Abstract
Stimuli-triggered drug delivery systems are primarily focused on the applications of the tumor microenvironmental or cellular physiological cues to enhance the release of drugs at the target site. In this study, we applied adenosine-5′-triphosphate (ATP), the primary “energy molecule”, as a trigger for enhanced release of preloaded drugs responding to the intracellular ATP concentration that is significantly higher than the extracellular level. A new ATP-responsive anticancer drug delivery strategy utilizing DNA-graphene crosslinked hybrid nanoaggregates as carriers was developed for controlled release of doxorubicin (DOX), which consists of graphene oxide (GO), two single-stranded DNA (ssDNA, denoted as DNA1 and DNA2) and ATP aptamer. The single-stranded DNA1 and DNA2 together with the ATP aptamer serve as the linkers upon hybridization for controlled assembly of the DNA-GO nanoaggregates, which effectively inhibited the release of DOX from the GO nanosheets. In the presence of ATP, the responsive formation of the ATP/ATP aptamer complex causes the dissociation of the aggregates, which promoted the release of DOX in the environment with a high ATP concentration such as cytosol compared with that in the ATP-deficient extracellular fluid. This supports the development of a novel ATP-responsive platform for targeted on-demand delivery of anticancer drugs inside specific cells.
Keywords: ATP responsive, graphene oxide, DNA, drug delivery, nanomedicine
1. Introduction
Stimuli-triggered delivery of anticancer drugs [1–3] is mainly focused on the applications of the tumor microenvironmental or cellular physiological characteristics including higher temperature [4–5], lower pH [6–8], higher redox potential [9–11], enzyme overexpression [12–13] and higher level of reactive oxygen species [14–15] to promote the release of drugs at the target site for cancer treatment. Numerous nanocarriers integrating with the responsive elements, such as liposomes, polymeric nanoparticles, protein/DNA nanostructures and inorganic nanovehicles, have been widely exploited to achieve controlled release of their cargoes within the tumor tissues or cells [16–22].
Recently, adenosine-5′-triphosphate (ATP), the primary energy molecule for cell function, is attracting extensive attentions, which can be used as a promising trigger for enhanced release of preloaded drugs form the carriers responding to the intracellular ATP concentration [23–26]. ATP has a very high concentration inside the cells (1–10 mM), which is much greater than that outside the cells (< 5 μM) [27, 28]. Such a pronounced gradient is the premise of design of the ATP-triggered intracellular drug delivery systems. However, the existing ATP-responsive anticancer drug delivery methods are often limited by complicated formulation design and relatively low loading capacity of drugs [23, 24].
Herein, we present a new ATP-responsive anticancer drug delivery strategy utilizing DNA-graphene crosslinked hybrid nanoaggregates as carriers (Fig. 1). This nanoaggregate consists of graphene oxide (GO), two single-stranded DNA (ssDNA, denoted as DNA1 and DNA2) and ATP aptamer. The GO nanosheet is applied to carry doxorubicin (DOX), a model small-molecule anticancer drug, with a high loading efficiency via supramolecular π-π stacking between GO and DOX [29–34]. The single-stranded DNA1 and DNA2 together with the ATP aptamer serve as the linkers upon hybridization for controlled assembly of the DNA-GO nanoaggregates (DNA-GA). Both DNA1 and DNA2 are composed of “head” and “tail” sequences, which are a target-specific sequence (the head one) complementary to the target ATP aptamer and a repeated GT sequence (the tail one) to facilitate the binding of DNA1 or DNA2 onto the GO nanosheets, respectively [35–37]. DNA1 and DNA2 are separately added to the DOX-loaded GO (DOX/GO) solution to form the DOX-loaded DNA-GO complex (DOX/DNA-GC) via strong interactions including van der Waals forces, π-π stacking and hydrogen bond [38, 39]. When the ATP aptamer is added into the mixture of DOX/DNA1-GC and DOX/DNA2-GC (DOX/DNA12-GC), the hybridization of the ATP aptamer with both DNA1 and DNA2 results in the assembly of the GO nanosheets to form the layered-structural DOX-loaded DNA-GO nanoaggregates (DOX/DNA-GA). Such aggregates, with an increased average size and a decreased specific surface area toward the surrounding medium [40, 41], can effectively inhibit DOX release from the GO nanosheets. The ATP aptamer has been widely used for ATP detection based on its specific and stable binding to ATP [42–44]. In the presence of ATP, the responsive formation of the ATP/ATP aptamer complex causes the dissociation of DOX/DNA-GA into DOX/DNA-GC that has a decreased size and an increased surface area exposing to the medium, which promotes the release of DOX in the environment with a high ATP concentration such as cytosol compared with that in the ATP-deficient extracellular fluid. This supports the development of a novel ATP-responsive platform for targeted on-demand delivery of anticancer drugs inside specific cells.
Fig. 1.
Schematic illustration of ATP-responsive DNA-GA for controlled drug delivery.
2. Materials and methods
2.1. Materials
All chemicals unless mentioned were purchased from Sigma-Aldrich Co. LLC. All the DNA samples (sequence from 5′ to 3′), including DNA1 (GTG TGT GTG TGT GTG TGT GTG TGT GTG TGT ACC TTC CTC CGC), DNA2 (ACT CCC CCA GGT GTG TGT GTG TGT GTG TGT GTG TGT GTG TGT), ATP aptamer (ACC TGG GGG AGT ATT GCG GAG GAA GGT), control aptamer (ATT CTT TTT TAC AAT ACT CCC CCA GGT) and Cy3-labeled DNA2 (Cy3-DNA2, Cy3-ACT CCC CCA GGT GTG TGT GTG TGT GTG TGT GTG TGT GTG TGT), were purchased from Integrated DNA Technologies, Inc. (Coralville, IA, USA). Doxorubicin hydrochloride was purchased from BIOTANG Inc. (Lexington, MA, USA).
