Abstract
Background
Published data showed that the susceptibility of autoimmune diseases (ADs) was associated with the polymorphism rs2910164 in microRNA-146a (miR-146a). However, the results remain controversial so far. Two meta-analyses published in 2013 and 2014 came to opposite conclusions. In order to derive a more precise estimation of the relationship, we performed this meta-analysis.
Methods
We searched the PubMed, OvidSP and CNKI databases (published prior to September 8th, 2014) and extracted data from eligible studies. The procedure of meta-analysis was performed by using the Stata 12.0 software. Random effect model or fixed effect model were chosen respectively, according to the between study heterogeneities.
Results
A total of 24 case-control studies, 11 more than previous meta-analysis on this topic, were involved. We took stratified analyses by different ethnicities and different types of diseases in different genetic models. In Caucasian subgroup, significant increased risks of GC genotype and GC+CC genotype with ADs susceptibility were found in heterozygote model (GC vs GG, OR = 1.38, 95% CI 1.04–1.83, p = 0.024) and dominant model (GC+CC vs GG, OR = 1.37, 95% CI 1.01–1.85, p = 0.041), respectively. Meanwhile, in other disease subgroup, significant increased risks of C allele, CC genotype and GC+CC genotype were found in allele model (C vs G, OR = 1.16, 95% CI 1.04–1.31, p = 0.010), homozygote model (CC vs GG, OR = 1.42, 95% CI 1.10–1.84, p = 0.006) and dominant model (GC+CC vs GG, OR = 1.25, 95% CI 1.04–1.51, p = 0.020), respectively.
Conclusions
MiR-146a rs2910164 G>C polymorphism was associated with the susceptibility of ADs.
Introduction
Autoimmune diseases (ADs) are initiated by abnormal immune response to self-antigen, and then come the results including immune-mediated tissue destruction and chronic disabilities [1]. There are more than 100 diseases and syndromes in ADs, which cause a heavy economic burden in the world, about more than $100 billion annually [2]. More and more evidence indicated that genetic backgrounds play an important role in the pathogenesis of ADs, which may be controlled by a common set of susceptibility genes [3, 4].
MicroRNAs (miRNAs) are non-coding single-stranded RNA molecules. By binding to 3’ un-translated regions (UTRs) of targeted messenger RNAs, the miRNAs can repress, degrade or silence the gene expression [5, 6]. Therefore, miRNAs have been demonstrated to affect various functions in both innate and adaptive immune response [7, 8]. Among them, the microRNA-146a (miR-146a) was reported to be involved in both innate and adaptive immunity, especially played an important role in autoimmune disease [8, 9].
Single nucleotide polymorphisms (SNPs) or mutations in miRNAs may alter the expression level of the gene and the susceptibility of some diseases [10–13]. Many studies on the relationship between miR-146a rs2910164 G>C polymorphism and susceptibility of ADs have been performed so far [14–34]. However, the results remain inconsistent. Moreover, two meta-analyses on this issue published in 2013 and 2014 also generated opposite conclusions [35, 36]. Based on the new case-control studies [27, 28, 30–34], we conducted this updated meta-analysis according the criteria PRISMA statement [37], in order to clarify the association between miR-146a rs2910164 G>C polymorphism and susceptibility of ADs.
Materials and Methods
Publication search
A systematic search was performed in PubMed, OvidSP and Chinese National Knowledge Infrastructure (CNKI) databases covering all papers published prior to September 8th, 2014. The searching strategy was as follow: (autoimmune OR autoimmune disease OR autoimmunity) AND (polymorphism OR polymorphisms OR variation OR variations OR mutation OR mutations OR variant OR variants) AND (Has-mir-146a OR miR146a OR microRNA-146a OR miR-146 OR miR-146a OR rs2910164). The references in the studies were also read to find additional publications on the topic. Articles included should meet the criteria below: (1) case-control study; (2) evaluation of miR-146a rs2910164 G>C polymorphism and risk of ADs; (3) available and usable data of genotype frequency. The articles were excluded if they meet the exclusion criteria below: (1) a case report, review or descriptive study; (2) a lack of normal population as controls; (3) not show the evaluation of miR-146a rs2910164; (4) duplicate data in the studies.
Date extraction
Two authors (CL and WF) independently extracted the data from eligible studies. Different data extracted by CL and WF were checked by the third author YZ. The remained disagreements were discussed and judged by these three authors. The following information was extracted: the first author, publication year, diseases, country, ethnicity, genotyping methods, number of cases and controls, the gender distribution of cases and controls, number of genotypes and alleles, Hardy-Weinberg equilibrium (HWE) in control subjects, frequency of G allele in controls. Ethnicities were categorized as Caucasian, East Asian, Latin-American and Mediterranean. Study qualities were judged according to the criteria modified from previous publications [38–40] (See S1 Table. “Scale for methodological quality assessment”).
Statistical analysis
Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated as a measure of the association between miR-146a rs2910164 G>C polymorphism and ADs risk. The widely reported genetic models, allele model, heterozygote model, homozygote model, dominant model and recessive model, were used. In addition to comparing the pooled effects among all subjects, the stratified comparisons were also conducted according to different ethnicities and different diseases. In order to evaluate the accumulation of evidence, the cumulative meta-analysis was performed. The between study heterogeneity was measured by Cochran’s (Q) and Higgins’s (I2) tests. If the heterogeneity was considered significant (p<0.05), the random effect model was used to estimate the pooled OR. Otherwise, the fixed effect model was conducted. Also, logistic meta-regression analysis was carried out, if there was obvious significant heterogeneity, to explore potential sources of heterogeneity. The examined characteristics include: publication years, countries, genotyping methods, number of alleles and genotypes, number of female and male in cases, and the frequency of G allele in controls. The HWE was examined using Chi-square test with significance set at p<0.05. Sensitivity analysis was performed to evaluate the effect of each study on the combined ORs by deleting each study in each time, and to evaluate the effect of studies with low quality or without HWE on the pooled ORs by deleting these studies. Potential publication bias was determined by using Funnel plots and Begg’s test. An asymmetric plot and the p value less than 0.05 was recognized as significance. All statistical analyses were performed by Stata 12.0 software (Stata Corp., College Station, TX).
