SUMMARY
Recent studies identified cyclic GMP-AMP (cGAMP) as a metazoan second messenger triggering an interferon response. cGAMP is generated from GTP and ATP by cytoplasmic dsDNA sensor cGAMP synthase (cGAS). We combined structural, chemical, biochemical, and cellular assays to demonstrate that this second messenger contains G(2′,5′)pA and A(3′,5′)pG phosphodiester linkages, designated c[G(2′,5′) pA(3′,5′)p]. We show that, upon dsDNA binding, cGAS is activated through conformational transitions, resulting in formation of a catalytically competent and accessible nucleotide-binding pocket for generation of c[G(2′,5′)pA(3′,5′)p]. We demonstrate that cyclization occurs in a stepwise manner through initial generation of 5′-pppG(2′,5′)pA prior to cyclization to c[G(2′,5′)pA(3′,5′)p], with the latter positioned precisely in the catalytic pocket. Mutants of cGAS dsDNA-binding or catalytic pocket residues exhibit reduced or abrogated activity. Our studies have identified c[G(2′,5′)pA(3′,5′)p] as a founding member of a family of metazoan 2′,5′-containing cyclic heterodinucleotide second messengers distinct from bacterial 3′,5′ cyclic dinucleotides.
INTRODUCTION
The importance of cyclic dinucleotides as bacterial second messengers is well established, with cyclic di-GMP (c-di-GMP) (Ross et al. 1987) now acknowledged as a universal bacterial second messenger (reviewed in Römling et al., 2013). This versatile molecule has been shown to play key roles in cell cycle and differentiation, motility, and virulence, as well as in the regulation of biofilm formation and dispersion. Advances in our understanding of c-di-GMP have emerged with the identification, structural characterization, and mechanistic understanding of the catalytic activities of the bacterial enzymes responsible for the synthesis and degradation of this second messenger (reviewed in Schirmer and Jenal, 2009). Crystal structures of c-di-GMP in the free state (Egli et al., 1990) and when bound to enzymes responsible for its synthesis and degradation (reviewed in Römling et al., 2013) have shown that this second messenger can adopt either monomeric or dimeric bis-intercalated folds. It appears that formation of c-(3′,5′)-di-GMP from two molecules of GTP occurs via a two-step reaction and formation of 3′,5′-phosphodiester linkages, with two molecules of pyrophosphate as byproducts of the cyclization reaction. Moreover, multiple receptors targeted by c-(3′,5′)-di-GMP and the diverse ways that bacteria signal through this second messenger have been identified (reviewed in Krasteva et al., 2012). In parallel studies, c-(3′,5′)-di-GMP-specific riboswitches have also been identified (Sudarsan et al., 2008), including ones that are involved in cyclic dinucleotide-induced RNA splicing (Lee et al., 2010).
There is currently much interest toward gaining a molecular and functional understanding of innate immunity sensors of higher metazoans that recognize nucleic acids in the cytoplasm and trigger type I interferon induction (reviewed in Hornung and Latz, 2010; Keating et al., 2011). Cytoplasmic dsDNA of pathogenic bacterial or viral origin, and perhaps also displaced nuclear or mitochondrial DNA following cellular stress, represent such a trigger. These events involving self-nucleic acid recognition, in turn, could trigger autoimmune diseases such as systemic lupus erythematosus and Sjögren syndrome. Indeed, in recent years, many cytoplasmic DNA sensors have been identified, including DAI (DNA-dependent activator of IFN-regulatory factor) (Takaoka et al., 2007), LRRFIP1 (leucine-rich repeat and flight-less I interacting protein 1) (Yang et al., 2010), DDX41 (DEAD box polypeptide 41) (Zhang et al., 2011), and members of the HIN-200 (hematopoietic interferon-inducible nuclear proteins) family such as AIM2 (absent in melanoma 2) (Hornung et al., 2009; Bürckstümmer et al., 2009; Fernandes-Alnemri et al., 2009) and IFI16 (interferon-inducible protein 16) (Unterholzner et al., 2010). Molecular information is available on the HIN domain family, as reflected in structures of their complexes with dsDNA (Jin et al., 2012). A requirement for multiple sensors may be a reflection of distinctive cell-type-specific activities. Cytoplasmic detection of dsDNA activates stimulator of interferon genes (STING) in the cytoplasm (Ishikawa and Barber, 2008), which in turn initiates a cascade of events by first activating kinases IKK (IκB kinase) and TBK1 (TANK-binding kinase 1), leading to phosphorylation and activation of the transcription factors NF-κB (nuclear factor κB) and IRF3 (interferon regulatory factor). These phosphorylated transcription factors translocate to the nucleus to target immune and inflammatory genes, leading to the production of cytokines and type I interferons and thereby triggering the host immune response (reviewed O’Neill, 2013).
It has been shown that VSP-1 (Vibrio 7th pandemic island 1) genes encode a class of dinucleotide cyclases that preferentially synthesize a cyclic-GMP-AMP (designated cGAMP) molecule, thereby expanding our horizon to cyclic GA dinucleotides (Davies et al., 2012). More recently, cyclic GMP-AMP synthase (cGAS) was identified as a cytoplasmic DNA sensor that activated the type I interferon pathway by synthesizing the second messenger cGAMP (Sun et al., 2013; Wu et al., 2013). cGAS was shown to be a member of the nucleotidyltransferase family and to be capable of generating cGAMP in vitro from GTP and ATP in the presence of dsDNA (but not dsRNA), whereas chemically synthesized cGAMP containing a pair of 3′,5′ linkages was shown to stimulate the production of interferon. The authors also demonstrated through experiments involving either overexpression or knockdown of cGAS that cGAMP bound to and activated STING, resulting in the activation of transcription factor IRF3 and subsequent induction of interferon β.
A critical inherent assumption in these otherwise seminal studies on cGAS was that cGAMP contained a pair of 3′,5′ linkages (Sun et al., 2013; Wu et al., 2013), in line with those reported previously for c-di-GMP in bacterial systems as outlined above. In this paper, we have combined structural, chemical, in vitro biochemical, and in vivo cellular assays to establish unequivocally that this second messenger contains 2′,5′ linkage at the GpA step and 3′,5′ linkage at the ApG step, designated c[G(2′,5′)pA(3′,5′)p], thus identifying the founding member of a new family of 2′,5′ linkage-containing metazoan second messengers regulating type I interferon induction in response to cytoplasmic DNA.
RESULTS
Structure of Cyclic GMP-AMP Synthase
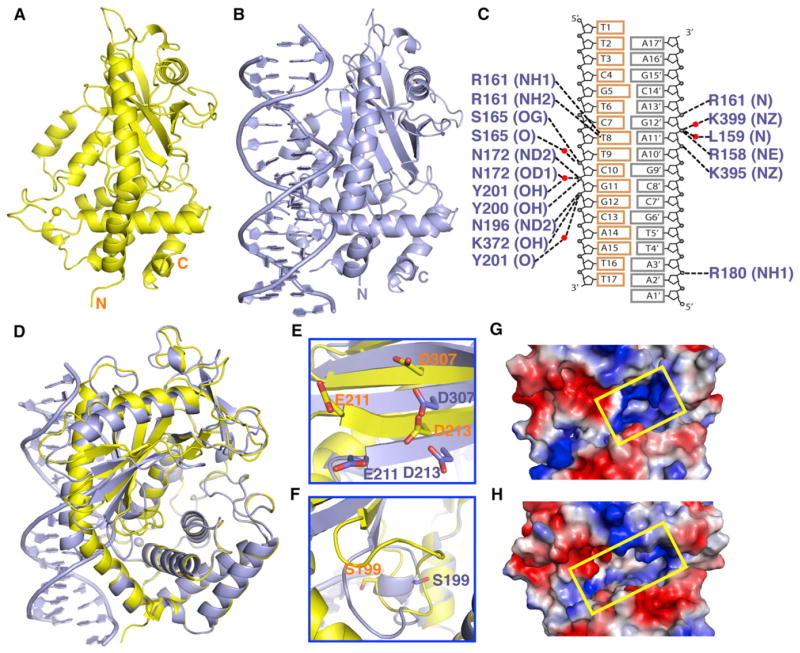
We have solved the 2.0 Å crystal structure of cyclic GMP-AMP synthase (cGAS; construct 147–507) (Figure S1A available online) in the free state (Figures 1A and S1B). The protein adopts a bilobal scaffold with mixed α/β topology (Figure 1A; X-ray statistics in Table S1) characteristic of members of the nucleotidyl-transferase superfamily. A DALI search identified ILF2/NF45, which contains a nucleotidyltransferase fold (PDB: 4AT8) (Wolkowicz and Cook, 2012), as most closely resembling the fold of cGAS, with a Z score of 15.1 and rmsd of 3.8 Å. In addition, the free state of human oligoadenylate synthetase 1 (OAS1) (PDB: 1PX5) (Hartmann et al., 2003) exhibited a Z score of 13.3 and a rmsd of 4.1 Å (Figure S1C).
