Abstract
Lipopolysaccharide (LPS) present on the outer membrane of Gram-negative bacteria is one of the most important pathogen-associated molecular patterns and a potent elicitor in innate immunity. In human, TLR4 (Toll-like receptor 4) and MD-2 (myeloid differiation-2) form a receptor complex to transduce the LPS signal into cells. However, in invertebrates, receptors that recognize LPS have not been determined. Here we report the purification, characterization and cDNA cloning of an ML (MD-2-related lipid-recognition) protein from the tobacco hornworm Manduca sexta. The full-length cDNA of this M. sexta ML protein, named MsML-1, is 532 bp with an open reading frame of 456 bp that encodes a polypeptide of 151 amino acids containing an ML domain. MsML-1 is a secreted glycoprotein and its mRNA is expressed in fat body and hemocytes. The expression level of MsML-1 mRNA in fat body and hemocytes as well as MsML-1 protein in hemolymph are not induced by immune challenge. Recombinant MsML-1 protein specifically binds to LPS from several Gram-negative bacteria and LPS Re mutant, as well as to lipid A, but not to KDO (2-keto-3-deoxyoctonate). Our results suggest that MsML-1 may function as a key accessory protein for LPS signaling in M. sexta against Gram-negative bacterial infection.
Keywords: ML domain, MD-2, Lipopolysaccharide, Lipid A, Innate immunity, Manduca sexta
1. Introduction
Lipopolysaccharide (LPS), an amphipathic molecule found exclusively in the outer membrane of Gram-negative bacteria, is one of most potent immune stimulating molecules in the innate immune system (Freudenberg and Galanos, 1990; Raetz and Whitfield, 2002). In human, LPS activates synthesis and release of the tumor necrosis factor (TNF) leading to toxic shocks. It is estimated that 20,000 people die each year in America as a result of septic shock caused by Gram-negative bacterial infection (Pinner et al., 1996). The signaling pathway that senses LPS in human has been well studied. LPS is first recognized by the serum LPS-binding protein (LBP), then LBP binds and delivers the LPS as a monomeric molecule to CD14, a co-receptor that contains leucine-rich repeats (LRRs) and exists either as a soluble extracellular protein (sCD14) or as a GPI-linked membrane protein (mCD14). CD14 helps to translocate the LPS molecule in the LBP–LPS complex into the hydrophobic pocket of another key accessory protein, the myeloid differentiation-2 (MD-2) (Mancek-Keber and Jerala, 2006; Wright et al., 1990; Yu et al., 1997). The LPS–MD-2 complex binds to the Toll-like receptor 4 (TLR4), causing the rearrangement of TLR4 and conformation change in the TIR (Toll-IL-1 receptor) domain of TLR4, which allows the recruitment of downstream adapter proteins leading to the production of numerous pro-inflammatory mediators, such as TNFα, interleukin-1 (IL-1) and IL-6 (Saitoh et al., 2004;West et al., 2006). In invertebrates, the immune deficiency (IMD) pathway in Drosophila has been identified as a signaling pathway that senses Gram-negative bacteria (Lemaitre et al., 1995). However, the core molecule eliciting the IMD signaling pathway is peptidoglycan but not LPS (Kaneko et al., 2004; Stenbak et al., 2004). Although LPS has also been shown to induce immune responses in Drosophila (Imler et al., 2000), receptors that can directly recognize LPS and transduce the signal have yet been determined.
LPS molecules exhibit a great compositional variation depending upon bacterial origins, but they all consist of a hydrophobic lipid A covalently connected to a hydrophilic polysaccharide. Lipid A is the most conserved moiety in LPS that triggers immune responses. The polysaccharide moiety in LPS is composed of a variable O-specific chain that determines the serological specificity of LPS and a core non-repeating hetero-oligosaccharide, which is linked to the lipid A through the acidic 2-keto-3-deoxyoctonate (KDO) (Freudenberg and Galanos, 1990; Raetz and Whitfield, 2002). LPS is always attached to LPS-binding proteins, either on the bacterial membrane or in the host biological fluid and cells. To understand how LPS is recognized and delivered as a signal to elicit immune responses, studies have been conducted on the general structures of LPS-binding proteins and interactions between LPS-binding proteins and LPS. Based on the sequence comparison and molecular modeling study on a number of LPS-binding proteins and peptides, an amphipathic cationic binding pattern, which has been identified in human LBP, bactericidal permeability-increasing protein (BPI), Limulus anti-LPS factor (LALF) and TLR4, is proposed to be responsible for specific binding of lipid A (Frecer et al., 2000). Co-crystallization of LPS and FhuA, an iron uptake receptor, reveals that the LPS recognition motif consists of a geometric arrangement of four cationic residues (Lys–Lys–Arg–Lys) that interact with the two phosphate groups of lipid A (Ferguson et al., 2000). However, both predictions are not stringent, because these motifs not always account for LPS binding (Chaby, 2004; Jerala, 2007). Generally, LPS-binding proteins include a lipid-binding pocket that can accommodate the apolar moiety (acyl chains) of LPS with low specificity and positively charged residues for electrostatic interactions with phosphorylated carbohydrate groups in LPS (Beamer et al., 1998; Chaby, 2004). In human MD-2, the antiendotoxic lipid IVa binds to a deep hydrophobic cavity and two positively charged residues in the vicinities of the cavity entrance help to attract negatively charged lipid IVa to MD-2 (Ohto et al., 2007).
ML (MD-2-related lipid-recognition) is a novel protein domain identified in mammalian MD-1, MD-2, GM2-activator (GM2A), Niemann-pick disease type-2 (Npc2) protein, mite allergen Der f2 (Dermatophagogoides farinae), and many other proteins with unknown physiological functions in animals, plants and fungi (Inohara and Nunez, 2002). All ML proteins possess a putative secretion signal peptide and two pairs of conserved cysteine residues. The ML proteins are ~150 residues and do not show any homology to non-lipid transfer proteins. Many members of the ML family have been shown to regulate lipid metabolism, host response to pathogenic components such as LPS, and other cellular functions involved in lipid recognition (Inohara and Nunez, 2002). In this paper, we report the purification, characterization and cDNA cloning of an ML protein from the tobacco hornworm, Manduca sexta. This M. sexta ML protein, named MsML-1, is a secreted glycoprotein in the hemolymph of M. sexta larvae, and its mRNA is mainly expressed in fat body and hemocytes. The expression level of MsML-1mRNAin fat body and hemocytes andMsML-1protein in hemolymph are not induced by immune challenge. Recombinant MsML-1 protein specifically binds to LPS from several Gram-negative bacteria and LPS Re mutant, as well as to lipid A, but not to KDO. Our results suggest that MsML-1 may function as a key accessory protein for LPS signaling in M. sexta against Gram-negative bacterial infection.
2. Materials and methods
2.1. Insects and plasma samples
M. sexta larvae were reared using an artificial diet (Dunn and Drake, 1983). Hemolymph from naïve, control (injected with saline), or induced (injected with microorganisms or microbial components) larvae was collected at 24 h post-injection as described previously (Yu and Kanost, 2000).
2.2. Microorganisms, LPS and microbial components
Gram-positive Micrococcus luteus and yeast (Saccharomyces cerevisiae) were purchased from Sigma. Escherichia coli strain XL1-blue was from Stratagene. Lipopolysaccharide (LPS) from E. coli strains 026:B6 and 0111:B4, Pseudomonas aeruginosa and Salmonella minnesota, LPS Re mutant from Salmonella minnesota (Re 595 mutant), mono- and di-phosphory lipid A, lipoteichoic acid (LTA) from Staphyloccocus aureus, laminarin, and KDO were also from Sigma.