Results
Study Characteristics
There were 406 articles matching the searching strategy, and additional 5 articles [20, 23, 25, 28, 31] were included by scanning the references of original papers. After step by step of screening the titles, abstracts and full-texts of the articles, as shown in Fig 1, 21 articles were recognized appropriate for this meta-analysis, including 24 studies for rs2910164, 11 studies more than the previous meta-analysis published in 2014 [36].
Fig 1. Flowchart for identification of studies included in the meta-analysis.
In 411 articles, 51 were found not related to ADs and 43 were found not related to miR-146a by scanning the titles. After that, 233 articles were recognized as reviews, 37 were found not related to human patients and 8 articles were repeated papers by reviewing the abstracts. The full-texts of the left 39 articles were carefully reviewed, in which 18 articles were found not about rs2910164. At last, 21 articles were remained for this meta-analysis, which included 24 case-control studies for rs2910164.
Within the 21 articles, five kinds of genotyping methods were used. There were 4 racial included, Caucasian, East Asian, Latin-American and Mediterranean. According the different types of diseases, these studies were divided into five subgroups. The studies on Rheumatoid Arthritis (RA), Psoriatic Arthritis (PsA), Juvenile RA (JRA) and Juvenile Idiopathic Arthritis-Enthesitis-Related Arthritis (JIA-ERA) were included into Inflammatory Arthritis (IA) subgroup. The studies on Ulcerative Colitis (UC) and Crohn’s Disease (CD) were included into Inflammatory Bowel Disease (IBD) subgroup. Among these studies, the patients with Behcet’s Disease (BD), Vogt-Koyanagi-Harada syndrome (VKH), Fuchs Uveitis Syndrome (FUS) and Pediatric Uveitis (PU) were all suffering uveitis, which was a common syndrome of ADs. So the Uveitis subgroup was formed. The remained studies, except that for Systemic Lupus Erythematosus (SLE), were included in other diseases subgroup. There were 5 studies not in HWE in control groups (p<0.05), but the p value of HWE were not less than 0.01. And there was not enough data in another study to generate the HWE in control. The detail characteristics are shown in Table 1.
Table 1. Characteristics of published studies of rs2910164.
| First author | Year | Diseases | Country | Ethnicity | Sample size | Female/Male | Genotyping methods | Case | Control | HWE of control (p value) | Frequency of G Allele in controls | Quality | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Allele | Genotype | Allele | |||||||||||||||||||
| case | control | case | control | GG | GC | CC | G | C | GG | GC | CC | G | C | |||||||||
| Chatzikyriakidou | 2010a | RA | Greece | Caucasian | 136 | 147 | 109/27 | 115/32 | PCR-RFLP | 73 | 53 | 10 | 199 | 73 | 80 | 53 | 14 | 213 | 81 | Y(0.240) | 0.72 | 7 |
| Chatzikyriakidou | 2010b | PsA | Greece | Caucasian | 29 | 66 | 10/19 | 30/36 | PCR-RFLP | 14 | 12 | 3 | 40 | 18 | 39 | 18 | 9 | 96 | 36 | N(0.011) | 0.73 | 5 |
| Luo | 2011 | SLE | China | East Asian | 816 | 1080 | Direct sequencing | 669 | 963 | 864 | 1296 | 0.40 | 8 | |||||||||
| Zhang | 2011 | SLE | China | East Asian | 213 | 209 | 201/12 | 195/14 | PCR-RFLP | 33 | 101 | 79 | 167 | 259 | 23 | 104 | 82 | 150 | 268 | Y(0.239) | 0.36 | 8 |
| Yang | 2011 | RA | China | East Asian | 208 | 240 | 172/36 | 195/45 | PCR-RFLP | 28 | 95 | 85 | 151 | 265 | 30 | 116 | 94 | 176 | 304 | Y(0.529) | 0.37 | 8 |
| Fenoglio | 2011 | MS | Italy | Caucasian | 346 | 339 | 240/106 | 207/132 | TaqMan | 181 | 135 | 30 | 497 | 195 | 195 | 115 | 29 | 505 | 173 | N(0.048) | 0.74 | 7 |
| Okubo | 2011 | UC | Japan | East Asian | 170 | 403 | 74/96 | 208/195 | PCR-RFLP | 28 | 67 | 75 | 123 | 217 | 74 | 178 | 151 | 326 | 480 | Y(0.095) | 0.40 | 7 |