Figure 1. Structures of cGAMP Synthase in the Free State and Bound to dsDNA.
(A) 2.0 Å crystal structure of cGAMP synthase (cGAS) in the free state. The backbone of the protein in a ribbon representation is colored in yellow.
(B) 2.1 Å crystal structure of cGAS bound to a complementary 16 bp DNA duplex (with one base 5′ overhang at each end). The protein and DNA are colored in blue in the binary complex.
(C) A schematic of intermolecular hydrogen bonds in the binary cGAS-DNA complex.
(D) Superposed structures of cGAS in the free state (yellow) and in the cGAS-DNA complex (blue).
(E and F) Large changes within the β sheet (E) and catalytic pocket (F) segments were observed for the transition from cGAS in the free state (yellow) to the binary complex with bound DNA (blue).
(G) Narrow entrance to the catalytic pocket in the structure of cGAS in the free state, with the protein in an electrostatic representation.
(H) Widened entrance to the catalytic pocket in the structure of the binary cGAS-DNA complex.
See also Figures S1 and S2 and Table S1.
Structure of Binary Complex of cGAS with Bound dsDNA
We have cocrystallized cGAS bound to a 16 bp complementary dsDNA (plus 1 nt 5′ overhang at either end) and solved the structure of the binary complex at 2.1 Å resolution (X-ray statistics in Table S1). The structure of the binary complex is shown in Figure 1B. The majority of the intermolecular contacts in the binary cGAS-dsDNA complex (summarized in Figure 1C) are between cGAS and the sugar-phosphate backbone of the DNA (Figures S2A and S2B), with only one base-specific contact (Figure S2B). The superposed structures of cGAS in the free (yellow) and dsDNA-bound (blue) states are shown in Figure 1D. There are large conformational changes on formation of the binary dsDNA complex (Figures S2C–S2E), as can be seen within a β sheet segment containing catalytic Glu211, Asp213, and Asp307 residues (Figure 1E), as well for loop and helical segments within the catalytic pocket containing Ser199 (Figure 1F). Equally important, a very narrow entrance leads to the catalytic pocket for cGAS in the free state (Figure 1G), whereas this entrance widened significantly in the binary complex with DNA (Figure 1H). The cGAS fold in the dsDNA-bound state is similar to that reported recently for the OAS1 in the dsRNA bound state plus 2′-dATP (PDB: 4IG8) (Donovan et al., 2013) (Z score, 18.2; rmsd, 3.2 Å; Figure S2D).
Structure of Ternary Complex of cGAS with dsDNA and Bound ATP
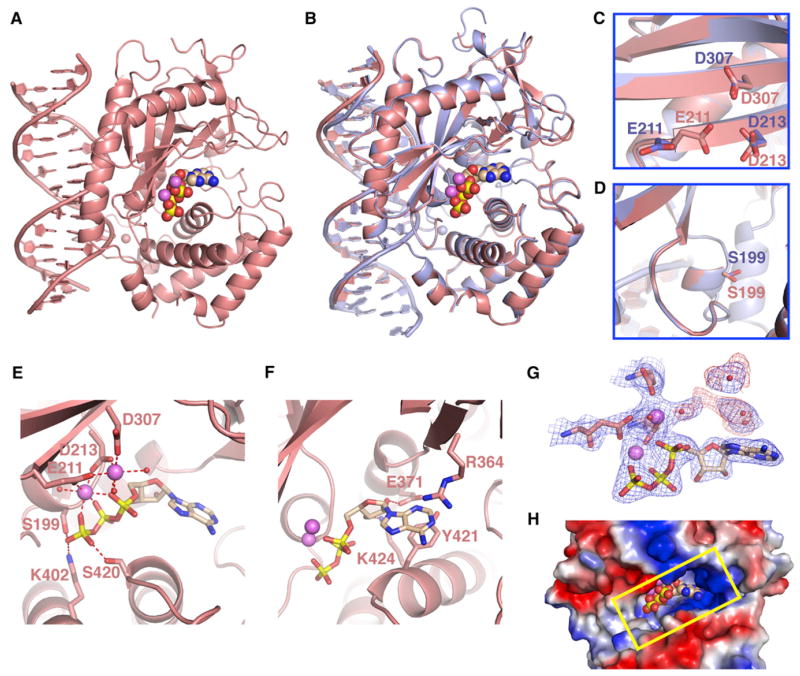
We have cocrystallized cGAS bound to dsDNA and ATP and have solved the structure of the ternary complex at 2.4 Å resolution. The ATP is bound in the catalytic pocket positioned within the interior of the cGAS in the ternary complex (Figure 2A). There is close superposition of the binary complex of cGAS and dsDNA (blue), with the ternary complex containing bound ATP (salmon) as shown in Figure 2B. Minimal conformational changes are observed within the β sheet segment carrying the catalytic acidic residues (Figure 2C) and the loop and helix segments forming the catalytic pocket (Figure 2D) on ternary complex formation. The only notable change is the movement of the side chain of Glu211 toward the other two acidic residues in the ternary complex (Figure 2C). The γ-phosphate of the triphosphate group of ATP is hydrogen bonded to polar side chains (Ser199, Ser420, and Lys402), and two bound cations (tentatively assigned to Mg2+) serve a bridging role for interactions between the α- and β-phosphate of the triphosphate and side chains of catalytic acidic residues (Glu211, Asp213, and Asp307) (Figure 2E). In addition, the adenine ring of bound ATP stacks over Tyr421 in one direction and partially over the guanidinium group of Arg364 in the other direction (Figure 2F).
Figure 2. Structure of the Ternary Complex of cGAS, dsDNA, and ATP.
(A) 2.4 Å crystal structure of the ternary complex of cGAS bound to dsDNA and ATP. The protein and dsDNA are colored in salmon in the ternary complex, with bound ATP in a space-filling representation.
(B) Superposed structures of the binary complex of cGAS and DNA (blue) and the ternary complex with added ATP (salmon).
(C and D) Absence of changes in the backbone within the β sheet (C) and catalytic pocket (D) segments were observed for the transition from the binary cGAS and dsDNA complex (blue) to the ternary complex with added ATP (salmon).
(E and F) Two alternate views of intermolecular contacts between ATP and catalytic pocket residues in the ternary complex. Two cations are shown as magenta spheres, with hydrogen bonds shown by dashed red lines.
(G) 2Fo-Fc electron density map contoured at 1.2σ (blue) and Fo-Fc map contoured at 3.0σ (red) of bound ATP and a pair of cations and coordinating residues in the catalytic pocket of the ternary complex. This map contains some weak unaccounted for electron density (red).
(H) View of bound ATP in a space-filling representation within the catalytic pocket, with the protein in an electrostatic representation.
See also Table S2.
It should be noted that we observe additional weak electron density (red contours in Figure 2G) that is unaccounted for at this time in the 2.4 Å structure of the ternary complex. The additional density could be either water molecules or an AMP molecule with modest (30%) occupancy. A view of the bound ATP looking into the catalytic pocket of the ternary complex is shown in Figure 2H.