2.3. Purification of M. sexta ML-1 (MsML-1) protein from hemolymph
When we were purifying hemolin from bacteria-induced hemolymph using a cation exchange chromatography (CM Bio-Gel A, Bio-Rad) followed by a hydrophobic interaction chromatography (methyl HIC, 5 mL, Bio-Rad) (Yu and Kanost, 1999), a protein at about 16-kDa was partially purified as a byproduct. This protein was further purified by a second cation exchange chromatography, and was eluted at 0.5M NaCl along with a major co-purified protein at ~46-kDa. Protein fractions were analyzed by 15% SDS-PAGE.
To further separate the 16-kDa protein from the 46-kDa major co-purified protein, fractions containing the 16-kDa protein from the second cation exchange column were pooled and dialyzed against 2 L of 10 mM sodium phosphate buffer (pH 7.2) overnight at 4 °C. The dialyzed fraction was then loaded onto an equilibrated hydroxylapatite column (Econo-Pac CHT-II, 5 mL, Bio-Rad) at a flow rate of 0.5 mL/min. The column was washed with 10 mM sodium phosphate (pH 7.2), and eluted with a 10–400 mM gradient of sodium phosphate (pH 7.2) (total 30 mL). Fractions of 1mL were collected and analyzed by SDS-PAGE. The 46-kDa protein was eluted at ~120 mM sodium phosphate, while the 16-kDa protein (named MsML-1 for M. sexta ML-1, see below and Section 3) was eluted at ~200 mM sodium phosphate.
2.4. Characterization of MsML-1 protein
MsML-1 and the 46-kDa protein (2 µg each) were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane (0.2µm, Bio-Rad). Protein bands were stained with amido black and destained in deionized water. Then both protein bands were cut out for amino-terminal sequencing by automated Edman degradation using an Applied Biosystems Protein Sequencing System (Model 473A) at the Biotechnology Core Facility of Kansas State University. The N-terminal sequences obtained from the Edman degradation were compared to the protein sequences in the databank. The 46-kDa protein was identified as leureptin (accession number: AAO21503), while MsML-1 is a new protein.
The mass of MsML-1 was determined by matrix-assisted laser desorption ionization (MALDI) mass spectrometry at the Proteomic Core Facility, School of Biological Sciences, University of Missouri-Kansas City.
Purified MsML-1 was also analyzed by SDS-PAGE under non-reducing and reducing conditions. MsML-1 (0.5µg each) was dissolved in the sample loading buffer in the presence (reducing) or absence (non-reducing) of β-mercaptoethanol and heated to 95 °C for 5 min. Protein samples were then analyzed by 15% SDS-PAGE, transferred to a nitrocellulose membrane, and MsML-1 was identified by immunoblotting using rabbit polyclonal antibody to recombinant MsML-1 (see below).
To determine glycosylation of MsML-1, MsML-1 (2.0 µg) purified from hemolymph was denatured by heating to 100 °C for 3 min in 20 mM sodium phosphate buffer, pH 7.2, then incubated with 1 U of N-glycosidase F (PNGase F) (Sigma) in 50µL of 50 mM phosphate buffer, pH 7.2, 0.1% SDS, 0.5% (v/v) Nonidet P-40 and 0.5% (v/v) 2-mercaptoethanol for 24 h at 37 °C. Both N-glycosidase F treated and untreated MsML-1 samples (1.0 µg each) were analyzed by 15% SDS-PAGE and the gel was stained with Coomassie Brilliant Blue.
2.5. Isolation of cDNA clones for MsML-1
A degenerate primer (5′-GAR GAR CAR GCI ATH TTY TAY AA-3′) was designed based on the amino-terminal sequence of MsML-1 (EEQAIFYN). Polymerase chain reaction (PCR) was performed using lambda phage cDNA from an E. coli-induced M. sexta larval fat body cDNA library in λ ZAPII (Stratagene) as a template and the degenerate primer (final concentration of 7.5 pmol) and T7 primer (final concentration of 0.5 pmol). PCR reactions were carried out as following: denaturing for 30 s at 94 °C, annealing for 30 s at 55 °C, and extension for 40 s at 72 °C for a total of 30 cycles. A PCR product of 500 bp was obtained, cloned into the plasmid vector pGEMR-T (Promega) and sequenced.
To obtain a full-length cDNA clone for MsML-1, an E. coliinduced M. sexta larval fat body cDNA library was screened using the cDNA fragment (digested from the recombinant pGEMR-T vector with Apa I and Spe I) as a probe. The positive cDNA clones were purified to homogeneity and subcloned by in vivo excision of pBluescript phagemids. The nucleotide sequences of the cDNA clone were determined from double-stranded plasmid DNA templates.
2.6. Analyses of sequence data and structural modeling
Sequence alignment was performed using ClustalW (http://www.ch.embnet.org/software/ClustalW.html), and identities between MsML-1 and other proteins were calculated using an align program (http://xylian.igh.cnrs.fr/bin/align-guess.cgi). Molecular weight and theoretical pI were calculated using the PeptideMass program (http://us.expasy.org/tools/peptidemass.html). Model structures of MsML1 were constructed using the PyMOL program.
2.7. Expression of recombinant MsML-1 in bacteria and production of polyclonal rabbit antiserum
The cDNA fragment encoding residues 20–151 was generated by PCR using primers Y62 (5′-TCG CCA TGG AGC AGG CCA TCT-3′) and Y63 (5′-TAG GCA TGC TTT ATT TCC TAA TC-3′). The PCR product was digested with Nco I and Spe I, recovered and cloned into Nco I/Spe I digested expression vector H6pQE-60 (Lee et al., 1994). Recombinant proteins were expressed in E. coli strain M15 (Stratagene) as described previously (Yu et al., 1999).
Recombinant MsML-1 was expressed as an inclusion body and purified under denaturing conditions in 8M urea by nickel-nitrilotriacetic acid (Ni-NTA) chromatograph (Qiagen). Purified recombinant MsML-1 (rMsML-1) in 8Murea was renatured by a three-step dialysis as described previously (Yu et al., 2005). Then purified rMsML-1 (1 mg) was run on a preparative 15% SDS-PAGE, and the gel slice containing rMsML-1 was cut out and used as an antigen for the production of polyclonal rabbit antiserum (Cocalico Biologicals, Inc.).
2.8. Tissue distribution of MsML-1
To investigate expression of MsML-1 mRNA in different tissues, total RNAs from hemocytes, fat body, Malpighian tubules, epidermis and midguts of day 2 fifth instar M. sexta naïve larvae were extracted, treated with DNase I and transcribed into cDNA as described previously (Ao et al., 2008). MsML-1 gene-specific primers (Y062/Y063) were used for PCR. Ribosomal protein S3 (RPS3) (Jiang et al., 1996) cDNA fragment was amplified with K12 (5′-TTA ATT CCG AGC ACT CCT TG-3′) and K13 (5′-GGA GCT GTA CGC TGA GAA AG-3′) primers as an internal control. All PCR reactions were performed as following: initial denaturing at 94 °C for 2 min, and then denaturing at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s for different numbers of cycles, followed by an extension at 72 °C for 10 min. Five microliters of the 1:10 diluted cDNA were used as a template for MsML-1 amplification for 32 and 35 cycles. One microliter of 1:10 diluted cDNA was used as a template for RPS3 amplification for 30 cycles.