| Lofgren | 2012 | SLE | Sweden | Caucasian | 1109 | 1428 | TaqMan | 623 | 422 | 64 | 1668 | 550 | 819 | 531 | 78 | 2169 | 687 | Y(0.503) | 0.76 | 7 | ||
| Jimenez-Morales | 2012 | SLE | Mexico | Latin-American | 367 | 531 | 304/63 | 299/232 | TaqMan | 163 | 167 | 37 | 493 | 241 | 236 | 229 | 66 | 701 | 361 | Y(0.369) | 0.66 | 8 |
| Jimenez-Morales | 2012 | JRA | Mexico | Latin-American | 210 | 531 | 125/85 | 299/232 | TaqMan | 102 | 80 | 28 | 284 | 136 | 236 | 229 | 66 | 701 | 361 | Y(0.369) | 0.66 | 8 |
| Qian | 2012 | RA | China | East Asian | 123 | 220 | 105/18 | 196/24 | PCR-LDR | 16 | 65 | 42 | 97 | 149 | 35 | 109 | 76 | 179 | 261 | Y(0.694) | 0.41 | 8 |
| Sakoguchi | 2012 | SSc | Japan | East Asian | 52 | 107 | PCR-RFLP | 1 | 28 | 23 | 30 | 74 | 3 | 53 | 51 | 59 | 155 | N(0.013) | 0.28 | 1 | ||
| Zhou | 2012 | FUS | China | East Asian | 219 | 612 | 102/117 | 262/350 | PCR-RFLP | 36 | 91 | 92 | 163 | 275 | 79 | 279 | 254 | 437 | 787 | Y(0.862) | 0.36 | 9 |
| Hashemi | 2013 | RA | Iran | Mediterranean | 104 | 110 | 91/13 | 70/40 | T-ARMS-PCR | 57 | 39 | 8 | 153 | 55 | 64 | 37 | 9 | 165 | 55 | Y(0.280) | 0.75 | 8 |
| EI-Shal | 2013 | RA | Egypt | Mediterranean | 217 | 245 | 217/0 | 245/0 | PCR-RFLP | 30 | 103 | 84 | 163 | 271 | 15 | 119 | 111 | 149 | 341 | N(0.021) | 0.30 | 7 |
| Gazouli | 2013 | UC | Greece | Caucasian | 210 | 300 | 109/101 | 159/141 | PCR-RFLP | 126 | 78 | 6 | 330 | 90 | 200 | 90 | 10 | 490 | 110 | Y(0.974) | 0.82 | 7 |
| Gazouli | 2013 | CD | Greece | Caucasian | 242 | 300 | 134/108 | 159/141 | PCR-RFLP | 105 | 113 | 24 | 323 | 161 | 200 | 90 | 10 | 490 | 110 | Y(0.974) | 0.82 | 7 |
| Zhou | 2014 | BD | China | East Asian | 809 | 1132 | 131/678 | 513/619 | PCR-RFLP | 131 | 440 | 238 | 702 | 916 | 154 | 518 | 460 | 826 | 1438 | Y(0.670) | 0.36 | 10 |
| Zhou | 2014 | VKH | China | East Asian | 613 | 1132 | 287/326 | 513/619 | PCR-RFLP | 64 | 273 | 276 | 401 | 825 | 154 | 518 | 460 | 826 | 1438 | Y(0.670) | 0.36 | 10 |
| Singh | 2014 | JIA-ERA | India | Mediterranean | 150 | 216 | 17/133 | 15/201 | PCR-RFLP | 75 | 56 | 19 | 206 | 94 | 112 | 91 | 13 | 315 | 117 | Y(0.327) | 0.73 | 8 |
| Lin | 2014 | IgAN | China | East Asian | 404 | 711 | 169/235 | 261/450 | PCR-LDR | 103 | 195 | 106 | 401 | 407 | 219 | 360 | 132 | 798 | 624 | Y(0.454) | 0.56 | 8 |
| Zhao | 2014 | ITP | China | East Asian | 280 | 270 | 181/99 | 156/114 | PCR-RFLP | 35 | 134 | 111 | 204 | 356 | 36 | 135 | 99 | 207 | 333 | Y(0.344) | 0.38 | 8 |
| Wei | 2014 | PU | China | East Asian | 520 | 1204 | 278/242 | 659/545 | PCR-RFLP | 113 | 248 | 159 | 474 | 566 | 163 | 553 | 488 | 879 | 1529 | Y(0.750) | 0.37 | 9 |
| Okada | 2014 | PM/DM | Japan | East Asian | 44 | 107 | 28/16 | PCR-RFLP | 0 | 29 | 15 | 29 | 59 | 3 | 53 | 51 | 59 | 155 | N(0.013) | 0.28 | 3 | |
Abbreviations: RA, Rheumatoid Arthritis; PsA, Psoriatic Arthritis; SLE, Systemic Lupus Erythematosus; MS, Multiple Sclerosis; UC, Ulcerative Colitis; JRA, Juvenile Rheumatoid Arthritis; SSc, Systemic Sclerosis; FUS, Fuchs Uveitis Syndrome; CD, Crohn’s Disease; BD, Behcet’s Disease; VKH, Vogt-Koyanagi-Harada syndrome; JIA-ERA, Juvenile Idiopathic Arthritis-Enthesitis-Related Arthritis; IgAN, Immunoglobulin A nephropathy; ITP, Immune Thrombocytopenia; PU, Pediatric Uveitis; PM/DM, Polymyositis/Dermatomyositis.
Association between miR-146a rs2910164 G>C polymorphism and ADs risk
First, we investigated the overall association between rs2910164 and susceptibility of ADs. No significant difference of the pooled OR was found in any genetic model. However, in terms of stratified analysis, the significance emerged. In Caucasian subgroup, significant increased risks of GC genotype and GC+CC genotype with ADs susceptibility were found in heterozygote model (GC vs GG, OR = 1.38, 95% CI 1.04–1.83, p = 0.024) and dominant model (GC+CC vs GG, OR = 1.37, 95% CI 1.01–1.85, p = 0.041), respectively (Table 2, Fig 2A and B).