Structure of Ternary Complex of cGAS with dsDNA and Bound 5′-pppG(2′,5′)pG
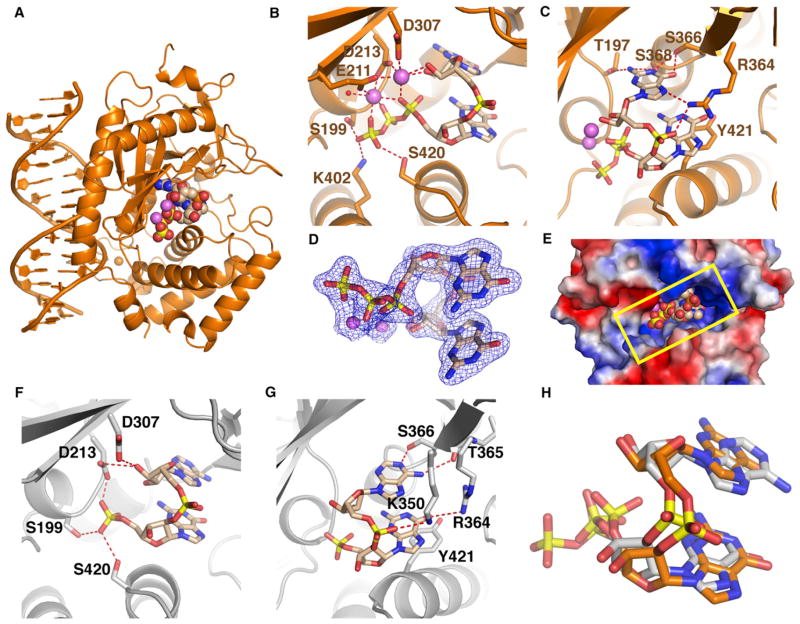
We have cocrystallized cGAS bound to dsDNA and GTP and solved the structure of the ternary complex at 1.9 Å resolution. The structure of the ternary complex is shown in Figure 3A (X-ray statistics in Table S2). Of note, phosphodiester bond formation has occurred in the catalytic pocket, yielding the bound ligand 5′-pppGpG (shown positioned in the catalytic pocket in space-filling representation in Figure 3A). Importantly, minimal conformational changes are associated with the transition from the binary complex to the ternary complex with bound 5′-pppGpG (Figures S3A–S3C).
Figure 3. Structure of the Ternary Complex of cGAS and dsDNA with Bound 5′-pppG(2′,5′)pG and 5′-pG(2′,5′)pA.
(A) 1.9 Å crystal structure of the ternary complex of cGAS bound to dsDNA and 5′-pppG(2′,5′)pG. The protein and DNA are colored in orange in the ternary complex, with bound 5′-pppG(2′,5′)pG in a space-filling representation.
(B and C) Two alternate views of intermolecular contacts between 5′-pppG(2′,5′)pG and catalytic pocket residues in the ternary complex. Two cations are shown as magenta spheres, with hydrogen bonds shown by dashed red lines.
(D) 2Fo-Fc electron density map contoured at 1.2σ (blue) of bound 5′-pppG(2′,5′)pG in the catalytic pocket of the ternary complex.
(E) View of bound 5′-pppG(2′,5′)pG in a space-filling representation within the catalytic pocket, with the protein in an electrostatic representation.
(F and G) Two alternate views of intermolecular contacts between 5′-pG(2′,5′)pA and catalytic pocket residues in the 2.3 Å ternary complex of cGAS, dsDNA, and GMP + ATP.
(H) Superposition of structures of bound 5′-pppG(2′,5′)pG (orange) and 5′-pG(2′,5′)pA (silver).
See also Figure S3 and Tables S2 and S3.
Strikingly, the GpG linkage of 5′-pppGpG is 2′,5′ rather than the anticipated 3′,5′, with the first and second G residues in addition adopting syn and anti glycosidic torsion orientations, respectively (Figure S3D). The triphosphate group of 5′-pppG(2′,5′)pG is coordinated to two cations (Figure 3B), with the first G stacked on Tyr421. The second G uses its Watson-Crick edge to hydrogen bond with polar side chains (Thr197, Ser366, and Ser368), and its Hoogsteen edge to hydrogen bond with Arg364 (Figure 3C). The observed 5′-pppG(syn)(2′,5′)pG(anti) topology can be traced with a high degree of confidence (2Fo-Fc map in Figure 3D, with two views of the Fo-Fc omit map shown in Figure S3E). A view of the bound 5′-pppG(2′,5′)pG looking into the catalytic pocket of the ternary complex is shown in Figure 3E. We have superposed the structures of bound 5′-pppG(2′,5′)pG (biscuit) and ATP (cyan) in their respective ternary complexes with cGAS and dsDNA and observe that the first G of the bound 5′-pppG(2′,5′)pG is positioned in the plane of the bound ATP (Figure S3F). The two bound cations have been tentatively assigned to Mg2+ based on omit maps (Figure S3G) and the octahedral coordination around each cation (Figures S3H and S3I).
We also grew crystals of the ternary complex with 3′-dGTP and observed formation of the related 5′-pppdG(2′,5′)pdG intermediate (cannot form a 3′,5′ linkage) in the 2.1 Å structure of this complex (X-ray statistics in Table S2).
Structure of Ternary Complex of cGAS with dsDNA and Bound 5′-pG(2′,5′)pA
We have also cocrystallized cGAS in the presence of dsDNA, GMP, and ATP and solved the structure of a complex at 2.3 Å resolution (structural statistics in Table S3). By using GMP rather than GTP, we hoped to trap the intermediate following formation of the first phosphodiester bond and indeed observed the bound linear product of 5′-pG(syn)(2′,5′)pA(anti) (Figures 3F and 3G). No cations were observed in the absence of a triphosphate moiety in the product. Of note, attempts at cocrystallization of cGAS with dsDNA, GTP, and AMP only yielded crystals that diffracted very poorly (12 Å resolution). We observed good superposition of the intermediates 5′-pppG(syn)(2′,5′)pG(anti) (in orange) and 5′-pG(syn)(2′,5′)pA(anti) (in silver), as shown in Figure 3H.
Structure of Ternary Complex of cGAS with dsDNA and Bound c[G(2′,5′)pA(3′,5′)p]
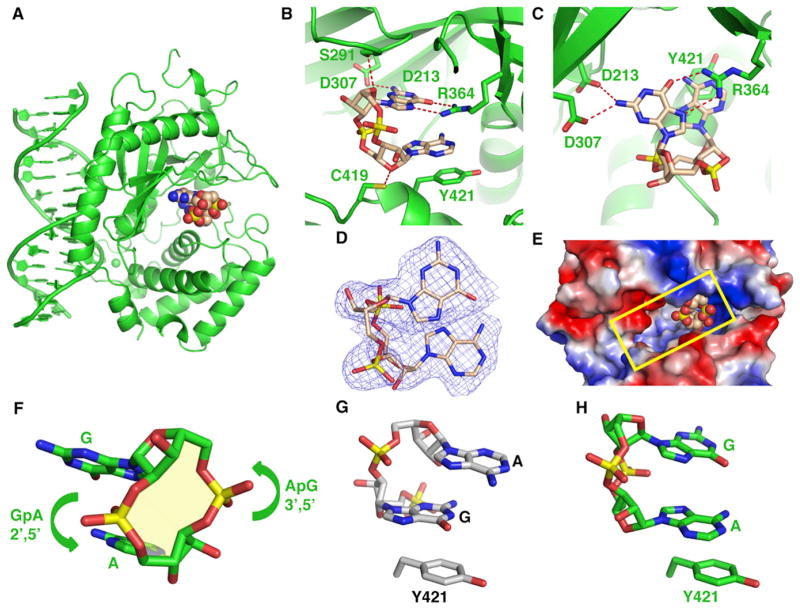
We have cocrystallized cGAS with dsDNA, GTP, and ATP and have solved the structure of the complex at 2.3 Å resolution. These crystals took 2 weeks to grow, unlike other crystals mentioned above that grew within a few days. The structure of the ternary complex is shown in Figure 4A (X-ray statistics in Table S3). Most unexpectedly, the bound small ligand shown in a space-filling representation in Figure 4A is a cyclic dinucleotide. Of note, no conformational changes are associated with the transition from the binary complex to the ternary complex with bound cyclic dinucleotide, with even the side chain of Glu211 adopting identical orientations (Figures S4A–S4C).
Figure 4. Structure of the Ternary Complex of cGAS and DNA with Bound Product c[G(2′,5′)pA(3′,5′)p].
(A) 2.3 Å crystal structure of the ternary complex of cGAS bound to dsDNA and product c[G(2′,5′)pA(3′,5′)p]. The protein and DNA are colored in green in the ternary complex, with bound product c[G(2′,5′)pA(3′,5′)p] in a space-filling representation.
(B and C) Two alternate views of intermolecular contacts between product c[G(2′,5′)pA(3′,5′)p] and catalytic pocket residues in the ternary complex.
(D) 2Fo-Fc electron density map contoured at 1.2σ (blue) of bound c[G(2′,5′)pA(3′,5′)p] in the catalytic pocket of the ternary complex.
(E) View of bound c[G(2′,5′)pA(3′,5′)p] in a space-filling representation positioned toward one end of the catalytic pocket, with the protein in an electrostatic representation.