2.9. Expression of MsML-1 mRNA in hemocytes and fat body and MsML-1 protein in hemolymph after immune challenge
Expression of MsML-1 mRNA in hemocytes and fat body after microbial challenge was determined by real-time PCR. Microbial challenge in M. sexta larvae, RNA extraction from fat body and hemocytes, as well as cDNA synthesis were performed as described previously (Ao et al., 2008). The real-time PCR was carried out in 25-µL reactions containing 12.5µL 2 × SYBR® Premix Ex Taq™ (Takara, USA), 0.5 µL ROX® Reference dye II and 5 pmol of each primer. One microliter of 1:10 diluted cDNA was used for RPS3 (endogenous control) and 5µL of 1:10 diluted cDNA were used for MsML-1 gene amplification (target). Real-time PCR program was 10 s at 95 °C, followed by 40 cycles of 95 °C for 5 s, 60 °C for 40 s. PCR products were detected by agarose gel analysis after each PCR reaction to confirm the specific gene amplification. Data from three replicas of each sample were analyzed by the ABI 7500 SDS software (Applied Biosystem, USA) using a comparative method (2−ΔΔCt). The baseline was set automatically by the software to maintain the consistency. cDNA samples from naïve hemocytes or fat body were designated as the calibrators. The expression level of MsML-1 mRNA in other samples was calculated by 2−ΔΔCt, which stands for the n-fold difference relative to the calibrator. All data were present as relative mRNA expression (means of three individual measurements ± S.E.M.).
To investigate MsML-1 protein expression in hemolymph after immune challenge, day 2 fifth instar M. sexta larvae were injected with 50 µL saline (as a control), or heat-killed E. coli XL-1 blue (108 cells/larva), M. luteus (100µg/larva), yeast (S. cerevisiae) (107 cells/larva), LPS (E. coli 026:B6, 20µg/larva), LTA (20µg/larva), or laminarin (20 µg/larva) (four larvae for each group). Hemolymph was collected from each larva at 0 and 24 h post-injection, and hemocytes were removed by centrifugation. Equal volume of plasma from each larva was mixed, and the mixed plasma was diluted with ddH2O(1:1) and added to 2 × SDS loading buffer. For electrophoresis, the diluted plasma samples (corresponding to 2µL of the original mixed plasma) were loaded to 15% SDS-PAGE, and MsML-1 in each sample was detected by rabbit polyclonal antiserum to recombinant MsML-1 (1:2000 dilution).
2.10. Binding of recombinant MsML-1 to immobilized LPS and lipid A
To test whether recombinant MsML-1 can bind to LPS, LPS from E. coli strains 0111:B4 and 026:B6, Pseudomonas aeruginosa and Salmonella minnesota, LPS Re mutant from Salmonella minnesota (Re 595 mutant), and KDO (each at 2µg), or mono- and di-phosphoryl lipid A (each at 1 µg) in 50µL deionized water were coated to each well of a flat-bottom microtiter plate (Maxisorp, Nunc). The plate was air-dried overnight at room temperature, heated to 60 °C for 30 min, and then blocked with 100 µL/well of 1 mg/mL Bovine serum albumin (BSA) in the binding buffer (50 mM Tris–HCl, 50 mM NaCl, pH 8.0) at 37 °C for 2 h, followed by rinsing with 200µLof binding buffer twice. Re-natured recombinant MsML-1 (rMsML-1) or CP36 (rCP36), an M. sexta cuticle protein (Suderman et al., 2003) used as a negative control protein, was diluted to 20µm in the binding buffer containing 0.1 mg/mL BSA and added to the coated plate at 50 µL/well. Binding of the recombinant proteins to the immobilized LPS or lipid A was allowed to occur at room temperature for 3 h. The plate was rinsed with Tris-buffered saline (TBS) (25 mM Tris–HCl, 137 mM NaCl and 3 mM KCl, pH 7.0) four times, each for 5 min. Then polyclonal rabbit antiserum to rMsML-1 or rCP36 (1:2000 dilution in TBS containing 0.1 mg/mL BSA) was added at 100µL/well and the plate was incubated at 37 °C for 2 h. The plate was rinsed again with TBS four times, each for 5 min. Alkaline phosphatase-conjugated goat anti-rabbit IgG (Sigma, 1:10,000 dilution in TBS containing 0.1 mg/mL BSA)was added at 100µL/well, and the plate was incubated for 2 h at 37 °C. Finally, the plate was rinsed with TBS twice followed by DEA buffer (10 mM diethanolamine, 0.5 mM MgCl2, pH 9.6) twice. Then p-nitro-phenyl phosphate (Sigma, 1 mg/mL in DEA buffer) was added at 50 µL/well, and the plate was incubated at room temperature for 20–30 min. Absorbance at 405 nm of each well was recorded by a microtiter plate reader (PowerWave XS, Bio-Tek Instrument, Inc.). As a blank control, the plate was coated with deionized water without LPS or lipid A, and the binding was performed exactly the same as described above. The OD405 values of the blank controls were used to blank the experimental data. Data from three replicas of each sample were analyzed. These experiments were repeated at least three times, and one representative set of data was used to make figures using the GraphPad Prism software.
To further confirm binding of rMsML-1 to lipid A, diphosphoryl lipid A from E. coli F598 Rd mutant (Sigma L5399) was used to coat the plate (1 µg/well). Then increasing concentrations (0–200 µm) of rMsML-1 or rCP36 were added to each well and the binding was performed the same as described above. These experiments were also repeated at least three times, and one representative set of data was used to make figures.
3. Results
3.1. Purification of MsML-1 protein from hemolymph
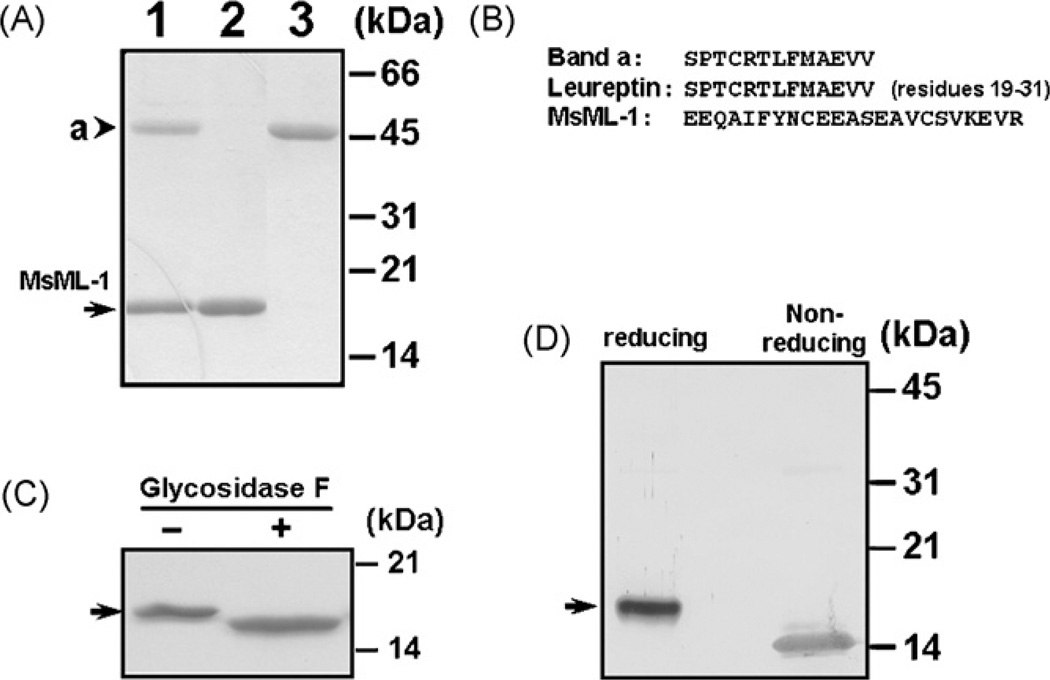
When we were purifying hemolin from bacteria-induced hemolymph (Yu and Kanost, 1999), two proteins with apparent molecular weights of ~16- and 46-kDa, respectively, were co-purified as by-products after a cation exchange chromatography followed by a hydrophobic interaction and a second cation exchange chromatographies (Fig. 1A, lane 1). The 16-kDa protein was then further separated from the 46-kDa protein by a hydroxylapatite column and both proteins were purified to homogeneity (Fig. 1A, lanes 2 and 3). The 46-kDa protein (lane 3) was eluted at ~120 mM sodium phosphate, while the 16-kDa protein (lane 2) was eluted at ~200 mM sodium phosphate. Then, the amino-terminal sequences of both the 16- and 46- kDa proteins were determined by Edman degradation (Fig. 1B). Thirteen residues were obtained from the amino-terminus of the 46-kDa protein, which perfectly matched residues 19–31 of the deduced amino acid sequence of leureptin (accession number: AAO21503). Twenty-three residues were obtained from the amino-terminus of the 16-kDa protein, which did not match any sequences in the Genbank database. We named the 16-kDa protein as M. sextaML-1(MsML-1) since it contains an ML-domain (see below).