Table 2. Stratified analysis of association between ADs risk and rs2910164.
| Stratify | Study(n) | Allele model (C vs G) | Heterozygote model (GC vs GG) | Homozygote model (CC vs GG) | Dominant model (GC+CC vs GG) | Recessive model (CC versus GG+GC) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effects size | Heterogeneity | Effect model | Effects size | Heterogeneity | Effect model | Effects size | Heterogeneity | Effect model | Effects size | Heterogeneity | Effect model | Effects size | Heterogeneity | Effect model | |||||||||||||
| OR | p | I 2 (%) | p | OR | p | I 2 (%) | p | OR | p | I 2 (%) | p | OR | p | I 2 (%) | p | OR | p | I 2 (%) | p | ||||||||
| (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | |||||||||||||||||||||||
| Total | 24 | 1.02 | 0.653 | 77.9 | 0.000 | Random | 1.04 | 0.556 | 58.6 | 0.000 | Random | 1.01 | 0.914 | 73.1 | 0.000 | Random | 1.04 | 0.622 | 70.7 | 0.000 | Random | 1.00 | 0.995 | 70.2 | 0.000 | Random | |
| (0.93–1.13) | (0.91–1.19) | (0.81–1.27) | (0.89–1.21) | (0.85–1.17) | |||||||||||||||||||||||
| Ethnicities | Caucasian | 6 | 1.25 | 0.083 | 79.5 | 0.000 | Random | 1.38 | 0.024 | 72.0 | 0.003 | Random | 1.28 | 0.306 | 61.9 | 0.022 | Random | 1.37 | 0.041 | 77.9 | 0.000 | Random | 1.14 | 0.511 | 44.7 | 0.107 | Random |
| (0.97–1.60) | (1.04–1.83) | (0.80–2.07) | (1.01–1.85) | (0.77–1.67) | |||||||||||||||||||||||
| East Asian | 13 | 0.97 | 0.663 | 79.3 | 0.000 | Random | 0.95 | 0.418 | 34.0 | 0.118 | Fixed | 0.96 | 0.817 | 78.8 | 0.000 | Random | 0.95 | 0.642 | 64.3 | 0.001 | Random | 0.96 | 0.731 | 80.0 | 0.000 | Random | |
| (0.85–1.10) | (0.84–1.07) | (0.71–1.32) | (0.77–1.18) | (0.78–1.19) | |||||||||||||||||||||||
| Latin-American | 2 | 0.94 | 0.440 | 0.0 | 0.897 | Fixed | 0.95 | 0.633 | 27.7 | 0.240 | Fixed | 0.88 | 0.466 | 0.0 | 0.579 | Fixed | 0.93 | 0.519 | 0.0 | 0.432 | Fixed | 0.91 | 0.550 | 0.0 | 0.331 | Fixed | |
| (0.81–1.10) | (0.76–1.18) | (0.63–1.23) | (0.76–1.15) | (0.66–1.25) | |||||||||||||||||||||||
| Mediterranean | 3 | 0.97 | 0.874 | 68.9 | 0.040 | Random | 0.81 | 0.417 | 61.9 | 0.072 | Random | 0.93 | 0.892 | 82.4 | 0.003 | Random | 0.83 | 0.519 | 72.4 | 0.027 | Random | 1.14 | 0.725 | 70.0 | 0.036 | Random | |
| (0.69–1.38) | (0.48–1.36) | (0.30–2.82) | (0.46–1.48) | (0.56–2.32) | |||||||||||||||||||||||
| Diseases | IA | 8 | 0.97 | 0.607 | 77.9 | 0.384 | Fixed | 0.92 | 0.384 | 28.2 | 0.204 | Fixed | 0.95 | 0.767 | 42.8 | 0.093 | Random | 0.94 | 0.496 | 29.8 | 0.190 | Fixed | 0.99 | 0.906 | 8.2 | 0.366 | Fixed |
| (0.87–1.09) | (0.77–1.11) | (0.67–1.35) | (0.79–1.12) | (0.82–1.19) | |||||||||||||||||||||||
| SLE | 4 | 0.98 | 0.625 | 79.5 | 0.622 | Fixed | 1.02 | 0.748 | 0.0 | 0.379 | Fixed | 0.91 | 0.468 | 5.2 | 0.348 | Fixed | 1.01 | 0.857 | 6.9 | 0.342 | Fixed | 0.93 | 0.544 | 0.0 | 0.567 | Fixed | |
| (0.90–1.06) | (0.89–1.17) | (0.71–1.17) | (0.89–1.16) | (0.75–1.16) | |||||||||||||||||||||||
| IBD | 3 | 1.48 | 0.057 | 83.5 | 0.002 | Random | 1.52 | 0.096 | 76.6 | 0.014 | Random | 1.82 | 0.190 | 76.3 | 0.015 | Random | 1.61 | 0.069 | 80.3 | 0.006 | Random | 1.57 | 0.178 | 63.3 | 0.066 | Random | |
| (0.99–2.21) | (0.93–2.49) | (0.74–4.46) | (0.96–2.68) | (0.82–3.02) | |||||||||||||||||||||||
| Uveitis | 4 | 0.87 | 0.271 | 90.5 | 0.000 | Random | 0.88 | 0.429 | 72.7 | 0.012 | Random | 0.75 | 0.262 | 88.8 | 0.000 | Random | 0.83 | 0.310 | 83.1 | 0.000 | Random | 0.83 | 0.293 | 89.8 | 0.000 | Random | |
| (0.67–1.12) | (0.65–1.20) | (0.46–1.24) | (0.57–1.20) | (0.58–1.18) | |||||||||||||||||||||||
| Others | 5 | 1.16 | 0.010 | 15.5 | 0.316 | Fixed | 1.19 | 0.092 | 0.0 | 0.884 | Fixed | 1.42 | 0.006 | 0.0 | 0.649 | Fixed | 1.25 | 0.020 | 0.0 | 0.942 | Fixed | 1.08 | 0.611 | 53.1 | 0.074 | Random | |
| (1.04–1.31) | (0.97–1.44) | (1.10–1.84) | (1.04–1.51) | (0.80–1.47) | |||||||||||||||||||||||
Fig 2. Forest plots of ADs risk associated with rs2910164.