(F) A view of c[G(2′,5′)pA(3′,5′)p] highlighting the 2′,5′ linkage at the GpA step and the 3′,5′ linkage at the ApG step.
(G) Stacking of the G residue of 5′-pG(2′,5′)pA on Tyr 421 in its ternary complex with cGAS and dsDNA.
(H) Stacking of the A residue of c[G(2′,5′)pA(3′,5′)p] on Tyr 421 in its ternary complex with cGAS and dsDNA.
Importantly, we can trace the GpA step in the bound cyclic dinucleotide without ambiguity (the 3′ OH of G can be traced) and establish that this linkage is 2′,5′ (Figure S4D). On the other hand, the linkage at the ApG step in the bound cyclic dinucleotide could be either 2′,5′ or 3′,5′ based on the observed density and cannot be assigned with certainty solely based on structure. We have undertaken the refinement with a 3′,5′ linkage at the ApG step based on evidence outlined later and have prepared the drawings in Figures 4 and S4 with 2′,5′ linkage at the GpA step and 3′,5′ linkage at the ApG step. We can distinguish G from A based on the observed density for the two-amino group of G and note that both adopt anti alignments in the bound cyclic dinucleotide c[G(2′,5′)pA(3′,5′)p] (Figure S4D). The A residue is stacked on Tyr421 (Figures 4B and 4C), whereas the G residue is anchored in place through hydrogen bonding to the side chains of Asp213, Asp307, and Arg364 (Figures 4B and 4C). Further, the A and G residues partially stack on each other. The 2Fo-Fc electron density for the bound c[G(2′,5′)pA(3′,5′)p] is shown in Figure 4D, with omit maps shown in Figure S4E. A view of the bound c[G(2′,5′)pA(3′,5′)p] looking into the catalytic pocket of the ternary complex is shown in Figure 4E, with the c[G(2′,5′) pA(3′,5′)p] bound toward one end of the opening. We also do not observe bound cations, given that c[G(2′,5′)pA(3′,5′)p] does not contain triphosphates, and the G base directly coordinates with Asp213 and Asp307 (Figure 4C). We have superposed the structures of bound c[G(2′,5′)pA(3′,5′)p] (biscuit) and ATP (cyan) in their respective ternary complexes with cGAS and dsDNA and observe that the A of the bound c[G(2′,5′)pA(3′,5′) p] is positioned in the plane of the bound ATP (Figure S4F).
A view of c[G(2′,5′)pA(3′,5′)p] highlighting the 2′,5′ linkage at the GpA step and the 3′,5′ linkage at the ApG step is shown in Figure 4F. We note that, in the ternary complex with 5′-pG(2′,5′)pA linear product, it is the G base that stacks on Tyr421 (Figures 3G and 4G), whereas in the ternary complex with c[G(2′,5′)pA(3′,5′)p] product, it is the A base that stacks on Tyr421 (Figures 4C and 4H). Thus, the linear product and cyclic final product are related by a flip-over within the catalytic pocket.
Biochemical Characterization of cGAS Activity
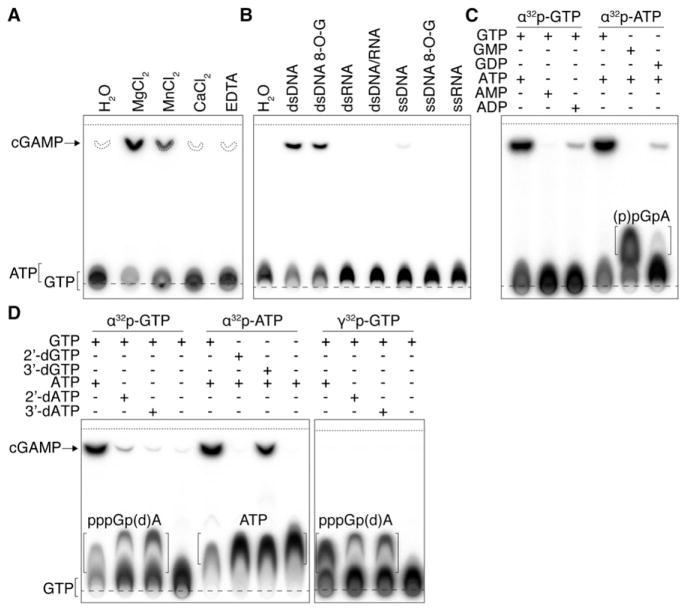
To validate the structural results, we established an activity assay using thin-layer chromatography (Figure S5) and monitored cyclic dinucleotide c[G(2′,5′)pA(3′,5′)p] formation from ATP and GTP using purified recombinant full-length and truncated cGAS proteins. cGAS required the presence of dsDNA and Mg2+ or Mn2+ for activity (Figures 5A–5B). We tested c[G(2′,5′)pA(3′,5′)p] formation as a function of dsDNA length and found that dsDNA of 36 bp or longer were optimal, yet the 16 bp dsDNA used for crystallography elicited some activity (Figure S6A). Double-stranded RNA, a DNA/RNA duplex, or single-stranded DNA or RNA did not stimulate cyclic dinucleotide formation (Figure 5B). The trace amount of c[G(2′,5′)pA(3′,5′)p] detected for the specific ssDNA used in this experiment was attributable to a stretch of sequence complementarity, and the substitution of G by 8-oxoguanine (8-O-G) was sufficient to destabilize its predicted interaction and eliminate the residual cGAS activity. Replacement of guanine by 8-O-G within the dsDNA did not alter cGAS activity.
Figure 5. Characterization of c[G(2′,5′) pA(3′,5′)p] Formation by cGAS.
Generation of c[G(2′,5′)pA(3′,5′)p] and linear products and intermediates were assayed by thin-layer chromatography (TLC) using purified recombinant truncated (A, amino acids 147–507, used in crystallization studies) and full-length cGAS (B–D, amino acids 1–507). Long- and short-dashed lines indicate the origin and solvent fronts, respectively.
(A) A 45 nt dsDNA was incubated with truncated cGAS in reaction buffer containing indicated divalent cation (or EDTA) and α32p-ATP and -GTP. Chemically synthesized cGAMP containing both 3′,5′ linkages was cospotted in every sample, and its migration, visualized by UV, is indicated (dashed outlines).
(B) cGAS was incubated with single (ss)-stranded or double (ds)-stranded DNA, RNA, DNA/RNA duplex, or 8-oxoguanine (8-O-G)-modified DNA of similar sequence, and c[G(2′,5′)pA(3′,5′)p] formation was monitored using α32p-ATP.
(C) Mono- and diphosphorylated adenosine and guanosine were used as substrates to determine the order of c[G(2′,5′)pA(3′,5′)p] formation. Slow-migrating 2′,5′-linked species was observed when cGAS and dsDNA is incubated with α32p-ATP and GMP (5′-pGpA) or GDP (5′-ppGpA).
(D) dsDNA-dependent cGAMP reaction intermediates were visualized by using 2′ or 3′ dATP and dGTP. Slow-migrating intermediate species, corresponding to pppGpA (lane 1) or pppGpdA (lanes 2 and 3), are seen by changing TLC mobile-phase composition. Intermediate species were confirmed using γ32p-GTP.
See also Figures S5 and S6 and Table S6.
We quantified the activity of cGAS to yield c[G(2′,5′)pA(3′,5′)p] under multiple turnover conditions. Over 78% (SD ± 2.6%, n = 5) of the original ATP and GTP provided was converted to c[G(2′,5′) pA(3′,5′)p] within 40 min, leading to an estimated observed rate constant of 0.19 min−1 and involving >750 turnovers per enzyme molecule.
To determine the order of intermediate formation, we first substituted GTP by GMP or GDP (Figure 5C). Both compounds led to the formation of the respective 5′-pG(2′,5′)pA product and 5′-ppG(2′,5′)pA intermediates, and only 5′-ppG(2′,5′)pA could react further to yield a reduced amount of the cyclic dinucleotide. Substitution of ATP by ADP or AMP resulted in no product or intermediate formation, with only ADP (with GTP) leading to the generation of reduced levels of c[G(2′,5′)pA(3′,5′)p].