Fig. 1.
Analysis of M. sexta ML-1 (MsML-1) protein purified from hemolymph. (A) SDS-PAGE analysis of MsML-1 purified from hemolymph. MsML-1 and a 46-kDa protein were purified to homogeneity from hemolymph after several chromatographies. Co-purified MsML-1 and the 46-kDa protein (lane 1, 3µg) and purified MsML-1 (lane 2, 2µg) and the 46-kDa protein (lane 3, 2µg) were analyzed by 15% SDS-PAGE, and the gel was stained by Coomassie Brilliant Blue. (B) Amino-terminal sequences of MsML-1 and the 46-kDa protein. The amino-terminal sequences of MsML-1 and the 46-kDa protein (band a) were determined by Edman degradation, and the sequence of leureptin (residues 19–31) was obtained from the Genbank database (accession number: AAO21503). (C) Deglycosylation of MsML-1. MsML-1 (2.0µg) purified from hemolymph was heated to 100 °C for 3 min in 20 mM sodium phosphate buffer, pH 7.2. The denatured protein was then incubated with or without 1U of N-glycosidase F (PNGase F) (Sigma) in 50µL of 50 mM phosphate buffer, pH 7.2, 0.1% SDS, 0.5% (v/v) Nonidet P-40 and 0.5% (v/v) 2-mercaptoethanol for 24 h at 37 °C. Both N-glycosidase F treated and untreated MsML-1 samples (1.0µg each) were analyzed by 15% SDS-PAGE and the gel was stained with Coomassie Brilliant Blue. (D) Immunoblotting analysis of MsML-1 under reducing and non-reducing conditions. MsML-1 (0.5µg each) purified from hemolymph was dissolved in the sample loading buffer in the presence (reducing) or absence (non-reducing) of β -mercaptoethanol and heated to 95 °C for 5 min. Protein samples were separated by 15% SDS-PAGE, and MsML-1 was identified by immunoblotting using rabbit polyclonal antiserum against recombinant MsML-1. The arrowhead indicates the 46-kDa protein (band a) while the arrows indicate native MsML-1.
3.2. cDNA cloning and sequence analysis of MsML-1
Using a degenerate primer designed based on the amino-terminal sequence of MsML-1 and T7 primer, a fragment of 500 bp was obtained by PCR. Then, an E. coli-induced larval fat body cDNA library was screened using the PCR fragment as a probe, and the nucleotide sequences of the positive clones were determined. The full-length cDNA sequence of MsML-1 is 532 bp long with an open reading frame of 453 bp, which encodes a 151-residue polypeptide (accession number: EU329722).
The deduced amino acid sequence of MsML-1 contains an 18-residue secretion signal peptide, which is confirmed by Edman degradation of the mature protein (Figs. 1B and 2). The mature MsML-1 has a predicted molecular weight of 15227 Da (monoisotopic) and a theoretical pI of 6.57. In the mature MsML-1, a potential N-linked glycosylation site is identified at Asn-58. Treatment of the MsML-1 purified from hemolymph with N-glycosidase F, which specifically cleaves N-linked glycans, resulted in a slightly smaller protein band (Fig. 1C). In addition, the mass of the purified MsML-1 determined by Mass Spectrometry (MALDI TOFF) is 16113.82 Da (data not shown), which is larger than the mass (15227 Da, monoisotopic) calculated from the deduced amino acid sequence. These results indicate that MsML-1 is N-glycosylated.
Fig. 2.
Alignment of MsML-1 with homologous insect proteins, mite allergen Der f2, and human and mouse MD-2. The deduced amino acid sequence of MsML-1 is aligned with homologous proteins from other insect species, D. melanogaster (CG11314 and CG11315), B. mori (accession number: AB030701), A. gambiae (accession number: EAA00385), A. aegypti (accession number: XP 001648436), A. mellifera (accession number: XP 392711), as well as with mite allergen Der f2 (accession number: CAI05848), human MD-2 (hMD-2) (accession number: BAA78717) and mouse MD-2 (mMD-2) (accession number: BAA93619) proteins. Cysteine residues that are conserved in all the proteins are labeled with asterisks above the alignment; two cysteine residues that are conserved only in insect proteins are labeled with filled diamonds; a cysteine residue that is conserved in insect proteins and mite allergen Der f2 but absent in MD-2 is labeled with a filled triangle; and two cysteine residues that are conserved only in MD-2 proteins are labeled with open triangles. Identities between MsML-1 and other proteins are also indicated in the parentheses.
Analysis of the deduced amino acid sequence indicates that MsML-1 contains an ML domain (Inohara and Nunez, 2002). The ML family proteins are single-domain proteins, including mammalian MD-1, MD-2, Niemann-pick disease type-2 (Npc2) and its orthologs, mite allergen Der f2 (Dermatophagogoides farinae), and GM2-activator (GM2A) (Inohara and Nunez, 2002). MsML-1 is most similar in amino acid sequence to a Bombyx mori hemolymph protein (48.4% identity) that promotes replication of the nucleopolyhedrovirus in vitro (Kanaya and Kobayashi, 2000) (Fig. 2). It is also similar to two proteins (CG11315 and CG11314) from Drosophila melanogaster (30.4% and 31.4% identity, respectively), and an Npc2-like protein from Anopheles gambiae (35.8%), Aedes aegypti (34.1%) and Apis mellifera (34.6%). However, MsML-1 shows a low similarity to mite allergen Der f2 (23.9% identity) and mammalian MD-2 (~20% identity) in overall amino acid sequence (Fig. 2).
Mature MsML-1 and its insect homologous proteins contain 8 cysteine residues, while mite allergen Der f2 and Npc2 proteins have 6 cysteines, but MD-2 has 7 cysteine residues (Fig. 2). Thus, MsML-1 and its insect homologous proteins may form 4 pairs of disulfide bonds, while mite allergen Der f2 and MD-2 may form only 3 pairs of disulfide bonds. Among these cysteine residues, five are highly conserved in insect proteins, MD-2 and mite Der f2 (Fig. 2, cysteines labeled with asterisks). Compared to mammalian MD-2, the extra cysteine residue in insect proteins is present in the amino-terminal region (Fig. 2). It has been shown that secreted mammalian MD-2 protein exists as a heterogeneous collection of disulfide-linked oligomers composed of stable dimers (Visintin et al., 2001). However, when native MsML-1purified from hemolymph was analyzed on SDS-PAGE under non-reducing conditions, noMsML-1dimers or oligomers were detected by immunoblotting with rabbit polyclonal antibody to recombinant MsML-1 (Fig. 1D).
