767–79. doi: 10.1016/j.celrep.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 131.Fullwood MJ, et al. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19(4):521–32. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang J, et al. ChIA-PET analysis of transcriptional chromatin interactions. Methods. 2012;58(3):289–99. doi: 10.1016/j.ymeth.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 133.Kieffer-Kwon KR, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155(7):1507–20. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30(3):271–7. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]