2.2. Preparation of DNA-GO complex (DNA-GC)
DNA-GC was prepared by mixing DNA1 or DNA2 with GO. After ultrasonication for 1 h, GO (0.2 mg/mL) was incubated with DNA1 or DNA2 (2 μM) in the HEPES buffer (10 mM, pH 7.4) containing magnesium chloride (MgCl2) (1 mM) at room temperature for 1 h. To remove the unbound DNA1 or DNA2, the solution was centrifuged at 21000 × g for 1 h and the supernatant was discarded. The precipitated DNA-GC was then resuspended in the HEPES buffer (10 mM, pH 7.4) containing sodium chloride (NaCl) (100 mM). The particle size of DNA-GC was measured by a Zetasizer (Nano ZS, Malvern).
2.3. Preparation of DNA-GO aggregate (DNA-GA)
DNA-GA was prepared by incubating DNA1-GC and DNA2-GC with the ATP aptamer. DNA1-GC and DNA2-GC (500 μL) were mixed at the ratio of 1:1 in the HEPES buffer (10 mM, pH 7.4) containing NaCl (100 mM), followed by adding the ATP aptamer (1 μM). The mixture solution was then incubated at room temperature for 24 h. The particle size of DNA-GA was measured by the Zetasizer. For atomic force microscope (AFM) characterization, the DNA-GA solution was dropped onto a mica substrate (Ted Pella). After air-dry, the sample was observed by AFM (NanoScope IIIa). To evaluate the ATP-responsive dissociation of DNA-GA, DNA-GA was incubated with different concentrations of ATP, cytidine triphosphate (CTP) or guanosine triphosphate (GTP). The particle size was measured by the Zetasizer.
2.4. Fluorescence-based determination of formation and dissociation of DNA-GA
To explore the formation and ATP-responsive dissociation of DNA-GO aggregate, the fluorescence quenching/recovery measurements were applied on the basis of the interaction between GO and Cy3-DNA2. To investigate the effect of electrolytes on the adsorption efficiency of DNA onto the GO surface, Cy3-DNA2 (1.2 μM) was mixed with GO (0.1 mg/mL) in the HEPES buffer containing different concentrations of NaCl or MgCl2 at room temperature for different time. The fluorescence intensity of Cy3 was measured at an emission wavelength of 580 nm with an excitation wavelength of 550 nm by a fluorescence microplate reader (Infinite M200 PRO, Tecan). The fluorescence quenching efficiency ((1−F/F0) × 100%) was determined, where F0 and F were the fluorescence intensities of Cy3 before and after Cy3-DNA2 was incubated with GO, respectively. To explore the formation of DNA-GA, DNA1 and Cy3-DNA2 (1.2 μM) was first mixed with GO (0.1 mg/mL) in the HEPES buffer containing MgCl2 (1 mM) at room temperature for 1 h to obtain DNA1-GC and Cy3-DNA2-GC, respectively. Subsequently, DNA1-GC and Cy3-DNA2-GC were mixed at the ratio of 1:1, followed by adding different concentrations of the ATP aptamer and incubating for different time. The fluorescence intensity of Cy3 was measured during the formation process of DNA-GA. The fluorescence recovery ratio (F/F0 × 100%) was determined, where F0 and F were the fluorescence intensities of Cy3 before and after the mixture of DNA1-GC and Cy3-DNA2-GC (Cy3-DNA12-GC) was incubated with the ATP aptamer, respectively. To trace the ATP-triggered dissociation of DNA-GA, Cy3-DNA-GA was incubated with different concentrations of ATP or CTP for 30 min. The fluorescence intensity of Cy3 was measured during the disassembly process of Cy3-DNA-GA. The fluorescence quenching efficiency ((1−F/F0) × 100%) was determined, where F0 and F were the fluorescence intensities of Cy3 before and after Cy3-DNA-GA was incubated with ATP or CTP, respectively.
2.5. Preparation of DOX-loaded DNA-GA (DOX/DNA-GA)
To prepare DOX/GO, 40 μL of the dimethyl sulphoxide (DMSO) solution containing triethanolamine-treated DOX (5 mg/mL) was added dropwise into 10 mL of the GO solution (0.2 mg/mL) under vigorous stirring. After incubation at room temperature for 24 h, the solution was dialyzed against the deionized (DI) water to remove the triethanolamine, DMSO and the unbound DOX. The fluorescence spectra of DOX were scanned at an excitation wavelength of 480 nm by the fluorescence microplate reader. Subsequently, the preparation of DOX/DNA-GA was similar to that of DNA-GA except that DOX/GO was used to substitute the blank GO. The particle sizes of DOX/GO, DOX/DNA-GC and DOX/DNA-GA were measured by the Zetasizer.
2.6. In vitro release of DOX
1 mL of DOX/DNA-GC or DOX/DNA-GA (20 μg/mL) was added into a dialysis tube (10K MWCO) (Slide-A-Lyzer, Thermo Scientific) against 14 mL of the HEPES buffer containing different concentrations of ATP, followed by gently shaking at 37 °C in a shaker (New Brunswick Scientific) at 100 rpm. At predetermined time intervals, the total buffer solution was sampled and replaced with 14 mL of the fresh buffer solution. The fluorescence intensity of DOX released was measured at an emission wavelength of 596 nm with an excitation wavelength of 480 nm by the fluorescence microplate reader.