3.3. MsML-1 mRNA is expressed in fat body and hemocytes
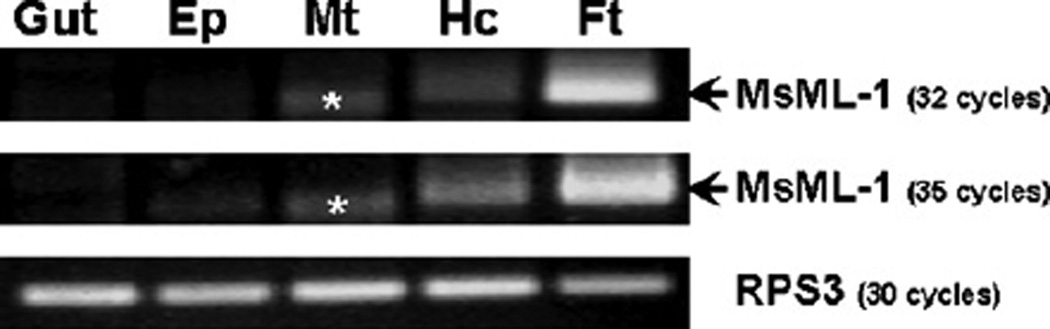
To determine tissue distribution of MsML-1, RT-PCR was performed. MsML-1 transcript was detected in hemocytes and fat body, but was not present in the midgut, epidermis, or Malpighian tubule of naïve larvae (Fig. 3). The PCR fragment in the Malpighian tubule sample (Fig. 3, lane Mt, labeled with the asterisks), which was slightly smaller than the predicted size of MsML-1, was not increased after 3 more cycles of amplification, suggesting that it was a non-specific amplification product. DNA sequencing also confirmed that this PCR fragment was not MsML-1 (data not shown). MsML-1mRNAwas much more abundant in fat body than in hemocytes (Fig. 3, lanes Ft and Hc), indicating that MsML-1 was mainly synthesized in fat body.
Fig. 3.
Tissue distribution of MsML-1. Total RNAs (1µg each) from midgut (Gut), epidermis (Ep), Malpighian tubule (Mt), hemocytes (Hc) and fat body (Ft) of M. sexta naïve larvae were reverse-transcribed into cDNA using Generacer™ Oligo-dT primer. The cDNA was used as a template for PCR reactions with MsML-1 specific primers (32 and 35 cycles) or RPS3 specific primers (30 cycles). PCR products were analyzed on an agarose gel and stained with ethidium bromide. The asterisks indicate a non-specific amplification product in the Malpighian tubule sample.
3.4. Expression of MsML-1 is not induced by microorganisms
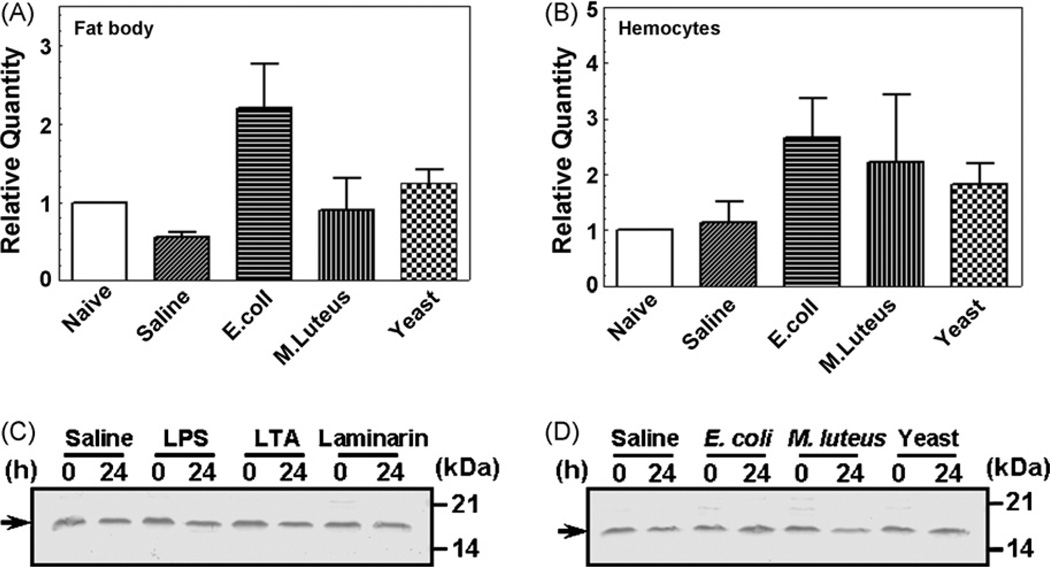
Expression of MsML-1 mRNA in hemocytes and fat body after immune-challenge was determined by real-time PCR. Compared to the naïve larvae, the MsML-1 mRNA in fat body was slightly decreased in the saline-injected larvae, increased about 1-fold in the E. coli-injected larvae, and remained at almost the same levels in the M. luteus- and yeast-injected larvae (Fig. 4A). MsML-1 mRNA in hemocytes remained at a similar level in the naïve and saline-injected larvae, but increased (up to ~2-fold) in all three groups of larvae injected with microorganisms (Fig. 4B). Despite the variations in the MsML-1 mRNA level in hemocytes and fat body of larvae injected with saline and microorganisms (at 24 h post-injection), these changes were not statistically significant (p > 0.05). Northern blot analysis also showed that MsML-1 mRNA was more abundant in fat body than in hemocytes of control larvae (injected with saline), and the mRNA level was not induced in fat body and hemocytes when larvae were injected with E. coli or M. luteus (data not shown). We then determined MsML-1 protein concentration in hemolymph of naïve larvae or larvae injected with saline (control), microorganisms or microbial components by immunoblotting. Our results showed that MsML-1 protein concentration remained almost constant in the hemolymph of naïve larvae (0 h post-injection) and larvae injected with microorganisms (E. coli, M. luteus and S. cerevisiae) or microbial components (LPS, LTA and laminarin) (24 h post-injection) (Fig. 4C and D). These results are consistent with those of the real-time PCR and Northern blot analysis, indicating that expression of MsML-1 is not induced by microorganisms.
Fig. 4.
Expression of MsML-1 mRNA in fat body and hemocytes and MsML-1 protein in hemolymph after immune challenge. Day 2 fifth instar M. sexta larvae were injected with heat-killed E. coli (108 cells/larva), M. luteus (100µg/larva), yeast (S. cerevisiae) (107 cells/larva) or saline (as a control). Hemocytes and fat body were collected at 24 h post-injection. Total RNAs were prepared from these hemocytes and fat body, and then transcribed into cDNAs using GeneRacer™ Oligo-dT primer. Expression of MsML-1 mRNA in microbial challenged hemocytes or fat body was determined by real-time PCR in three replicas (A and B). The bars represent the mean of three individual measurements±S.E.M. For MsML-1 protein expression in hemolymph after immune challenge (C and D), day 2 fifth instar M. sexta larvae were injected with saline (as a control), heat-killed E. coli, M. luteus or yeast (S. cerevisiae) as described above, or with LPS (E. coli 026:B6), LTA or laminarin (20µg each per larva) (four larvae for each group). Hemolymph was collected from each larva at 0 and 24 h after injection and equal volumes of the plasma samples were mixed, diluted (1:1 in water), and then added to the 2 × SDS loading buffer. For electrophoresis, diluted plasma samples (each corresponding to 2 µL of the original mixed plasma) were loaded to 15% SDS-PAGE. MsML-1 in each plasma sample was detected by immunoblotting using rabbit polyclonal antibody to recombinant MsML-1. The arrows indicate MsML-1 protein in hemolymph.