To determine the involvement of 2′ or 3′-hydroxyl (OH) groups of GTP and ATP for formation of c[G(2′,5′)pA(3′,5′)p], we tested 2′ and 3′ deoxyguanosine triphosphate and 2′ and 3′ deoxyadenosine triphosphate as substrates for cGAS (Figures 5D and S6B). 2′-dGTP was unable to form a cyclic dinucleotide, unlike 3′-dGTP, indicating that the 2′ OH of guanosine was required for the formation of the linkage with the α-phosphate at the 5′ position of adenosine. In contrast, both 2′- and 3′-dATP led to markedly reduced formation of cyclic GA dinucleotides, although 2′-dATP + GTP yielded noticeably more product. In both cases, we observed accumulation of 5′-pppG(2′,5′)pdA reaction intermediates (Figure 5D, lanes 2 and 3), which migrated slightly faster than the all-ribose intermediate (5′-pppG(2′,5′)pA, lane 1).
Identification of c[G(2′,5′)pA(3′,5′)p] as the Product Formed by dsDNA-Dependent cGAS Activity
The syntheses and purification of the three isomeric cGAMP molecules, c[G(2′,5′)pA(2′,5′)p] (2′,5′ linkages at GpA and ApG steps) 6, c[G(2′,5′)pA(3′,5′)p] (2′,5′ at GpA step and 3′,5′ at ApG step) 11, and c[G(3′,5′)pA(3′,5′)p] (3′,5′ at GpA and ApG steps) 15 shown in Figure S7 were carried out using procedures previously reported by the Jones laboratory (Gaffney et al., 2010; Gaffney and Jones, 2012). The identity of the three isomeric cGAMP molecules 6, 11, and 15 (Figure S7) was validated from heteronuclear NMR analysis using through-bond connectivities. The experimental NMR data for c[G(2′,5′)pA(3′,5′)p] are outlined in Figure S8, with the proton and carbon chemical shifts for all three isomeric cGAMP molecules listed in Table S4.
We analyzed the product generated by dsDNA-dependent cGAS activity using reverse-phase high-performance liquid chromatography (HPLC) and compared its elution profile to chemically synthesized c[G(3′,5′)pA(3′,5′)p], c[G(2′,5′)pA(2′,5′) p], and c[G(2′,5′)pA(3′,5′)p] compounds (Figures 6A and S6D). A prominent peak consistently eluted from the HPLC system at precisely 23.5 min, which corresponded to the elution profile of c[G(2′,5′)pA(3′,5′)p]. Coinjection with c[G(3′,5′)pA(3′,5′)p] or c[G(2′,5′)pA(2′,5′)p] demonstrated that the cGAS reaction product does not coelute, unlike coinjection with chemically synthesized c[G(2′,5′)pA(3′,5′)p].
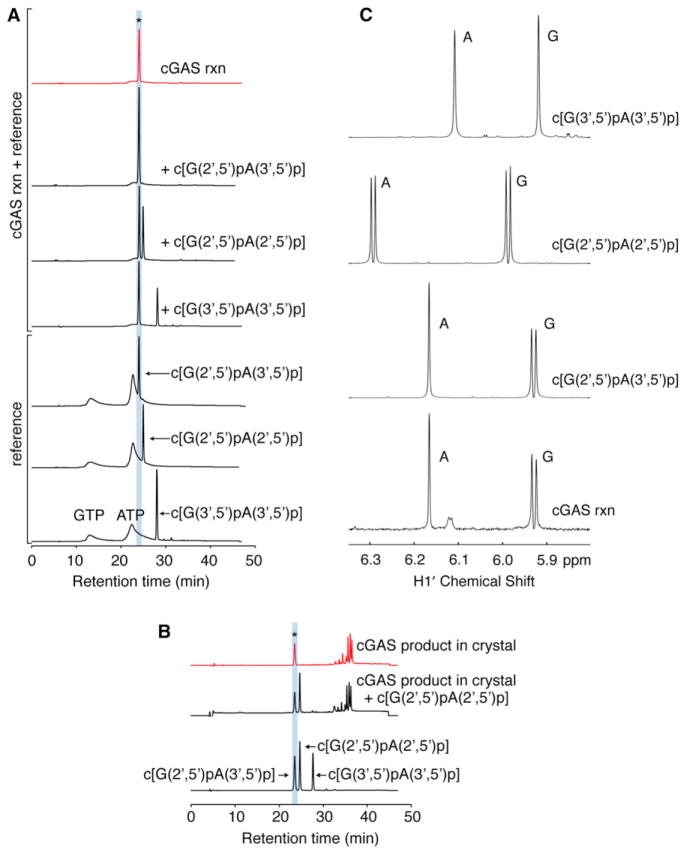
Figure 6. Definitive Identification of c[G(2′,5′)pA(3′,5′)p] as the Enzymatic Product of cGAS.
(A) UV 260 nm chromatographs of GTP, ATP, c[G(2′,5′)pA(2′,5′)p], c[G(3′,5′)pA(3′,5′)p], c[G(2′,5′) pA(3′,5′)p], and cGAS reaction (rxn, asterisk) solutions from reverse-phase HPLC analyses. cGAS reaction samples were injected alone (red) or with addition of indicated reference standards. Shaded region shows the retention time corresponding to the elution of c[G(2′,5′)pA(3′,5′)p].
(B) UV 260 nm chromatographs from HPLC analysis of the cGAS product obtained from dissolved crystals when injected alone (top trace, red) or coinjected with c[G(2′,5′)pA(2′,5′)p] reference compound (middle trace). Additional unidentified peaks were present in the dissolved crystal solution but elute later. The three reference cGAMP compounds were coinjected due to a change (0.5 s) in the retention time of c[G(2′,5′)pA(3′,5′)p] as a result of applying the dissolved crystal solution to the column.
(C) NMR spectra of the sugar H1′ proton region of three chemically synthesized cGAMP reference compounds with the cGAS rxn in 99.9% D2O in 10 mM K2HPO4-KH2PO4 (pH 6.6) buffer. The NMR spectrum of the cGAS rxn corresponds to c[G(2′,5′)pA(3′,5′)p] reference compound. The H1′ proton is a doublet (3JHH = 9 Hz) when the phosphate is attached to the 2′ position but a singlet when the phosphate is attached to the 3′ position, reflecting the different puckers of the five-membered sugar ring dependent on the position of the attached phosphate group.
See also Figures S7 and S8 and Table S4.
To demonstrate that the in-vitro-produced cyclic dinucleotide matched the molecule determined crystallographically, we analyzed by HPLC the dissolved cGAS crystals that had been co-incubated with DNA, ATP, and GTP (Figure 6B). A peak corresponding to c[G(2′,5′)pA(3′,5′)p] was observed, distinct from the c[G(2′,5′)pA(2′,5′)p] coinjected reference molecule as before. Additional unidentified peaks of longer retention times were also seen. Presumably, these unidentified compounds originating from the crystallization buffer and/or additives were not completely removed despite washing the crystals prior to HPLC analysis.
In addition, cGAS-generated c[G(2′,5′) pA(3′,5′)p] was purified by HPLC and subjected to one-dimensional NMR analysis. Its NMR spectrum in the sugar H1′ region is identical to that of chemically synthesized standard c[G(2′,5′)pA(3′,5′)p] and distinct from chemically synthesized c[G(2′,5′)pA(2′,5′)p] and c[G(3′,5′) pA(3′,5′)p] (Figure 6C). Thus, both HPLC (Figure 6A) and NMR (Figure 6C) independently validate that the product generated by cGAS is c[G(2′,5′)pA(3′,5′)p].
Functional Analysis of cGAS Mutant Proteins
We next assayed the biochemical and functional consequences of mutations on cGAS in its capacity to form c[G(2′,5′)pA(3′,5′)p] in vitro and to stimulate the type I interferon pathway in cells. We generated alanine-substitution mutants of the corresponding amino acid residues shown to be involved either in dsDNA binding or in cGAS activity in the crystal structures of the binary and ternary complexes. For in vitro cGAS activity assays, we generated and purified six recombinant mutant cGAS forms; four were predicted to eliminate dsDNA binding, and two point mutant proteins were substituted with alanine at potentially key catalytic residues. Incubation of DNA with mutant cGAS proteins led to little or no c[G(2′,5′) pA(3′,5′)p] formation for all but two mutants (R161A and S199A; Figure 7A).