(A-B) Forest plots of ADs risk associated with rs2910164 stratified analyzed by ethnicities. (A) Heterozygote model, GC vs GG, Caucasian subgroup, random model. (B) Dominant model, GC+CC vs GG, Caucasian subgroup, random model. (C-E) Forest plots of ADs risk associated with rs2910164 stratified analyzed by diseases. (C) Allele model, C vs G, Other diseases subgroup, fixed model. (D) Homozygote model, CC vs GG, Other diseases subgroup, fixed model. (E) Dominant model, GC+CC vs GG, Other diseases subgroup, fixed model. OR: odds ratio; 95% CI: 95% confidence interval.
Meanwhile, in other disease subgroup, significant increased risks of C allele, CC genotype and GC+CC genotype with ADs were found in allele model (C vs G, OR = 1.16, 95% CI 1.04–1.31, p = 0.010), homozygote model (CC vs GG, OR = 1.42, 95% CI 1.10–1.84, p = 0.006) and dominant model (GC+CC vs GG, OR = 1.25, 95% CI 1.04–1.51, p = 0.020), respectively (Table 2, Fig 2C, D, E). There was not any significance in IA, SLE, IBD and Uveitis subgroup. However, a trend of increased disease risk of C allele could be found in allele model (C vs G, OR = 1.48, 95% CI 0.99–2.21, p = 0.057) in IBD subgroup (Table 2).
The cumulative meta-analysis showed that the significance of ORs emerged after the studies published in 2013 enrolled in Caucasian subgroup (Fig 3A and B), while after the studies published in 2014 enrolled in other disease subgroup (Fig 3C, D, and E).
Fig 3. Cumulative meta-analysis of the association between rs2910164 and ADs risk.
Every rhombus represents the pooled OR when studies accumulated over time, and the horizontal line represents the 95% CI of the pooled ORs. (A) Heterozygote model, GC vs GG, Caucasian subgroup, random model. (B) Dominant model, GC+CC vs GG, Caucasian subgroup, random model. (C) Allele model, C vs G, Other diseases subgroup, fixed model. (D) Homozygote model, CC vs GG, Other diseases subgroup, fixed model. (E) Dominant model, GC+CC vs GG, Other diseases subgroup, fixed model.
Evaluation of heterogeneity
The heterogeneities among studies were obvious in the overall comparisons (I2 = 77.9%, Tau2 = 0.044, p = 0.000). The meta-regression analysis was conducted to further explore sources of heterogeneity. We assessed allele comparison by potential sources of publication year, country, genotyping methods, number of genotypes and alleles, and number of female and male in cases. None of the potential sources above could explain the heterogeneity by meta-regression analysis. However, when we compared the frequencies of G allele in controls, we found that the heterogeneity could partly (Adjusted R2 = 32.7%) explained by the variation of frequencies of G allele in controls.
Sensitivity and publication bias analysis
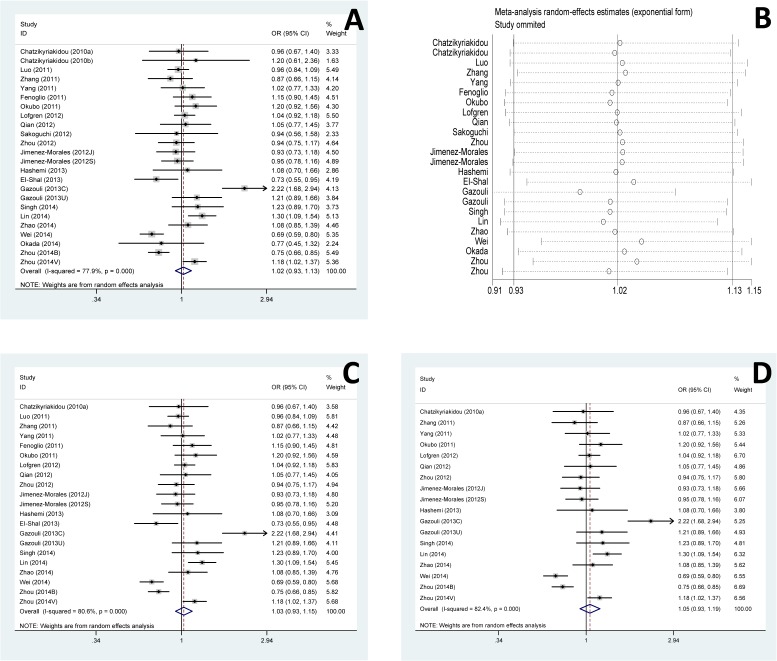
We performed the sensitivity analysis to test the influence of a single study on the overall meta-analysis by deleting each study once a time. As a result, the pooled estimate didn’t show significant difference, which indicated that the results in this meta-analyses were statistically reliable. Moreover, the pooled ORs did not vary much even after the three studies [15, 24, 34] with low quality or six studies [15, 16, 19, 24, 27, 34] without HWE removed (Fig 4). No evidence of publication bias was found in current meta-analysis, identified by the Funnel plots, Egger’s test (p = 0.261) and Begg’s test (p = 0.862) (Fig 5).