2.7. Cell culture
The human cervical adenocarcinoma (HeLa) cells were cultured in DMEM with FBS (10%, v:v), penicillin (100 U/mL) and streptomycin (100 μg/mL) in a cell culture incubator (Thermo Scientific) at 37 °C under a condition of 5% CO2 and 90% relative humidity. The cells were sub-cultivated approximately every 3 days at 80% confluence using trypsin (0.25%, w:v) at a split ratio of 1:5.
2.8. Cellular uptake
HeLa cells (1 × 105 cells/well) were seeded in 6-well plates. After culture for 48 h, the cells were incubated with DOX/DNA-GC or DOX/DNA-GA (4 μg/mL) at 4 and 37 °C for 2 h, respectively. The cells were then washed by ice-cold PBS twice and lyzed using the cell lysis buffer (Pierce, Thermo Scientific) at room temperature for 10 min. The cell lysate was collected and centrifuged at 14000 × g for 5 min. The fluorescence intensity of DOX in the cell lysate was measured at an emission wavelength of 596 nm with an excitation wavelength of 480 nm by the fluorescence microplate reader. The total protein concentration in the cell lysate was quantified using the BCA protein assay kit (Pierce, Thermo Scientific). The cellular uptake of DOX was calculated as UDOX = QDOX/Qprotein, where QDOX and Qprotein were the amounts of DOX and protein in the cells, respectively. The relative uptake efficiency was determined as the ratio of UDOX at 4 °C to that at 37 °C.
2.9. Intracellular release of DOX
HeLa cells (1 × 105 cells/well) were seeded in 6-well plates. After culture for 48 h, the cells were incubated with DOX/DNA-GA (4 μg/mL) at 37 °C for 2 h. The excessive DOX/DNA-GC or DOX/DNA-GA was removed and the cells were incubated with the fresh FBS free culture medium at 4 or 37 °C for additional 1 or 2 h. The cells were washed by ice-cold PBS twice and harvested. The fluorescence intensity of DOX in the cells was measured using flow cytometry (BD FACSCalibur). The intracellular release rate of DOX was indicated as (Ft/F0 × 100%), where F0 is the fluorescence intensity of DOX in the cells after the first 2 h incubation with DOX/DNA-GA, and Ft is the fluorescence intensity of DOX in the cells after incubation for addition time periods (1 or 2 h) following the first 2 h incubation.
2.10. Intracellular distribution
HeLa cells (1 × 105 cells/well) were seeded in a confocal microscopy dish (MatTek). After culture for 24 h, the cells were incubated with DOX/DNA-GA (4 μg/mL) at 37 °C. At predetermined time intervals (2 and 6 h), the cells were washed by ice-cold PBS twice and then stained by Hoechst 33342 (1 μg/mL) (Life Technologies) at 37 °C for 10 min. The cells were then washed by ice-cold PBS twice and immediately observed using the confocal laser scanning microscope (CLSM) (LSM 710, Zeiss).
2.11. Cytotoxicity
HeLa cells (1 × 104 cells/well) were seeded in 96-well plates. After culture for 24 h, the cells were incubated with the DOX solution, DOX/DNA-GC or DOX/DNA-GA for 24 or 48 h, followed by adding 20 μL of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (5 mg/mL). After 4 h of incubation, the medium was removed, and the cells were mixed with 150 μL of DMSO. The absorbance was measured at a test wavelength of 570 nm and a reference wavelength of 630 nm by the microplate reader (Infinite M200 PRO, Tecan).
3. Results and discussion
3.1. DNA-mediated assembly and ATP-triggered dissociation of GA
To explore the DNA-mediated assembly and ATP-triggered dissociation of DNA-GA, the particle size was determined using the dynamic laser scanning (DLS) measurement [35, 45–47] (Fig. 2). After ultrasonication and purification, we obtained a well-dispersed GO sheet in water, which had an average particle size of 180 nm (Fig. 2a). DNA-GC was prepared by incubating the as-prepared GO nanosheets with DNA1 or DNA2 at a high salt condition [48], followed by centrifugation to remove the free DNA. The presence of DNA was able to enhance the stability of GO in a wider range of ionic strength. GO without DNA showed an increased particle size with the ionic strength, while DNA-GC exhibited a good stability even in the HEPES buffer solution containing a high concentration of NaCl (100 mM).
Fig. 2.
(a) The particle size of GO, DNA1-GC, DNA2-GC and DNA12-GC after incubation in the HEPES buffer containing different concentrations of NaCl for 24 h. (b) The particle size of the mixture of DNA1-GC and DNA2-GC after incubation with different concentrations of the ATP or control aptamer in the HEPES buffer containing 100 mM NaCl for 24 h. (c) The AFM images of DNA-GC and DNA-GA. The scale bars indicate 200 nm. (d) The particle size of DNA-GA after incubation with ATP, CTP and GTP in the HEPES buffer containing 100 mM NaCl for 0.5 h.
After incubating the mixture of DNA1-GC and DNA2-GC (DNA12-GC) with different concentrations of the ATP aptamer, the hybridization of DNA1 and DNA2 with the ATP aptamer resulted in an ATP aptamer concentration-dependent formation of nanoaggregates (Fig. 2b). DNA-GA showed a concomitant increase in the particle size with the concentration of the ATP aptamer. The average particle sizes of DNA-GA with the ATP aptamer concentration of 0.2 and 1 μM were 290 and 550 nm, respectively. To confirm that the interaction of the ATP aptamer with both DNA1 and DNA2 played an essential role in the assembly of DNA-GA, we applied a mismatched DNA as a control aptamer to examine the specificity of this process. Upon mixing DNA12-GC with the control aptamer, no significant changes were determined in the particle size of the mixture when varying the concentrations of the control aptamer. The mixture remained the particle size at 200 nm as the concentration of the control aptamer increased, suggesting that the ATP aptamer with the specific sequence could hybridize with DNA1 and DNA2, which associatively acted as a bridge to connect DNA-GC together for the formation of DNA-GA. We further applied DLS to monitor the DNA-mediated GO assembly process, which showed close association with the relative physical sizes of GA [35, 45–47].