3.5. MsML-1 specifically binds to immobilized LPS and lipid A
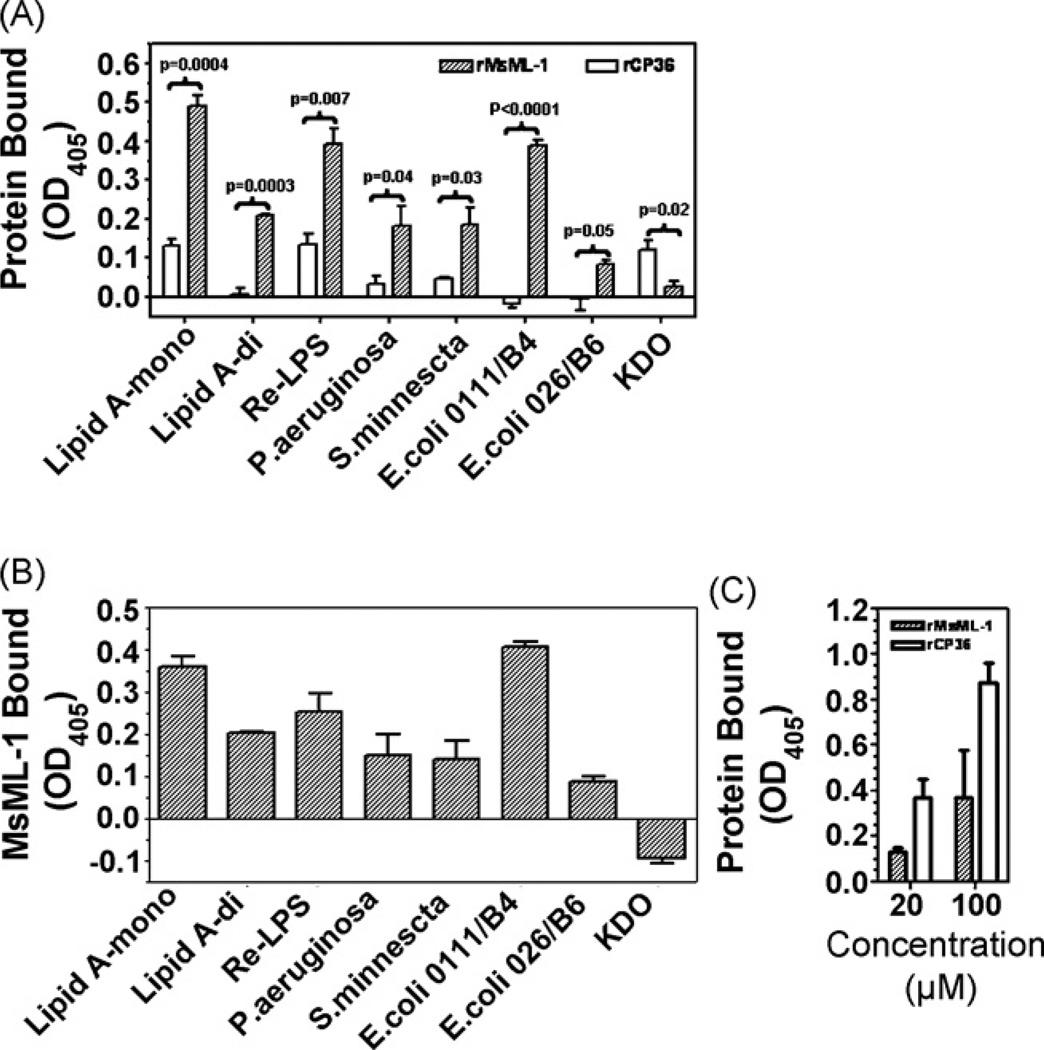
Bacterial smooth LPS molecules consist of three moieties: the biologically active lipid A, the variable O-specific antigen, and the inner core oligosaccharide composed of KDO and heptoses. To investigate direct binding of MsML-1 to LPS, a 96-well microtiter plate was coated with smooth LPS from different Gram-negative bacteria (E. coli, P. aeruginosa and S. minnesota), LPS Re mutant, mono- or di-phosphoryl lipid A, or KDO, and recombinant MsML-1 (rMsML-1) or CP36 (rCP36), an M. sexta cuticle protein (Suderman et al., 2003) used as a control protein, was added to the LPS (or lipid A)-coated plate. Binding of recombinant proteins to the immobilized LPS or lipid A was detected by rabbit polyclonal antibody to rMsML-1 or rCP36. We showed that at 20 µm of proteins, the total binding of rMsML-1 to LPS and lipid A was significantly higher (p = 0.05 to <0.0001) than that of rCP36, but the total binding of rMsML-1 to KDO was lower than that of rCP36 (p = 0.02) (Fig. 5A). Since the reaction between anti-rCP36 antibody and rCP36 was stronger than that between anti-rMsML-1 antibody and rMsML-1 when same concentrations of recombinant proteins were directly coated to a plate (Fig. 5C), the total binding of rCP36 (at the same concentrations as rMsML-1) to immobilized LPS or lipid A can be considered as non-specific binding. Thus, specific binding of rMsML-1 to LPS or lipid A (Fig. 5B) was calculated by subtracting the total binding of rCP36 from the total binding of rMsML-1. The results showed that most rMsML-1 specifically bound to LPS of E. coli strain 0111:B4 and monophosphoryl lipid A, while least rMsML-1 bound to LPS of E. coli strain 026:B6, but no rMsML-1 bound to KDO (Fig. 5B). Similar high amounts of rMsML-1 bound to LPS from P. aeruginosa, S. minnesota, LPS Re mutant and di-phosphoryl lipid A (Fig. 5B).
Fig. 5.
Recombinant MsML-1 binds to immobilized LPS and lipid A. A 96-well microtiter plate was coated with LPS (E. coli strains 0111:B4 and 026:B6, P. aeruginosa, S. minnesota, Re mutant of S. minnesota Re595) (50 µL/well, 2 µg/well), mono- or di-phosphoryl lipid A (1 µg/well), or KDO (2 µg/well) (panels A and B), and renatured recombinant MsML-1 (rMsML-1) or CP36 (rCP36), diluted to 20 µm in the binding buffer (50 mM Tris–HCl, 50 mM NaCl, pH 8.0) containing 0.1 mg/mL BSA, was added to the LPS (or lipid A)-coated plate (50 µL/well) and incubated at room temperature for 3 h. A 96-well microtiter plate was also directly coated with renatured rMsML-1 or rCP36 (50 µL/well, 20 and 100 µm) (panel C). Then, the total binding (panel A) of recombinant proteins and the specific binding of MsML-1 (panel B) to immobilized LPS or lipid A were determined by ELISA using the rabbit polyclonal antiserum against rMsML-1 or rCP36. The specific binding of recombinant MsML-1 to LPS and lipid A was calculated by subtracting the total binding of rCP36 from the total binding of rMsML-1. Reaction of antibodies to recombinant proteins directly coated on the plate (panel C)was also measured by ELISA. Each point represents the mean of three individual measurements±S.E.M. Significance of difference was calculated using a Student’s T-test.