Fig 4. Sensitivity analysis of association of rs2910164 and ADs risk.
(A) Pooled analysis of association of rs2910164 and ADs risk. Allele model, C vs G. (B) Sensitivity analysis by iteratively removing one study at a time. (C) Sensitivity analysis by removing three studies with low quality. (D) Sensitivity analysis by removing six studies without HWE.
Fig 5. Publication bias on the rs2910164 polymorphism and ADs risk.
Discussion
Solid evidences had shown that miRNAs played important roles in the regulation of cell differentiation, proliferation, metabolism, apoptosis and tumorigenesis [41]. MiR-146a was one of the first miRNAs identified to be involved in innate immune response, and further demonstrated to be related to several types of cancers [42–44].
One SNP of miR-146a, rs2910164 G>C, was found not only related to cancer, but also related to some kinds of ADs [29]. Accumulating evidences emerged to distinguish if there was any relationship between rs2910164 and susceptibility of ADs. However, the results were inconsistent. A meta-analysis published in 2013 showed that there was not any association between rs2910164 and ADs risk [35]. While they did not include a study published by Luo et al [16] in 2011. Maybe due to there was only frequency data of allele and was not any data of each genotype in Luo’s study, which did not meet the inclusion criteria of this meta-analysis [35]. On the contrary, another meta-analysis published in 2014 did find some association there [36], however, some already published studies [15, 20, 25] prior their search date at April 2013 were not included although they should be. Moreover, the only significances of ORs were found in GG+CC/GC genetic model [36], not in the five well used genetic models as shown in the previous meta-analysis [35]. After that, other researchers published several new case-control studies about this topic in 2013 and 2014 [27, 28, 30–34]. So, it was needed to do an updated meta-analysis which included all the published manuscripts, in order to make a clarified conclusion.
In this meta-analysis, we enrolled 24 studies and pooled the corresponding data including 7591 cases and 9677 controls, and all data of these samples were available in the original publications. Our analysis revealed that there was not any significance of pooled OR. However, in stratified analyses, we found GC genotype and GC+CC genotype were significantly related to the increased susceptibility of ADs in Caucasian subgroup. As in other diseases subgroup, significantly increased risk was observed to be associated with C allele, CC genotype and GC+CC genotype in allele model, homozygote model and dominant model, respectively. All these differences were not found in the two previous meta-analyses [35, 36]. These differences between our data and that in previous meta-analyses may be explained by the different study number and different sample size enrolled in these analyses. As we included more studies and an enlarged sample size, our data should be recognized more powerful.
In cumulative meta-analysis, we found the significance of ORs after new case-control studies published in 2013 and 2014 enrolled. These data demonstrated the correlation of rs2910164 and susceptibility of ADs. In addition, not only sensitivity analysis by iteratively removing one study at a time, but also analysis removing three studies [15, 24, 34] with low quality or six studies [15, 16, 19, 24, 27, 34] without HWE, showed similar and consistent results. Thus, the results of cumulative meta-analysis and sensitivity analysis indicated the robustness of our data. There were still some limitations in our studies. First, although there were 24 studies included, the studies for some stratified analyses were limited. For example, there were only two studies for Latin-American subgroup. Second, there were obvious heterogeneities between different groups for some genetic model. Although the meta-regression and sensitivity analyses were conducted and we found that the variation of G allele frequency in controls could partly explain some heterogeneity, the results still needed to be treated with caution. Third, although there was some significance of ORs in some genetic models, the summary ORs were not very high even after the new studies enrolled in the cumulate meta-analyses. So, future well-designed large studies on this topic will be welcomed to confirm this conclusion. Finally, only rs2910164 in miR-146a was included in this study. However, there were more other SNPs in miR-146a and more other genes could also contribute to susceptibility of ADs. Not only the effect of the SNPs or genes, but also the interaction or network among these genetic locations, should be studied in the future. Furthermore, our data did not study the gene-environment interactions based on the lack of this interaction from the original case-control studies. So, in the future, studies investigating the gene-environment interactions would also help to make clear of the role of the genetic locations in the pathogen of ADs.
Conclusions
Taken together, our data demonstrated that the miR-146a rs2910164 G>C polymorphism was related to the susceptibility of ADs and our findings should be validated by future well-designed large studies.
Supporting Information
(DOC)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Natural Science Foundation of China (NO. 31100661), Specialized Research Fund for the Doctoral Program of Higher Education (NO. 20114433120019, NO. 20124433120002) and the Natural Science Foundation of Guangdong Province (NO. S2011040002748, NO. S2012040006972). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Marrack P, Kappler J, Kotzin BL. Autoimmune disease: why and where it occurs. Nat Med. 2001;7:899–905. [DOI] [PubMed] [Google Scholar]
- 2.American Autoimmune Related Diseases Association, National Coalition of Autoimmune Patient Groups. The Cost Burden of Autoimmune Disease: The Latest Front in the War on Healthcare Spending. 2011. Available: http://www.diabetesed.net/page/_files/autoimmune-diseases.pdf Accessed: 2014 Dec 31.