The AFM imaging was applied to directly observe the structure conversion from DNA-GC to DNA-GA (Fig. 2c). From the AFM images, DNA-GC displayed a clear single-layer structure with the thickness of 2 nm, whereas the thickness of DNA-GA was 5–15 nm, with a well-packed layered structure. Such a notable increase in the thickness of GO was indicated to be closely associated with the DNA hybridization.
To investigate the ATP-responsive disassembly of DNA-GA, we incubated DNA-GA with different concentrations of ATP and monitored the particle size change (Fig. 2d). After incubating with ATP, as expected, the particle size of DNA-GA effectively reduced with the increase of the ATP concentration. In the existence of 3 mM ATP, DNA-GA showed an average particle size of 270 nm compared with DNA-GA without ATP of 550 nm. In addition, there was a remarkable difference in the particle size of DNA-GA after incubation with 3 and 0.3 mM ATP. To further evaluate the selectivity of the DNA-GA dissociation, the ATP analogues, CTP and GTP, were added into the DNA-GA solution, followed by the size measurement. The particle size change of DNA-GA at 3 mM ATP was significantly greater than that at 3 mM CTP or GTP. It was suggested that when DNA-GA was exposed to a high level of ATP, the formation of ATP/ATP aptamer complex led to the dissociation of DNA-GA into DNA-GC with decreased particle sizes.
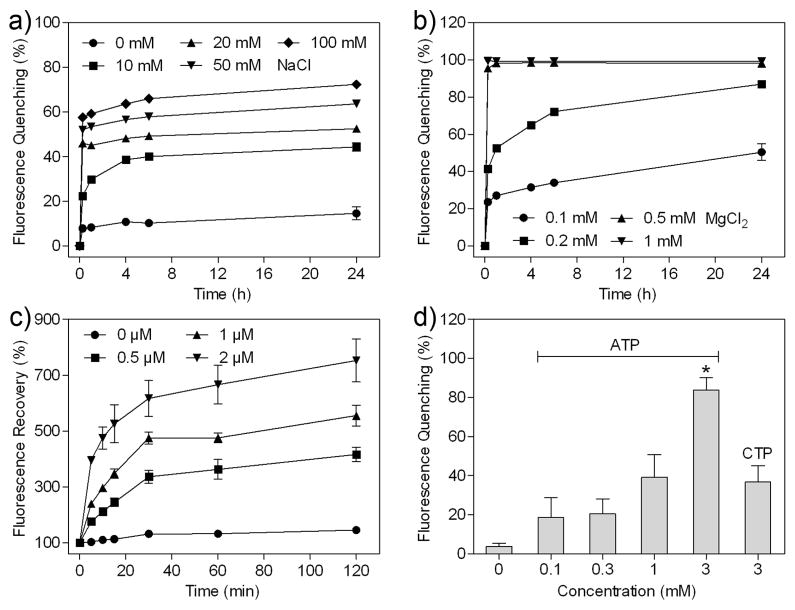
Furthermore, we utilized the fluorescence quenching and recovery measurements, based on the interaction between GO and Cy3-DNA2, to explore the ATP-aptamer-directed assembly and ATP-triggered disassembly of DNA-GA (Fig. 3) [49, 50]. The electrolytes including NaCl and MgCl2 were applied to screen the electrostatic repulsion between the polyanionic DNA and the negatively charged GO at neutral condition, which made DNA close to the GO surface for binding. After Cy3-DNA2 was incubated with the GO nanosheets at different concentrations of NaCl or MgCl2, the fluorescence of Cy3-DNA2 presented different degrees of quenching due to the emission from Cy3 to GO by förster resonance energy transfer (FRET), which indicated the adsorption of DNA onto the surface of the GO nanosheets (Fig. 3a and b). This quenching effect was enhanced by the increasing salt concentrations. The quenching efficiency of Cy3-DNA2 was less than 15% even at 24 h post-incubation with GO in the absence of the electrolytes, but about 58% at 100 mM NaCl and nearly 100% at 1 mM MgCl2 only after 15 min of incubation with GO. MgCl2 presented a stronger quenching capability than NaCl, which was attributed to the more effective capability of divalent metal ions than monovalent ones to shield charges for the connection of two negatively charged molecules [48].
Fig. 3.
(a, b) The fluorescence quenching of Cy3-DNA2 after incubating Cy3-DNA2 with GO in the HEPES buffer containing different concentrations of NaCl (a) and MgCl2 (b) for different time. (c) The fluorescence recovery of Cy3-DNA2 after incubating the mixture of DNA1-GC and Cy3-DNA2-GC with different concentrations of the ATP aptamer in the HEPES buffer containing 1 mM MgCl2 for different time. (d) The fluorescence quenching of Cy3-DNA2 after incubating Cy3-DNA-GA with ATP and CTP in the HEPES buffer containing 1 mM MgCl2 for 0.5 h. *P < 0.05, ATP (3 mM) vs CTP (3 mM).