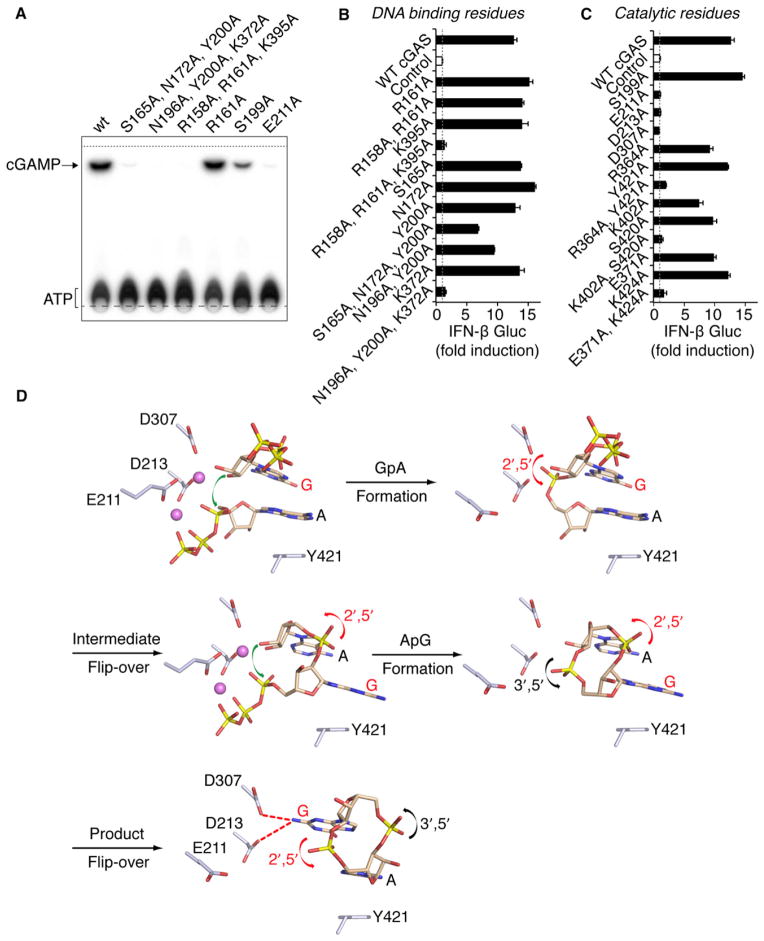
Figure 7. Functional Analysis of cGAS Mutants and Model for Two-Step Generation of c[G(2′,5′)pA(3′,5′)p].
(A) Levels of c[G(2′,5′)pA(3′,5′)p] formation by cGAS full-length WT and indicated mutants were compared by TLC analysis. Long- and short-dashed lines indicate the origin and solvent fronts, respectively.
(B and C) Expression vectors of murine cGAS WT or carrying single and multiple alanine mutations of DNA binding (B) and catalytic (C) residues were transiently transfected into HEK293 cells together with an IFN-β Gluc reporter and constitutive STING and Firefly luc expression plasmids. In this setting, expressed cGAS is engaged in the cytosol by the cotransfected DNA plasmids. Gluc values were determined in triplicate 36 hr after transfection, were normalized to Firefly luc, and are shown as fold induction over control plasmid (as mean ± SEM). Data in (B) and (C) are representative of three to five independent experiments for each mutant.
(D) A schematic representation of a proposed model associated with a two-step generation of c[G(2′,5′)pA(3′,5′)p] within the single catalytic pocket of cGAS. In this model, the first step involves formation of a 5′-pppGpA intermediate followed by formation of c[G(2′,5′)pA(3′,5′)p]. Note also that the bound ligand is predicted to undergo two flip-overs on the pathway to c[G(2′,5′)pA(3′,5′)p] formation.
To assess the impact on cGAS function in cells, we generated additional alanine mutants of cGAS for expression in mammalian cells. The full-length cGAS mutants together with STING and an IFN-β luciferase reporter were transiently expressed in HEK293 cells. In this assay, cGAS is engaged by the cotransfected DNA plasmids, and WT cGAS expression resulted in close to 15-fold enhanced luciferase activity compared to a control plasmid (Figure 7B). Single mutations of DNA-binding residues, including Arg161 responsible for the only direct interaction with a DNA base, were not sufficient to impair cGAS activity. However, ablation of interactions with two or three consecutive phosphodiesters in either DNA strand (Figures 1C, S2B, and S2C) resulted in diminished or entirely abrogated cGAS function (Figure 7B). At the catalytic site, single mutants Glu211, Asp213, or Asp307 affecting the binding of divalent cations (Figures 2E and 3B) all resulted in nonfunctional cGAS (Figure 7C). Furthermore, abrogation of cGAS activity required mutation of both amino acid residues involved in: (1) the binding of ATP (or GTP) γ phosphate (Lys402, Ser420; Figures 2E and 3B), (2) the binding of ATP adenosine (Glu371, Lys424; Figure 2F), or (3) the base stacking of ATP and c[G(2′,5′)pA(3′,5′)p] (Arg364, Tyr 421; Figures 2F and 4B); single mutants of these residues only slightly impaired cGAS function (Figure 7C). Gly198 and Ser199 are highly conserved residues that were found to undergo significant conformational changes upon ligand binding (Figures 1F and 2D). Nevertheless, single mutations G198A and S199A did not severely impair cGAS function, but the double mutant of these positions resulted in loss of the majority of its activity (Figure S6E). Similarly, conversion of the highly mobile Gly198 to sterically restricted proline abrogated cGAS activity (Figure S6E).
DISCUSSION
Conformational Transitions in cGAS on Complex Formation
Our structural studies highlight the fact that cGAS undergoes a pronounced conformational change upon binding of dsDNA (Figure 1D), whereby it repositions catalytic residues Glu211, Asp213, Asp307, and Ser199 (Figures 1E and 1F) while simultaneously opening access to the catalytic pocket (Figures 1G and 1H). In essence, cGAS adopts a catalytically competent conformation only when engaging dsDNA, thereby accounting for its role as a cytosolic dsDNA sensor. By contrast, only minimal conformational changes that are restricted to the side chain of Glu211 are observed when proceeding from the binary complex of cGAS and dsDNA to the ternary complex with ATP (Figures 2B–2D) and GTP (where the pocket contains off-pathway intermediate 5′-pppG(2′,5′)G; Figures S3A–S3C), with no change observed even for the side chain of Glu211 on formation of the ternary complex with ATP + GTP (where the pocket contains product c[G(2′,5′)pA(3′,5′)p]; Figure S4A–S4C).
Phosphodiester Bond Formation in Catalytic Pocket
The structural and functional experiments established phosphodiester bond formation in the catalytic pocket after binding of dsDNA to cGAS in the absence of any additional components. The structural studies on cGAS incubated with dsDNA and GTP identified accumulation of the linear reaction intermediate 5′-pppG(2′,5′)pG (Figures 3B and 3C). Related structural studies on the ternary complex in the presence GMP and ATP identified accumulation of the linear reaction product 5′-pG(2′,5′)pA (Figures 3F and 3G). We predict that the former intermediate is off-pathway and thus impaired for the formation of the second phosphodiester bond to form a cyclic product, given that the first G is syn, and the distance is long between the 2′ OH (or 3′ OH) of the second G and the α phosphate of the triphosphate moiety. Nevertheless, these results suggest that formation of c[G(2′,5′) pA(3′,5′)p] is likely to occur in a stepwise manner, involving formation of sequential phosphodiester bonds to yield the cyclic dinucleotide product. By contrast, structural studies on cGAS in the presence of dsDNA, GTP, and ATP resulted in formation of c[G(2′,5′)pA(3′,5′)p] (Figures 4B and 4C), without accumulation of an intermediate and consistent with an on-pathway reaction involving formation of a pair of sequential phosphodiester linkages.
Positioning of G and A Residues of Bound c[G(2′,5′) pA(3′,5′)p]
The G and A residues of c[G(2′,5′)pA(3′,5′)p] adopt distinct positions in the structure of the ternary complex with cGAS and dsDNA. The A residue of the bound c[G(2′,5′)pA(3′,5′)p] is stacked on Tyr421 (Figure 4B) and occupies the position of the adenine ring in the ATP complex (Figure 2F) and the first base in the 5′-pppG(2′,5′)pG (Figure 3C) and 5′-pG(2′,5′)pA (Figure 3G) complexes. The A residue of bound c[G(2′,5′)pA(3′,5′)p] is not involved in any intermolecular hydrogen bonds and hence could potentially be replaced by even a pyrimidine (C or U) residue. By contrast, the G residue of bound c[G(2′,5′)pA(3′,5′)p], which is partially stacked over the A residue, forms a network of intermolecular hydrogen bonds involving its Watson-Crick and Hoogsteen edges (Figures 4B and 4C) and cannot be replaced by any of the other three bases (C, A, and U). Thus, the cGAS-binding pocket has distinct recognition elements that distinguish between G and A and thus can bind c[G(2′,5′)pA(3′,5′)p] in a unique orientation.