- 3. Goris A, Liston A. The immunogenetic architecture of autoimmune disease. Cold Spring Harb Perspect Biol 2012,4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller FW. Environmental agents and autoimmune diseases. Adv Exp Med Biol. 2011;711:61–81. [DOI] [PubMed] [Google Scholar]
- 5. Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. 10.1038/nature07758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–194. 10.1016/j.jaut.2009.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rusca N, Monticelli S. MiR-146a in Immunity and Disease. Mol Biol Int. 2011;2011:437301 10.4061/2011/437301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu WD, Lu MM, Pan HF, Ye DQ. Association of MicroRNA-146a with autoimmune diseases. Inflammation. 2012;35:1525–1529. 10.1007/s10753-012-9467-0 [DOI] [PubMed] [Google Scholar]
- 10. Iizuka T, Sawabe M, Takubo K, Liu M, Homma Y, Suzuki M, et al. hTERT promoter polymorphism, -1327C>T, is associated with the risk of epithelial cancer. Springerplus. 2013;2:249 10.1186/2193-1801-2-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suzuki M, Liu M, Kurosaki T, Suzuki M, Arai T, Sawabe M, et al. Association of rs6983561 polymorphism at 8q24 with prostate cancer mortality in a Japanese population. Clin Genitourin Cancer. 2011;9:46–52. 10.1016/j.clgc.2011.04.004 [DOI] [PubMed] [Google Scholar]
- 12. Liu M, Suzuki M, Arai T, Sawabe M, Enomoto Y, Nishimatsu H, et al. A replication study examining three common single-nucleotide polymorphisms and the risk of prostate cancer in a Japanese population. Prostate. 2011;71:1023–1032. 10.1002/pros.21317 [DOI] [PubMed] [Google Scholar]
- 13. Liu M, Kurosaki T, Suzuki M, Enomoto Y, Nishimatsu H, Arai T, et al. Significance of common variants on human chromosome 8q24 in relation to the risk of prostate cancer in native Japanese men. BMC Genet. 2009;10:37 10.1186/1471-2156-10-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. A polymorphism in the 3'-UTR of interleukin-1 receptor-associated kinase (IRAK1), a target gene of miR-146a, is associated with rheumatoid arthritis susceptibility. Joint Bone Spine. 2010;77:411–413. 10.1016/j.jbspin.2010.05.013 [DOI] [PubMed] [Google Scholar]
- 15. Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. The role of microRNA-146a (miR-146a) and its target IL-1R-associated kinase (IRAK1) in psoriatic arthritis susceptibility. Scand J Immunol. 2010;71:382–385. 10.1111/j.1365-3083.2010.02381.x [DOI] [PubMed] [Google Scholar]
- 16. Luo X, Yang W, Ye DQ, Cui H, Zhang Y, Hirankarn N, et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011;7:e1002128 10.1371/journal.pgen.1002128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J, Yang B, Ying B, Li D, Shi Y, Song X, et al. Association of pre-microRNAs genetic variants with susceptibility in systemic lupus erythematosus. Mol Biol Rep. 2011;38:1463–1468. 10.1007/s11033-010-0252-6 [DOI] [PubMed] [Google Scholar]
- 18. Yang B, Zhang JL, Shi YY, Li DD, Chen J, Huang ZC, et al. Association study of single nucleotide polymorphisms in pre-miRNA and rheumatoid arthritis in a Han Chinese population. Mol Biol Rep. 2011;38:4913–4919. 10.1007/s11033-010-0633-x [DOI] [PubMed] [Google Scholar]
- 19. Fenoglio C, Cantoni C, De Riz M, Ridolfi E, Cortini F, Serpente M, et al. Expression and genetic analysis of miRNAs involved in CD4+ cell activation in patients with multiple sclerosis. Neurosci Lett. 2011;504:9–12. 10.1016/j.neulet.2011.08.021 [DOI] [PubMed] [Google Scholar]
- 20. Okubo M, Tahara T, Shibata T, Yamashita H, Nakamura M, Yoshioka D, et al. Association study of common genetic variants in pre-microRNAs in patients with ulcerative colitis. J Clin Immunol. 2011;31:69–73. 10.1007/s10875-010-9461-y [DOI] [PubMed] [Google Scholar]
- 21. Lofgren SE, Frostegard J, Truedsson L, Pons-Estel BA, D'Alfonso S, Witte T, et al. Genetic association of miRNA-146a with systemic lupus erythematosus in Europeans through decreased expression of the gene. Genes Immun. 2012;13:268–274. 10.1038/gene.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jimenez-Morales S, Gamboa-Becerra R, Baca V, Del Rio-Navarro BE, Lopez-Ley DY, Velazquez-Cruz R, et al. MiR-146a polymorphism is associated with asthma but not with systemic lupus erythematosus and juvenile rheumatoid arthritis in Mexican patients. Tissue Antigens. 2012;80:317–321. 10.1111/j.1399-0039.2012.01929.x [DOI] [PubMed] [Google Scholar]
- 23. Qian L, Gao D, Wang G, Li X, Li X, Chen J, et al. Relationship between the single nucleotide polymorphisms in pre-miR-146a rs2910164 and expression of miR-146a in rheumatoid arthritis. Chin J Microbiol Immunol. 2012;32:253–257. [Google Scholar]