The incubation of Cy3-DNA12-GC with the ATP aptamer attenuated the quenching effect of GO, thereby yielding the fluorescence recovery of Cy3 in a concentration-dependent manner (Fig. 3c), which was ascribed to the desorption of the dye from the GO surface upon the hybridization of the Cy3-labeled head sequence of DNA2 with the complementary one of the ATP aptamer. The fluorescence intensity of Cy3 significantly increased after treatment with different concentrations of the ATP aptamer in comparison with the untreated one, implying the assembly of DNA-GA. After the addition of ATP, Cy3-DNA-GA presented a reduction in the fluorescence intensity of Cy3 accompanied by the increase in the ATP concentration (Fig. 3d), since the efficient binding of ATP to the ATP aptamer resulted in the dehybridization between the ATP aptamer and Cy3-DNA2, followed by the re-adsorption of Cy3-DNA2 onto the surface of GO and the fluorescence re-quenching of Cy3 by GO. The fluorescence quenching efficiency of Cy3-DNA-GA with ATP was noticeably higher than that with the same concentration of CTP. Taken together, the fluorescence quenching, recovery and re-quenching of Cy3 validated the assembly and disassembly of DNA-GA in part.
3.2. Preparation and characterization of DOX/GA
DOX was efficiently loaded on the GO surface to obtain DOX/GO by the π-π stacking effect, the main interactions between the conjugated structure of the GO sheet and the quinone portion of DOX as well as the hydrophobic force between them [29–34]. The DOX-loading capacity was about 10% as a mass ratio of DOX to GO. DOX/GO displayed a remarkable quenching effect of the DOX emission compared with free DOX after excitation at the same wavelength due to the FRET process along the interface of GO and DOX (Fig. 4a). DOX loading had no influence on the formation of DNA-GA, which was also determined by monitoring the particle size change (Fig. 4b). The average particle size of DOX/DNA-GA showed an ATP aptamer concentration-dependent increase, which was 585 nm at the ATP aptamer concentration of 1 μM, significantly higher than 271 nm in the absence of the ATP aptamer. In addition, no obvious change in the particle size was observed in the presence of the untargeted control aptamer. The particle size remained about 275 nm regardless of the concentration of the control aptamer. When exposed to high concentration of ATP (3 mM), the particle size of DOX/DNA-GA sharply dropped to 298 nm, while neither CTP nor GTP with the same concentration of ATP showed such a strong effect on contributing to the dissociation of DOX/DNA-GA (Fig. 4c).
Fig. 4.
(a) The fluorescence spectra of the DOX solution, DOX/GO and GO at the same excitation wavelength of 480 nm. (b) The particle size of the mixture of DOX/DNA1-GC and DOX/DNA2-GC after incubation with different concentrations of the ATP or control aptamer in the HEPES buffer containing 100 mM NaCl for 24 h. (c) The particle size of DOX/DNA-GA after incubation with ATP, CTP and GTP in the HEPES buffer containing 100 mM NaCl for 0.5 h. (d) The in vitro release profile of DOX/DNA-GA at different concentrations of ATP.
3.3. ATP-triggered DOX release
To assess the ATP-triggered DOX release property of DOX/DNA-GA, we determined the in vitro release profiles of DOX from DOX/DNA-GA in the presence and absence of different concentrations of ATP (Fig. 4d). Only 4.7% of DOX was released from DOX/DNA-GA within 24 h in the absence of ATP, which was due to the significantly decreased specific surface area interacting with surrounding medium compared with that of the original GO. The relatively low concentration of ATP did not accelerate the release of DOX. In the presence of 0.3 mM ATP, the 24 h cumulative DOX release ratio is about 5.4%. However, the release DOX was notably promoted, when DOX/DNA-GA was incubated in the same buffer solution containing 3 mM ATP that was comparable to the intracellular ATP concentration. DOX/DNA-GA exhibited approximately 6.1% of DOX released in the first 6 h and more than 10% of DOX released within 24 h, which was similar to the DOX release behavior of DOX/DNA-GC. It was suggested that the ATP-responsive disassembly of DOX/DNA-GA to DOX/DNA-GC resulted in the decreased particle size along with the increased specific surface area, thereby facilitating the release of DOX from the GO surface.
3.4. Intracellular ATP-responsive DOX release
To demonstrate the cellular internalization capability of DOX/DNA-GA, the uptake of DOX/DNA-GA at 37 and 4 °C were compared on HeLa cells. As shown in Fig. 5a, the cellular uptake of both DOX/DNA-GA and DOX/DNA-GC were dramatically inhibited at 4 °C. The relative cellular uptake efficiency of DOX/DNA-GA was about 16%, indicating that DOX/DNA-GA entered into the cells via a typical endocytotic process [51, 52]. We next investigated the intracellular DOX release from DOX/DNA-GA by monitoring the fluorescence intensity of DOX in the cells (Fig. 5b). HeLa cells were incubated with DOX/DNA-GA for 2 h. After completely removing the excess formulations, the cells were incubated with the fresh culture medium for an additional 1 or 2 h. The amount of released DOX was determined by monitoring the fluorescence intensity of the whole cells using flow cytometry. DOX/DNA-GA showed a concomitant increase of fluorescence intensity with the incubation time. The cells presented significant increase in the fluorescence intensity of DOX after another 1 and 2 h of incubation, respectively. To further confirm that the increased DOX release from DOX/DNA-GA was mediated by the intracellular ATP concentration, we impeded the ATP synthesis of the cells by reducing the temperature (4 °C), and subsequently recorded the fluorescence intensity of the treated cells, after 2 h of incubation with DOX/DNA-GA. There was no significant increase in the release of DOX in the cells at 4 °C, which suggested the intracellular ATP-enhancing DOX release ability of DOX/DNA-GA.
Fig. 5.