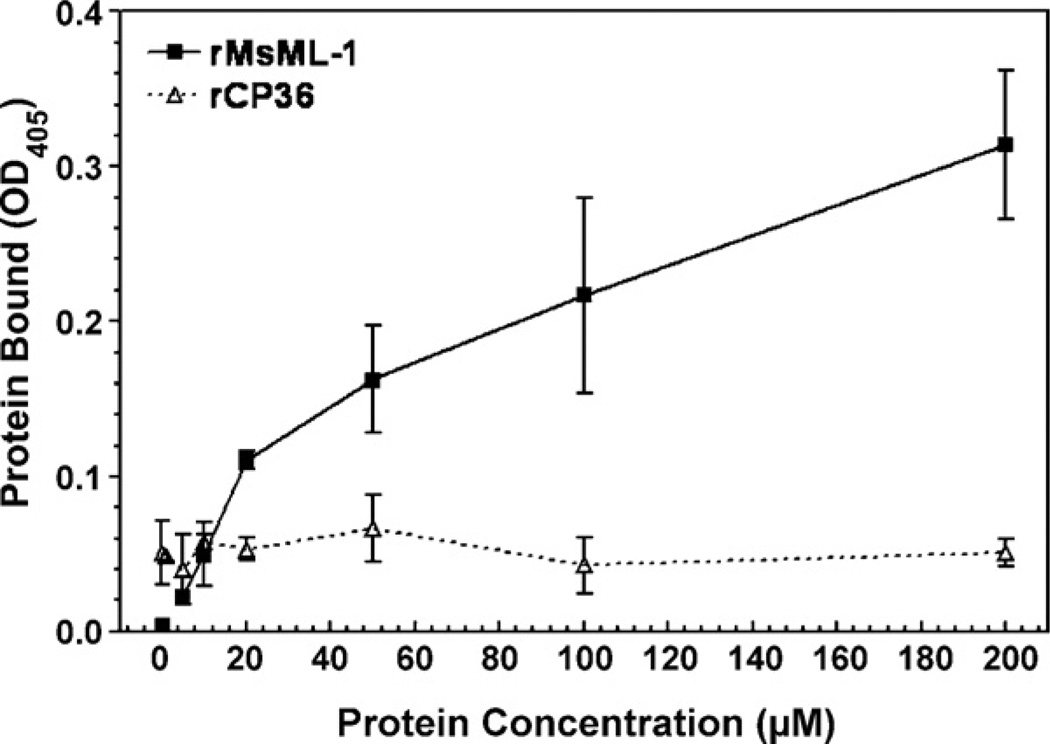
To further confirm that binding of rMsML-1 to LPS is via interaction with the lipid A moiety, di-phosphoryl lipid A (from E. coli F598) was used to coat the plate, and increasing concentrations (0–200 µm) of rMsML-1 or rCP36 were added to the lipid A-coated plate. The results showed that at low concentrations (0–10 µm) of recombinant proteins, little rMsML-1 bound to lipid A. When the protein concentrations were increased (20–200 µm), increasing amounts of rMsML-1 bound to lipid A, and the binding was still not saturated at 200 µm (Fig. 6). However, very little rCP36 bound to lipid A at all the protein concentrations (0–200 µm) tested (Fig. 6). These results suggest that rMsML-1 specifically binds to the lipid A moiety of LPS.
Fig. 6.
Recombinant MsML-1 specifically binds to immobilized lipid A. A 96-well microtiter plate was coated with diphosphoryl lipid A (E. coli F598 Rd mutant) (50 µL/well, 1 µg/well) and blocked with BSA. Renatured rMsML-1 or rCP36, diluted in the binding buffer at different concentrations (0–200 µm), was added at 50 µ/well and incubated at room temperature for 3 h. Binding of the recombinant proteins to the immobilized lipid A was determined by ELISA as described above in Fig. 5. Each point represents the mean of three individual measurements ± S.E.M.
4. Discussion
The ML family includes a variety of proteins, such as mammalian MD-1, MD-2, GM2-activator, Npc2, mite allergen Der f2, and several D. melanogaster and Caenorhabditis elegans proteins (Inohara and Nunez, 2002). Mammalian MD-2 binds bacterial LPS and plays an essential role in formation of a receptor complex with TLR4 to activate the LPS-induced signaling pathway (Nagai et al., 2002; Schromm et al., 2001; Shimazu et al., 1999; Viriyakosol et al., 2001). Although D. melanogaster and C. elegans have eight and five ML proteins, respectively (Inohara and Nunez, 2002), little is known about their direct interactions with LPS or lipid A. We isolated a protein from the hemolymph of M. sexta larvae and cloned its cDNA (Figs. 1 and 2). This Manduca protein belongs to the ML family and thus is named M. sexta ML-1 (MsML-1). MsML-1 is most similar to a B. mori protein (48.4% identity), which is also isolated from the hemolymph and has activity to promote in vitro replication of the nucleopolyhedrovirus (Kanaya and Kobayashi, 2000). However, it is unknown whether the B. mori protein can bind to LPS or not. We showed that recombinant MsML-1 (rMsML-1) specifically bound to LPS and lipid A (Figs. 5 and 6). To our knowledge, this is the first report of an insect member of the ML family that can directly bind to LPS via the lipid A moiety.
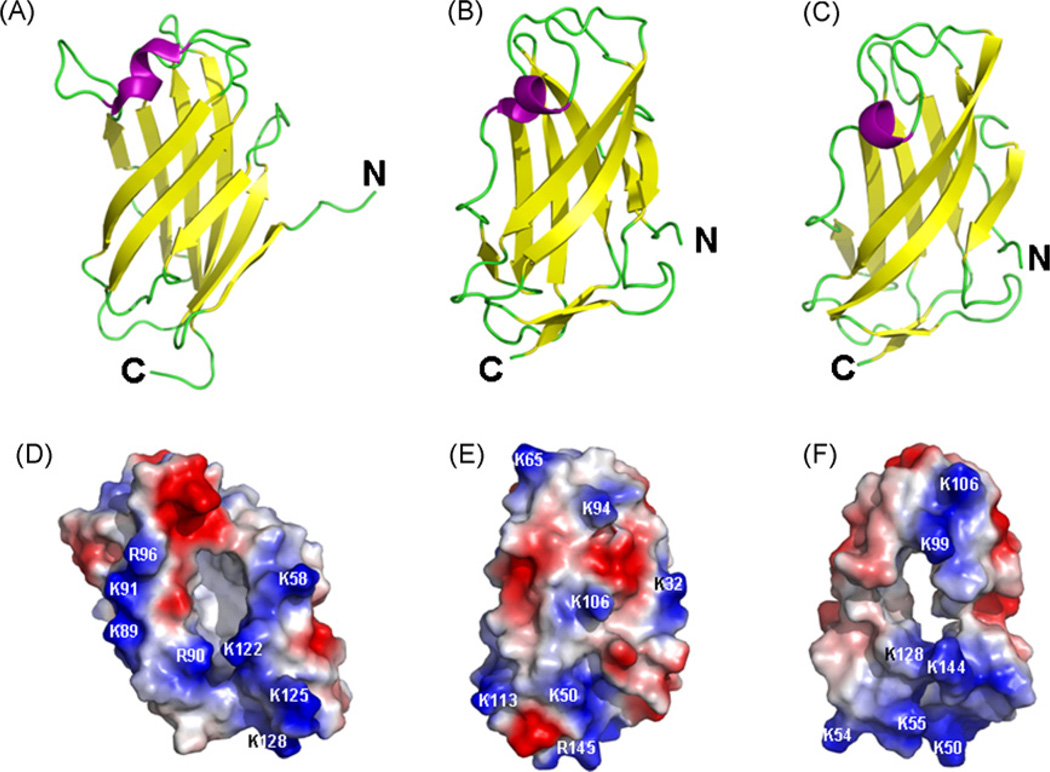
In the crystal structure of human MD-2, a deep hydrophobic cavity sandwiched by two β sheets accounts for its binding to the four acyl chains of the lipid IVa (Ohto et al., 2007) (Fig. 7A and D). The entrance of the hydrophobic cavity is positively charged and the inside of the cavity is lined with hydrophobic residues (Ohto et al., 2007) (Fig. 7D). Among a total of 18 positively charged residues (thirteen Lys and five Arg residues) in human MD-2, only Lys122 and Arg90 are located in the vicinities of the cavity entrance (Fig. 7D), and they help to attract the negatively charged lipid IVa to the cavity. The 3-dimensional structure of mite allergen Der f2 has similar two β sheets, but it does not have a deep hydrophobic cavity (Fig. 7B and E). We built a model structure of MsML-1 based on the structure of mite allergen Der f2, since Kim et al. (2007) reported that “the MD-2 structure could not be superimposed on any of the ML family proteins”. The model structure of MsML-1 (Fig. 7C) also contains two antiparallel β sheets like those in mite allergen Der f2, but MsML-1 has a deep hydrophobic cavity (Fig. 7F) similar to the one in human MD-2 (Fig. 7D), which is not observed in the mite allergen Der f2 (Fig. 7E). Among the 17 positively charged residues (fifteen Lys and two Arg residues) in MsML-1, Lys144 and Lys99 are in the vicinities of the cavity entrance (Fig. 7F). Although MsML-1 and human MD-2 have only ~20% identity in the amino acid sequences (Fig. 2), they both can bind to LPS and lipid A (Viriyakosol et al., 2001) (Figs. 5 and 6) because both proteins contain a deep hydrophobic cavity (Fig. 7D and F). Thus, the binding ability of an ML protein to LPS or lipid A may depend upon whether it contains a deep hydrophobic cavity or not.