In agreement with the crystallographic data, the biochemical results indicate a strong preference for GTP, consistent with the elaborate amino acid interactions observed in the structure between cGAS and this base. Incubation of cGAS with GTP alone, as well as GTP plus either CTP or UTP, can lead to cyclical dinucleotide formation (Figure S6C). Though ATP alone does not yield any cyclic or intermediate products, incubation with UTP results in some cyclic product formation, suggesting that UTP can also substitute for GTP, albeit at a much reduced reaction rate. Together, these findings indicate that cGAS may have more relaxed requirements for the second nucleotide compared to the first guanosine.
Structural Comparison of Linear 5′-pG(2′,5′)pA and Cyclic [G(2′,5′)pA(3′,5′)p] Product
We observe a striking difference in alignment within the catalytic pocket between the off-pathway linear 5′-pG(syn)(2′,5′)pA(anti) product (Figures 3F and 3G) and cyclic 5′-pG(anti)(2′,5′)A(anti) product (Figures 4B and 4C). In the former case, it is the G base that stacks over Tyr421 (Figure 3G), whereas in the latter, the A base stacks over Tyr421 (Figure 4C). The two alignments are compared in stereo in Figures 4G and 4H, with the linear 5′-pG(2′,5′)pA in silver and the cyclic c[G(2′,5′)pA(3′,5′)p] in green. This implies that the intermediate would have to rearrange its orientation by a complete flip-over within the catalytic pocket prior to the cyclization reaction. This may not be surprising, given that there is only a single set of catalytic residues, as reflected in the three basic (Glu211, Asp213, and Asp307) and one polar (Ser199) amino acid lining the catalytic pocket. Hence, following the first phosphodiester bond formation, the intermediate will need to realign so as to facilitate the second phosphodiester bond formation to complete cyclization.
Phosphodiester Linkages
A clear assumption in earlier studies that led to the identification of cyclic GAMP as a second messenger generated by the cytoplasmic dsDNA sensor cGAS (Sun et al., 2013; Wu et al., 2013) was that both phosphodiester linkages were of a 3′,5′ nature. Such 3′,5′ linkages have been observed previously in structures of bacterial second messenger c-di-GMP bound to both STING (Yin et al., 2012; Ouyang et al., 2012; Huang et al., 2012; Shu et al., 2012) and riboswitches (Smith et al., 2009; Kulshina et al., 2009). Nevertheless, the mass spectroscopic approach utilized (Wu et al., 2013) cannot distinguish between 3′,5′ and 2′,5′ linkages for one or both phosphodiester bonds of cyclic GAMP.
The first indication of a 2′,5′ linkage emerged from the structure of cGAS, dsDNA, and GTP, wherein an off-pathway product formed in the catalytic pocket exhibited a 5′-pppG(2′,5′)pG linkage (Figures 3B and 3C). In addition, pG(2′,5′)pA was observed in the structure of cGAS, dsDNA, and GMP + ATP (Figures 3F and 3G). More importantly, a 2′,5′ linkage was also observed for the GpA step of the bound c[G(2′,5′)pA(3′,5′)p] product in the catalytic pocket in the structure of cGAS, dsDNA, and GTP + ATP (Figures 4B and 4C).
Our initial biochemical analyses indicated that the 2′,5′ linkage between GTP and ATP occurs first, prior to the cyclization of the adenosine back to guanosine. This evidence was further supported by the observation that incubation of 2′-dATP with 2′-dGTP could not react to form any cyclic reaction products (Figure S6). In the second step, formation of a cyclical dinucleotide via the 2′ or 3′ OH of adenosine can proceed even when the other position is blocked through removal of oxygen, although there was an observable preference for utilization of 2′-dATP. Cyclic dinucleotide production in either case was very inefficient, suggesting that both positions may participate in the formation of a transition state for efficient phosphate hydrolysis and cyclization. This perplexing result, combined with ternary structural data concerning the connection from adenosine to guanosine, prompted us to further examine whether cGAS would ultimately have a preference for generating a 2′,5′ or 3′,5′ link for cyclization.
We observed a single HPLC peak, distinct from two cyclic-GA dinucleotide reference molecules (either both 2′,5′ or both 3′,5′ linkages) and coincident with c[G(2′,5′)pA(3′,5′)p] as the product of dsDNA-dependent cGAS activity (Figure 6A). This conclusion was validated by an independent NMR study (Figure 6C). Though biological production of cyclic dinucleotides appears evolutionarily conserved from prokaryotes to eukaryotes, their formation based on the chemical linkages is distinct. In the case of cGAS, formation of c[G(2′,5′)pA(3′,5′)p] appears similar to the 2′,5′ oligoadenylates generated by OAS1 (Donovan et al., 2013).
Potential Benefits of 2′,5′ and 3′,5′ Linkages in c[G(2′,5′) pA(3′,5′)p]
It is not clear why cGAS prefers to generate both a 2′,5′ (GpA step) and 3′,5′ (ApG step) cyclic GA dinucleotide. A 2′,5′ phosphodiester bond is uncommon, and few nucleases are known to be able to hydrolyze such a linkage (Kubota et al., 2004). Thus, 2′,5′ linkages might promote greater stability in cells to enable effective transduction of the second messenger, but the 3′,5′ linkage may facilitate its breakdown by numerous conventional endonucleases to prevent prolonged interferon response. Taken together, our structure and functional studies have clarified the chemical nature of metazoan cGAMP, highlighting the role of 2′,5′ linkages in second messengers that activate the type I interferon pathway.
Implications of cGAS dsDNA-Binding Mutants
Structural studies have identified intermolecular protein-DNA contacts on formation of the cGAS-dsDNA complex (Figure 1C). Because the interactions are primarily of an electrostatic nature and involve nonspecific recognition of the DNA phosphodiester backbone, they have been classified into three sets of triple mutants. The in vitro (Figure 7A) and cellular assays (Figure 7B) demonstrate complete loss of activity and ability to stimulate interferon production for the S165A, N172A, K372A triple mutant; the N196A, Y200A, K372A triple mutant; and the R158A, R161A, K395A triple mutant. We observed a partial loss of activity for the S165A, N172A, Y200A triple mutant. This reinforces the importance of complex formation between cGAS and dsDNA for the catalytic activity of cGAS.
Implications of cGAS Catalytic Pocket Mutants
Structural studies of ternary complexes of cGAS and dsDNA with bound NTPs have identified Glu211, Asp213, and Asp307 as important catalytic residues for phosphodiester bond formation. All three catalytic acidic residues are functionally dead on replacement by Ala, as observed in either in vitro (Figure 7A; Glu211A) or cellular (Figure 7C, all three catalytic residues) assays, whereas the S199A mutation retained substantial activity. Tyr421 is involved in stacking interactions with A, and Arg364 is hydrogen bonded with G in the cGAMP ternary complex (Figures 4B and 4C). Dual mutation of Y421A, R364A resulted in loss of the majority of its activity in cellular assays (Figure 7C).
Role of Divalent Cations
Structural studies of ternary complexes of dsDNA-bound cGAS with ligands have shown that the triphosphate moieties of ATP (Figures 2E and 2F) and 5′-pppG(2′,5′)pG (Figures 3B and 3C) are coordinated to a pair of cations (tentatively assigned to Mg2+). Indeed, functional studies have highlighted the importance of divalent cations to phosphodiester bond formation. Omission of divalent cations or use of EDTA prevented c[G(2′,5′) pA(3′,5′)p] formation, whereas Mg2+ and Mn2+ promoted cGAS activity (Figure 5A).
Comparison with Cytoplasmic dsRNA Sensor OAS1
In a parallel study to our contribution, structural studies and biochemical assays have been recently reported on the characterization of the dsRNA sensor human oligoadenylate synthetase 1 (OAS1), which polymerizes ATP into linear 2′,5′-linked oligoadenylate (Donovan et al., 2013). The crystallographic studies unequivocally demonstrated conformational changes in OAS1 associated with the transition from the free state (Hartmann et al., 2003) to the ternary complex with bound dsRNA and 2′-dATP (Donovan et al., 2013), which follow a similar pattern to those that we observed in this study for complex formation of cGAS with dsDNA and bound ligands. Thus, three catalytic Glu residues of OAS1 are brought into close proximity on formation of the ternary complex with dsRNA and 2′-dATP, thereby creating the coordination geometry for binding of two Mg2+ ions and 2′-dATP (Donovan et al., 2013), similar to what we observe for the cGAS system.