(a) The relative cellular uptake efficiency of DOX/DNA-GC and DOX/DNA-GA on HeLa cells at 4 °C compared with 37 °C. **P < 0.01, 4 °C vs 37 °C. (b) The release of DOX from DOX/DNA-GA in HeLa cells obtained using flow cytometry. The DOX release ratio is determined by comparing the fluorescence intensity of DOX after 2 h of incubation with that after an additional 1or 2 h of incubation at 37 °C or 4 °C. (c) The CLSM images of HeLa cells after incubation with DOX/DNA-GC for 2 and 6 h. The nuclei were stained with Hoechst 33342. The scale bars indicate 20 μm. (d) The in vitro cytotoxicity of the DOX solution, DOX/DNA-GC and DOX/DNA-GA against HeLa cells after incubating for 48 h. (e) The in vitro cytotoxicity of DOX/DNA-GA against HeLa cells after incubating for 24 and 48 h. *P < 0.05, **P < 0.01, 24 h vs 48 h.
3.5. Intracellular distribution and cytotoxicity
The intracellular distribution was visualized using CLSM (Fig. 5c). DOX and the stained nuclei displayed red and blue fluorescence, respectively. After 2 h of incubation, DOX/DNA-GA was endocytosed by the cancer cells and evenly distributed within the cells. As the incubation time was prolonged to 6 h, DOX was efficiently released from GO, and the released DOX was specifically accumulated into the nuclei for subsequently inducing cytotoxicity, as observed by the magenta fluorescence. The in vitro cytotoxicity of DOX/DNA-GA toward the cancer cells was evaluated using the MTT assay (Fig. 5d and e). DOX/DNA-GA presented an efficient and comparable cytotoxicity to DOX/DNA-GC against HeLa cells, indicating that the intracellular high ATP concentration resulted in the disintegration from DOX/DNA-GA to DOX/DNA-GC and therefore supported the parallel DOX release of DOX/DNA-GA with DOX/DNA-GC (Fig. 5d). Nevertheless, the free DOX solution showed the highest toxicity to HeLa cells under the same condition due to the partial inefficient release of DOX from the GO surface. The cytotoxicity of DOX/DNA-GA significantly increased when the incubation time increased (Fig. 5e), suggesting that the sustainedly released DOX from DOX/DNA-GA allowed the enhanced cytotoxic effect toward the cancer cells.
4. Conclusion
In summary, we have developed a new ATP-mediated controlled drug release system comprised of a 2D nanomaterial (GO) assembled nanoaggregates crosslinked by ATP-responsive DNA strands. The straightforward formulation design, high loading capacity of drugs and capability of site-specifically promoting drug release render this formulation as a promising strategy for enhanced therapeutic efficacy in cancer treatment. It also provides innovative insights for taking advantages of 2D nanostructures for drug delivery through a reversible assembly fashion, which can be programmed by metabolic elements.
Acknowledgments
This work was supported by NC TraCS (grant 550KR51307), NIH’s Clinical and Translational Science Awards (CTSA, 1UL1TR001111) at UNC-CH, the NC State Faculty Research and Professional Development Award, and the start-up package from the Joint BME Department of UNC-CH and NCSU to Z.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Motornov M, Roiter Y, Tokarev I, Minko S. Stimuli-responsive nanoparticles, nanogels and capsules for integrated multifunctional intelligent systems. Prog Polym Sci. 2010;35:174–211. [Google Scholar]
- 2.Mura S, Nicolas J, Couvreur P. Stimuli-responsive nanocarriers for drug delivery. Nat Mater. 2013;12:991–1003. doi: 10.1038/nmat3776. [DOI] [PubMed] [Google Scholar]
- 3.Ge Z, Liu S. Functional block copolymer assemblies responsive to tumor and intracellular microenvironments for site-specific drug delivery and enhanced imaging performance. Chem Soc Rev. 2013;42:7289–325. doi: 10.1039/c3cs60048c. [DOI] [PubMed] [Google Scholar]
- 4.Chen KJ, Liang HF, Chen HL, Wang Y, Cheng PY, Liu HL, et al. A thermoresponsive bubble-generating liposomal system for triggering localized extracellular drug delivery. ACS Nano. 2013;7:438–46. doi: 10.1021/nn304474j. [DOI] [PubMed] [Google Scholar]
- 5.May JP, Ernsting MJ, Undzys E, Li SD. Thermosensitive liposomes for the delivery of gemcitabine and oxaliplatin to tumors. Mol Pharm. 2013;10:4499–508. doi: 10.1021/mp400321e. [DOI] [PubMed] [Google Scholar]
- 6.Takemoto H, Miyata K, Hattori S, Ishii T, Suma T, Uchida S, et al. Acidic pH-responsive siRNA conjugate for reversible carrier stability and accelerated endosomal escape with reduced IFNα-associated immune response. Angew Chem Int Ed Engl. 2013;52:6218–21. doi: 10.1002/anie.201300178. [DOI] [PubMed] [Google Scholar]
- 7.Ju C, Mo R, Xue J, Zhang L, Zhao Z, Xue L, et al. Sequential intra-intercellular nanoparticle delivery system for deep tumor penetration. Angew Chem Int Ed Engl. 2014;53:6253–8. doi: 10.1002/anie.201311227. [DOI] [PubMed] [Google Scholar]