- 24. Sakoguchi A, Jinnin M, Makino T, Kajihara I, Makino K, Honda N, et al. The miR-146a rs2910164 C/G polymorphism is associated with telangiectasia in systemic sclerosis. Clin Exp Dermatol. 2013;38:99–100. 10.1111/j.1365-2230.2012.04453.x [DOI] [PubMed] [Google Scholar]
- 25. Zhou Q, Kijlstra A, Hou S, Yu H, Zhang X, Li X, et al. Lack of association of miR-146a and Ets-1 gene polymorphisms with Fuchs uveitis syndrome in Chinese Han patients. Mol Vis. 2012;18:426–430. [PMC free article] [PubMed] [Google Scholar]
- 26. Hashemi M, Eskandari-Nasab E, Zakeri Z, Atabaki M, Bahari G, Jahantigh M, et al. Association of pre-miRNA-146a rs2910164 and premiRNA-499 rs3746444 polymorphisms and susceptibility to rheumatoid arthritis. Mol Med Rep. 2013;7:287–291. 10.3892/mmr.2012.1176 [DOI] [PubMed] [Google Scholar]
- 27. El-Shal AS, Aly NM, Galil SM, Moustafa MA, Kandel WA. Association of microRNAs genes polymorphisms with rheumatoid arthritis in Egyptian female patients. Joint Bone Spine. 2013;80:626–631. 10.1016/j.jbspin.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 28. Gazouli M, Papaconstantinou I, Stamatis K, Vaiopoulou A, Zeglinas C, Vassiliou I, et al. Association study of genetic variants in miRNAs in patients with inflammatory bowel disease: preliminary results. Dig Dis Sci. 2013;58:2324–2328. 10.1007/s10620-013-2640-y [DOI] [PubMed] [Google Scholar]
- 29. Zhou Q, Hou S, Liang L, Li X, Tan X, Wei L, et al. MicroRNA-146a and Ets-1 gene polymorphisms in ocular Behcet's disease and Vogt-Koyanagi-Harada syndrome. Ann Rheum Dis. 2014;73:170–176. 10.1136/annrheumdis-2012-201627 [DOI] [PubMed] [Google Scholar]
- 30. Singh S, Rai G, Aggarwal A. Association of microRNA-146a and its target gene IRAK1 polymorphism with enthesitis related arthritis category of juvenile idiopathic arthritis. Rheumatol Int. 2014;34:1395–1400. 10.1007/s00296-014-3001-7 [DOI] [PubMed] [Google Scholar]
- 31. Lin J, Huang Y, Zhang X, Chen J, Sheng H. Association of miR-146a rs2910164 with childhood IgA nephropathy. Pediatr Nephrol. 2014;29:1979–1986. 10.1007/s00467-014-2818-3 [DOI] [PubMed] [Google Scholar]
- 32. Zhao H, Zhang Y, Xue F, Xu J, Fang Z. Has-mir-146a rs2910164 polymorphism and risk of immune thrombocytopenia. Autoimmunity. 2014;47:173–176. 10.3109/08916934.2014.883503 [DOI] [PubMed] [Google Scholar]
- 33. Wei L, Zhou Q, Hou S, Bai L, Liu Y, Qi J, et al. MicroRNA-146a and Ets-1 gene polymorphisms are associated with pediatric uveitis. PLoS One. 2014;9:e91199 10.1371/journal.pone.0091199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okada Y, Jinnin M, Makino T, Kajihara I, Makino K, Honda N, et al. MIRSNP rs2910164 of miR-146a is associated with the muscle involvement in polymyositis/dermatomyositis. Int J Dermatol. 2014;53:300–304. [DOI] [PubMed] [Google Scholar]
- 35. Chen HF, Hu TT, Zheng XY, Li MQ, Luo MH, Yao YX, et al. Association between miR-146a rs2910164 polymorphism and autoimmune diseases susceptibility: a meta-analysis. Gene. 2013;521:259–264. 10.1016/j.gene.2013.03.073 [DOI] [PubMed] [Google Scholar]
- 36. Yang Y, Zhang K, Zhou R. Meta-analysis of pre-miRNA polymorphisms association with susceptibility to autoimmune diseases. Immunol Invest. 2014;43:13–27. 10.3109/08820139.2013.822389 [DOI] [PubMed] [Google Scholar]
- 37. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li K, Tie H, Hu N, Chen H, Yin X, Peng C, et al. Association of two polymorphisms rs2910164 in miRNA-146a and rs3746444 in miRNA-499 with rheumatoid arthritis: a meta-analysis. Hum Immunol. 2014;75:602–608. 10.1016/j.humimm.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 39. Guo J, Jin M, Zhang M, Chen K. A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PLoS One. 2012;7:e30585 10.1371/journal.pone.0030585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng RL, Zhang H, Jiang WL. Tumor necrosis factor-alpha 308G>A polymorphism and risk of rheumatic heart disease: a meta-analysis. Sci Rep. 2014;4:4731 10.1038/srep04731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–369. 10.1038/nrg3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meisgen F, Xu Landen N, Wang A, Rethi B, Bouez C, Zuccolo M, et al. MiR-146a negatively regulates TLR2-induced inflammatory responses in keratinocytes. J Invest Dermatol. 2014;134:1931–1940. 10.1038/jid.2014.89 [DOI] [PubMed] [Google Scholar]
- 43. Yue C, Wang M, Ding B, Wang W, Fu S, Zhou D, et al. Polymorphism of the pre-miR-146a is associated with risk of cervical cancer in a Chinese population. Gynecol Oncol. 2011;122:33–37. 10.1016/j.ygyno.2011.03.032 [DOI] [PubMed] [Google Scholar]
- 44. Xu W, Xu J, Liu S, Chen B, Wang X, Li Y, et al. Effects of common polymorphisms rs11614913 in miR-196a2 and rs2910164 in miR-146a on cancer susceptibility: a meta-analysis. PLoS One. 2011;6:e20471 10.1371/journal.pone.0020471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.