In addition to similarities mentioned above, there are also differences in protein-nucleic acid recognition principles between the cGAS dsDNA sensor (our study) and the OAS1 dsRNA sensor (Donovan et al., 2013). cGAS targets the sugar-phosphate backbone of dsDNA within a central segment of the dsDNA duplex (Figure 1C), and OAS1 targets the sugar-phosphate backbone of dsRNA by contacting two minor groove segments separated by 30 Å (Donovan et al., 2013). This is not surprising, as the helical parameters of dsRNA and dsDNA are very distinct, and different recognition principles are used in protein-dsRNA (reviewed in Lunde et al., 2007) and protein-dsDNA (reviewed in Huffman and Brennan, 2002) complexes. Nevertheless, common principles are utilized to generate the critical catalytic site architecture, which in turn couples nucleic acid recognition (dsRNA or dsDNA in the cytoplasm) with the cascade of downstream events, leading to an antiviral state including type I interferon response (cGAS) and RNase L activation (OAS1).
Further, the formation of linear 2′,5′-linked oligoadenylate RNA mediated by OAS1 parallels the formation of c[G(2′,5′)pA(3′,5′)p] containing 2′,5′ linkage at the GpA step by cGAS. Thus, unlike the sole emphasis on 3′,5′ linkages observed previously for bacterial second messenger c-di-GMP (reviewed in Römling et al., 2013), we highlight that the metazoan second messenger c[G(2′,5′)pA(3′,5′)p] utilizes mixed linkages involving 2′,5′ at the GpA step and 3′,5′ at the ApG step.
cGAS Contains a Single Active Site for Stepwise Phosphodiester Bond Formation
Previous studies have established that diguanylate cyclase PleD forms a head-to-tail homodimer to form a reaction center at its interface so that the intermediate does not have to change its orientation on the pathway to formation of c-di-GMP (Chan et al., 2004). By contrast, in our current studies of ternary complexes of cGAS, dsDNA, and bound ligands, we observe no evidence for dimer or higher-order oligomer formation of cGAS in the crystal. Further, the ligand-binding pocket in our structures is buried within the cGAS topology and is not located on the surface, as it is in PleD (Chan et al., 2004).
Indeed, cGAS contains a single active site for the sequential phosphodiester bond formation steps, a feature that is quite remarkable given that the ligands are GTP and ATP and that the GpA linkage that forms first is 2′,5′ and the ApG linkage that forms second is 3′,5′, resulting in generation of c[G(2′,5′) pA(3′,5′)p]. We outline a model for formation of c[G(2′,5′) pA(3′,5′)p] from GTP and ATP within the single catalytic pocket of dsDNA-bound cGAS in Figure 7D. In this model, the first step involves formation of a 5′-pppGpA intermediate followed in the second step by formation of c[G(2′,5′)pA(3′,5′)p]. Note also that the bound ligand is predicted to undergo two flip-overs on the pathway to c[G(2′,5′)pA(3′,5′)p] formation. It should be kept in mind that the model as shown by the schematics in Figure 7D will require future validation or modification when additional data become available.
Implications of the Current Study for the Dinucleotide Cyclase DncV
In an earlier study, the bacterial dinucleotide cyclase DncV was shown to generate cyclic cGAMP (Davies et al., 2012). This first report on formation of cGAMP raises the interesting question as to the nature of the pair of phosphodiester linkages in this bacterial system. The answer is currently unavailable, but it will be very interesting in the future to elucidate the nature of phosphodiester linkages formed by DncV.
Future Opportunities
In summary, our identification of a family of metazoan second messengers involving cyclic GA dinucleotide c[G(2′,5′)pA(3′,5′)p] containing a 2′,5′ linkage (at the GpA step) opens new opportunities for functional studies in the field of cellular signaling and immune surveillance in metazoans, as well as in certain forms of human disease.
EXPERIMENTAL PROCEDURES
Experimental Procedures are described in detail in the Extended Experimental Procedures. These include sections on structural biology protocols listing protein expression and purification, crystallization and structure determination, and refinement and NMR analysis of cGAMP isomers. Biochemical protocols list sections on preparation of oligonucleotides for thin-layer chromatography assays, TLC analysis of c[G(2′,5′)pA(3′,5′)p] formation, and preparation of cGAS products for high-performance liquid chromatography. Cellular assay protocols describe the generation of point mutants and luciferase-based assays to monitor interferon induction. Finally, the chemical protocol is included for the synthesis of cyclic GA dinucleotides containing various combinations of 2′,5′ and 3′,5′ linkages as standards for identifying the cyclic GA dinucleotide product, c[G(2′,5′)pA(3′,5′)p], of the reaction of cGAS, dsDNA, and GTP + ATP.
Supplementary Material
Acknowledgments
We thank the synchrotron beam line staffs at the Brookhaven and Argonne National laboratories for their assistance. D.J.P. is supported by grants from the Abby Rockefeller Trust and the Maloris and STARR Foundations. T.T. is supported by HHMI. W.B. and G.H. are members of the DFG Excellence Cluster ImmunoSensation. W.B. is supported by DFG grant BA3544/1-1, G.H. by SFB670, and both by SFB704. R.A.J. is supported by NIH GM79760.
The structural research was performed by P.G., Y.W., and Y. L. under the supervision of D.J.P.; the in vitro biochemical studies were performed by M.A. and A.A.S. under the supervision of T.T.; the cellular assays were performed by W.B. and T.Z. under the supervision of G.H.; and the chemical synthesis was performed by B.L.G. under the supervision of R.A.J. All participants participated in the writing of the paper and agree with the contents.
Footnotes
ACCESSION NUMBERS
The following coordinates have been deposited in the RCSB Protein Data Bank. cGAS in the free state (PDB: 4K8V); binary complex of cGAS + dsDNA (PDB: 4K96); ternary complex of cGAS + dsDNA + ATP (PDB: 4K97); ternary complex of cGAS + dsDNA + 5′-pppG(2′,5′)pG (PDB: 4K98); ternary complex of cGAS + dsDNA + 5′-pppdG(2′,5′)pdG (PDB: 4K99); ternary complex of cGAS + dsDNA + 5′-pG(2′,5′)pA (PDB: 4K9A); ternary complex of cGAS + dsDNA + c[G(2′,5′)pA(3′,5′)p] (PDB: 4K9B).
Supplemental Information includes Extended Experimental Procedures, eight figures, and six tables can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.04.046.
References
- Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci USA. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli M, Gessner RV, Williams LD, Quigley GJ, van der Marel GA, van Boom JH, Rich A, Frederick CA. Atomic-resolution structure of the cellulose synthase regulator cyclic diguanylic acid. Proc Natl Acad Sci USA. 1990;87:3235–3239. doi: 10.1073/pnas.87.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney BL, Jones RA. One-flask syntheses of cyclic diguanosine monophosphate (c-di-GMP) Curr Protoc in Nucleic Acid Chem. 2012;14:14.8.1–14.8.7. doi: 10.1002/0471142700.nc1408s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney BL, Veliath E, Zhao J, Jones RA. One-flask syntheses of c-di-GMP and the [Rp,Rp] and [Rp,Sp] thiophosphate analogues. Org Lett. 2010;12:3269–3271. doi: 10.1021/ol101236b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Huffman JL, Brennan RG. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. 2002;12:98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating SE, Baran M, Bowie AG. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32:574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Krasteva PV, Giglio KM, Sondermann H. Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci. 2012;21:929–948. doi: 10.1002/pro.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Nakahara K, Ohtsuka T, Yoshida S, Kawaguchi J, Fujita Y, Ozeki Y, Hara A, Yoshimura C, Furukawa H, et al. Identification of 2′-phosphodiesterase, which plays a role in the 2–5A system regulated by interferon. J Biol Chem. 2004;279:37832–37841. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- Kulshina N, Baird NJ, Ferré-D’Amaré AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA. Immunology. Sensing the dark side of DNA. Science. 2013;339:763–764. doi: 10.1126/science.1234724. [DOI] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkowicz UM, Cook AG. NF45 dimerizes with NF90, Zfr and SPNR via a conserved domain that has a nucleotidyltransferase fold. Nucleic Acids Res. 2012;40:9356–9368. doi: 10.1093/nar/gks696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.