- 8.Liu D, Zhang H, Herranz-Blanco B, Mäkilä E, Lehto VP, Salonen J, et al. Microfluidic assembly of monodisperse multistage pH-responsive polymer/porous silicon composites for precisely controlled multi-drug delivery. Small. 2014;10:2029–38. doi: 10.1002/smll.201303740. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Cai X, Hu J, Shao N, Wang F, Zhang Q, et al. Glutathione-triggered “off-on” release of anticancer drugs from dendrimer-encapsulated gold nanoparticles. J Am Chem Soc. 2013;135:9805–10. doi: 10.1021/ja402903h. [DOI] [PubMed] [Google Scholar]
- 10.Zhao M, Hu B, Gu Z, Joo KI, Wang P, Tang Y. Degradable polymeric nanocapsule for efficient intracellular delivery of a high molecular weight tumor-selective protein complex. Nano Today. 2013;8:11–20. [Google Scholar]
- 11.Chen W, Huang Q, Ou W, Hao Y, Wang L, Zeng K, et al. Self-reporting liposomes for intracellular drug release. Small. 2014;10:1261–5. [Google Scholar]
- 12.Gu Z, Yan M, Hu B, Joo KI, Biswas A, Huang Y, et al. Protein nanocapsule weaved with enzymatically degradable polymeric network. Nano Lett. 2009;9:4533–8. doi: 10.1021/nl902935b. [DOI] [PubMed] [Google Scholar]
- 13.Jiang T, Mo R, Bellotti A, Zhou J, Gu Z. Gel-liposome-mediated co-delivery of anticancer membrane-associated proteins and small-molecule drugs for enhanced therapeutic efficacy. Adv Funct Mater. 2014;24:2295–304. [Google Scholar]
- 14.Shim MS, Xia Y. A reactive oxygen species (ROS)-responsive polymer for safe, efficient, and targeted gene delivery in cancer cells. Angew Chem Int Ed Engl. 2013;52:6926–9. doi: 10.1002/anie.201209633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Gracia Lux C, Joshi-Barr S, Nguyen T, Mahmoud E, Schopf E, Fomina N, et al. Biocompatible polymeric nanoparticles degrade and release cargo in response to biologically relevant levels of hydrogen peroxide. J Am Chem Soc. 2012;134:15758–64. doi: 10.1021/ja303372u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2:751–60. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 17.Gu Z, Biswas A, Zhao M, Tang Y. Tailoring nanocarriers for intracellular protein delivery. Chem Soc Rev. 2011;40:3638–55. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 18.Chow EK, Ho D. Cancer nanomedicine: from drug delivery to imaging. Sci Transl Med. 2013;5:216rv4. doi: 10.1126/scitranslmed.3005872. [DOI] [PubMed] [Google Scholar]
- 19.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Gold nanoparticles for biology and medicine. Angew Chem Int Ed Engl. 2010;49:3280–94. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cobley CM, Chen J, Cho EC, Wang LV, Xia Y. Gold nanostructures: a class of multifunctional materials for biomedical applications. Chem Soc Rev. 2011;40:44–56. doi: 10.1039/b821763g. [DOI] [PubMed] [Google Scholar]
- 21.Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7:389–93. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang T, Sun W, Zhu Q, Burns NA, Khan SA, Mo R, et al. Furin-mediated sequential delivery of anticancer cytokine and small-molecule drug shuttled by graphene. Adv Mater. 2015 doi: 10.1002/adma.201404498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mo R, Jiang T, DiSanto R, Tai W, Gu Z. ATP-triggered anticancer drug delivery. Nat Commun. 2014;5:3364. doi: 10.1038/ncomms4364. [DOI] [PubMed] [Google Scholar]
- 24.Mo R, Jiang T, Gu Z. Enhanced anticancer efficacy by ATP-mediated liposomal drug delivery. Angew Chem Int Ed Engl. 2014;53:5815–20. doi: 10.1002/anie.201400268. [DOI] [PubMed] [Google Scholar]
- 25.Naito M, Ishii T, Matsumoto A, Miyata K, Miyahara Y, Kataoka K. A phenylboronate-functionalized polyion complex micelle for ATP-triggered release of siRNA. Angew Chem Int Ed Engl. 2012;51:10751–5. doi: 10.1002/anie.201203360. [DOI] [PubMed] [Google Scholar]
- 26.Biswas S, Kinbara K, Niwa T, Taguchi H, Ishii N, Watanabe S, et al. Biomolecular robotics for chemomechanically driven guest delivery fuelled by intracellular ATP. Nat Chem. 2013;5:613–20. doi: 10.1038/nchem.1681. [DOI] [PubMed] [Google Scholar]
- 27.Gribble FM, Loussouarn G, Tucker SJ, Zhao C, Nichols CG, Ashcroft FM. A novel method for measurement of submembrane ATP concentration. J Biol Chem. 2000;275:30046–9. doi: 10.1074/jbc.M001010200. [DOI] [PubMed] [Google Scholar]
- 28.Gorman MW, Feigl EO, Buffington CW. Human plasma ATP concentration. Clin Chem. 2007;53:318–25. doi: 10.1373/clinchem.2006.076364. [DOI] [PubMed] [Google Scholar]
- 29.Yang K, Feng L, Shi X, Liu Z. Nano-graphene in biomedicine: theranostic applications. Chem Soc Rev. 2013;42:530–47. doi: 10.1039/c2cs35342c. [DOI] [PubMed] [Google Scholar]
- 30.Yang X, Zhang X, Liu Z, Ma Y, Huang Y, Chen Y. High-efficiency loading and controlled release of doxorubicin hydrochloride on graphene oxide. J Phys Chem C. 2008;112:17554–8. [Google Scholar]
- 31.Zhang L, Xia J, Zhao Q, Liu L, Zhang Z. Functional graphene oxide as a nanocarrier for controlled loading and targeted delivery of mixed anticancer drugs. Small. 2010;6:537–44. doi: 10.1002/smll.200901680. [DOI] [PubMed] [Google Scholar]
- 32.Wang F, Liu B, Ip AC, Liu J. Orthogonal adsorption onto nano-graphene oxide using different intermolecular forces for multiplexed delivery. Adv Mater. 2013;25:4087–92. doi: 10.1002/adma.201301183. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Xu P, Shu Z, Wu M, Wang L, Zhang S, et al. Multifunctional graphene oxide-based triple stimuli-responsive nanotheranostics. Adv Funct Mater. 2014;24:4386–4396. [Google Scholar]
- 34.Feng L, Li K, Shi X, Gao M, Liu J, Liu Z. Smart pH-responsive nanocarriers based on nano-graphene oxide for combined chemo- and photothermal therapy overcoming drug resistance. Adv Healthcare Mater. 2014;3:1261–1271. doi: 10.1002/adhm.201300549. [DOI] [PubMed] [Google Scholar]
- 35.Tang L, Wang Y, Liu Y, Li J. DNA-directed self-assembly of graphene oxide with applications to ultrasensitive oligonucleotide assay. ACS Nano. 2011;5:3817–22. doi: 10.1021/nn200147n. [DOI] [PubMed] [Google Scholar]
- 36.Lv W, Guo M, Liang MH, Jin FM, Cui L, Zhi L, et al. Graphene-DNA hybrids: self-assembly and electrochemical detection performance. J Mater Chem. 2010;20:6668–73. [Google Scholar]
- 37.Jin H, Jeng ES, Heller DA, Jena PV, Kirmse R, Langowski J, et al. Divalent ion and thermally induced DNA conformational polymorphism on single-walled carbon nanotubes. Macromolecules. 2007;40:6731–9. [Google Scholar]
- 38.Varghese N, Mogera U, Govindaraj A, Das A, Maiti PK, Sood AK, et al. Binding of DNA nucleobases and nucleosides with graphene. Chemphyschem. 2009;10:206–10. doi: 10.1002/cphc.200800459. [DOI] [PubMed] [Google Scholar]
- 39.Patil AJ, Vickery JL, Scott TB, Mann S. Aqueous stabilization and self-assembly of graphene sheets into layered bio-nanocomposites using DNA. Adv Mater. 2009;21:3159–64. [Google Scholar]
- 40.Stankovich S, Dikin DA, Piner RD, Kohlhaas KA, Kleinhammes A, Jia Y, et al. Synthesis of graphene-based nanosheets via chemical reduction of exfoliated graphite oxide. Carbon. 2007;45:1558–65. [Google Scholar]
- 41.Zhao B, Song J, Liu P, Xu W, Fang T, Jiao Z, et al. Monolayer graphene/NiO nanosheets with two-dimension structure for supercapacitors. J Mater Chem. 2011;21:18792–8. [Google Scholar]
- 42.Liu JW, Lu Y. Smart nanomaterials responsive to multiple chemical stimuli with controllable cooperativity. Adv Mater. 2006;18:1667–71. [Google Scholar]
- 43.Wang Y, Li Z, Hu D, Lin CT, Li J, Lin Y. Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J Am Chem Soc. 2010;132:9274–6. doi: 10.1021/ja103169v. [DOI] [PubMed] [Google Scholar]
- 44.Meng HM, Zhang X, Lv Y, Zhao Z, Wang NN, Fu T, et al. DNA dendrimer: an efficient nanocarrier of functional nucleic acids for intracellular molecular sensing. ACS Nano. 2014;8:6171–81. doi: 10.1021/nn5015962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong BJ, Compton OC, An Z, Eryazici I, Nguyen ST. Successful stabilization of graphene oxide in electrolyte solutions: enhancement of biofunctionalization and cellular uptake. ACS Nano. 2012;6:63–73. doi: 10.1021/nn202355p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao Y, Lin Y, Ren J, Qu X. Self-assembled, functionalized graphene and DNA as a universal platform for colorimetric assays. Biomaterials. 2013;34:4810–7. doi: 10.1016/j.biomaterials.2013.03.039. [DOI] [PubMed] [Google Scholar]
- 47.Lin B, Sun Q, Liu K, Lu D, Fu Y, Xu Z, et al. Label-free colorimetric protein assay and logic gates design based on the self-assembly of hemin-graphene hybrid nanosheet. Langmuir. 2014;30:2144–51. doi: 10.1021/la4048769. [DOI] [PubMed] [Google Scholar]
- 48.Wu M, Kempaiah R, Huang PJ, Maheshwari V, Liu J. Adsorption and desorption of DNA on graphene oxide studied by fluorescently labeled oligonucleotides. Langmuir. 2011;27:2731–8. doi: 10.1021/la1037926. [DOI] [PubMed] [Google Scholar]
- 49.Huang PJ, Liu J. DNA-length-dependent fluorescence signaling on graphene oxide surface. Small. 2012;8:977–83. doi: 10.1002/smll.201102156. [DOI] [PubMed] [Google Scholar]
- 50.Yang H, Paek K, Kim BJ. Efficient temperature sensing platform based on fluorescent block copolymer-functionalized graphene oxide. Nanoscale. 2013;5:5720–4. doi: 10.1039/c3nr01486j. [DOI] [PubMed] [Google Scholar]
- 51.Lu J, Liong M, Sherman S, Xia T, Kovochich M, Nel AE, et al. Mesoporous silica nanoparticles for cancer therapy: energy-dependent cellular uptake and delivery of paclitaxel to cancer cells. Nanobiotechnology. 2007;3:89–95. doi: 10.1007/s12030-008-9003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, et al. The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nat Mater. 2011;10:389–97. doi: 10.1038/nmat2992. [DOI] [PMC free article] [PubMed] [Google Scholar]