Fig. 7.
Structures of human MD-2, mite allergen Der f2 and MsML-1. Cartoon representations of human MD-2 (PDB: 2e56) (A) and mite allergen Der f2 (PDB: 1wrf) (B) structures and a model structure of MsML-1 (C) based on the structure of mite allergen Der f2. The electrostatic potential surfaces of human MD-2 (D), mite allergen Der f2 (E) and MsML-1 (F) were also calculated using the PyMOL program and positive and negative potentials are shown in blue and red, respectively. The potential surface of MD-2 (D) is viewed from a 90° rotation with respect to (A), whereas (E) and (F) are the same views of (B) and (C), respectively.
The crystal structure of the TLR4–MD-2 complex shows that MD-2 interacts with TLR4 via an extensive network of charge-mediated hydrogen bonds, and binding of LPS to MD-2 may induce a structural change in TLR4 to promote dimerization of TLR4 receptors to trigger the signaling pathway (Kim et al., 2007). In D. melanogaster, peptidoglycan from Gram-positive bacteria induces activation of a serine protease cascade leading to cleavage of a cytokine-like ligand Spatzle, and active Spatzle binds to the Toll receptor to activate antimicrobial peptide genes, while peptidoglycan from Gram-negative bacteria binds to peptidoglycan recognition proteins (PGRPs) to activate the IMD pathway (Hoffmann and Reichhart, 2002; Kaneko and Silverman, 2005). However, it has also been shown that bacterial LPS can induce expression of antimicrobial peptides in Drosophila (Imler et al., 2000). It is unclear how LPS triggers a signaling pathway in insects. In M. sexta, several antimicrobial peptide genes have been identified, including cecropin, attacin, gloverin, moricin and lysozyme (Gorman et al., 2004). Some antimicrobial peptide genes, such as gloverin and moricin, are identified only in lepidopteran insects but not in dipteran species like Drosophila and mosquitoes (Axen et al., 1997; Furukawa et al., 1999; Hara and Yamakawa, 1995; Kawaoka et al., 2008; Lundstrom et al., 2002; Mackintosh et al., 1998; Oizumi et al., 2005). It is not known whether these lepidopteran antimicrobial peptide genes are also regulated by the Toll or IMD pathway.
Recently, we have identified a Toll receptor from the hemocytes of M. sexta larvae, and expression of this M. sexta Toll mRNA in hemocytes is induced by Gram-negative E. coli, but not by Gram-positive M. luteus or yeast (S. cerevisiae) (Ao et al., 2008). We also found that injection of E. coli and LPS into M. sexta larvae significantly induced expression of gloverin and moricin genes in hemocytes (Yu X-Q, unpublished data). Thus, it is possible that additional signaling pathways for regulation of antimicrobial peptide genes exist in insects. MsML-1 may serve as a key accessory protein to bind and deliver LPS to a receptor. Our future work is to investigate whether M. sexta Toll receptor and MsML-1 are involved in an LPS-triggered signaling pathway to regulate expression of antimicrobial peptide genes in M. sexta.
Acknowledgements
The authors thank Zhonghua Wang, a Ph. D. candidate in the School of Biological Sciences, University of Missouri-Kansas City, for his help in protein structure modeling.
This work was supported by the National Institutes of Health Grant GM66356. The nucleotide sequence reported in this paper has been submitted to the Genbank™/EBI Data Bank with the accession number EU329722.
Abbreviations
- IMD
immune deficiency
- KDO
2-keto-3-deoxyoctonate
- LBP
LPS-binding protein
- LPS
lipopolysaccharide
- LTA
lipoteichoic acid
- ML
MD-2-related lipid-recognition
- MD-2
myeloid differentiation-2
- MsML-1
M. sexta ML-1
- rMsML-1
recombinant MsML-1
- rCP36
recombinant CP36
- Npc2
Niemann-pick disease type-2 protein
- PAGE
polyacrylamide gel electrophoresis
- RPS3
ribosomal protein S3
- RT-PCR
reverse transcription-polymerase chain reaction
- SDS
sodium dodecyl sulfate
- TLR4
Toll-like receptor 4
- TNF
tumor necrosis factor
- TBS
Tris-buffered saline.
References
- Ao JQ, Ling E, Yu XQ. A Toll receptor from Manduca sexta is in response to Escherichia coli infection. Mol. Immunol. 2008;45:543–552. doi: 10.1016/j.molimm.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Axen A, Carlsson A, Engstrom A, Bennich H. Gloverin, an antibacterial protein from the immune hemolymph of Hyalophora pupae. Eur. J. Biochem. 1997;247:614–619. doi: 10.1111/j.1432-1033.1997.00614.x. [DOI] [PubMed] [Google Scholar]
- Beamer LJ, Carroll SF, Eisenberg D. The BPI/LBP family of proteins: a structural analysis of conserved regions. Protein Sci. 1998;7:906–914. doi: 10.1002/pro.5560070408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaby R. Lipopolysaccharide-binding molecules: transporters, blockers and sensors. Cell. Mol. Life Sci. 2004;61:1697–1713. doi: 10.1007/s00018-004-4020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn PE, Drake D. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm, Manduca sexta. J. Invertebr. Pathol. 1983;41:77–85. [Google Scholar]
- Ferguson AD, Welte W, Hofmann E, Lindner B, Holst O, Coulton JW, Diederichs K. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Structure. 2000;8:585–592. doi: 10.1016/s0969-2126(00)00143-x. [DOI] [PubMed] [Google Scholar]
- Frecer V, Ho B, Ding JL. Interpretation of biological activity data of bacterial endotoxins by simple molecular models of mechanism of action. Eur. J. Biochem. 2000;267:837–852. doi: 10.1046/j.1432-1327.2000.01069.x. [DOI] [PubMed] [Google Scholar]
- Freudenberg MA, Galanos C. Bacterial lipopolysaccharides: structure, metabolism and mechanisms of action. Int. Rev. Immunol. 1990;6:207–221. doi: 10.3109/08830189009056632. [DOI] [PubMed] [Google Scholar]
- Furukawa S, Tanaka H, Nakazawa H, Ishibashi J, Shono T, Yamakawa M. Inducible gene expression of moricin, a unique antibacterial peptide from the silkworm (Bombyx mori) Biochem. J. 1999;340(Pt 1):265–271. [PMC free article] [PubMed] [Google Scholar]
- Gorman MJ, Kankanala P, Kanost MR. Bacterial challenge stimulates innate immune responses in extra-embryonic tissues of tobacco hornworm eggs. Insect Mol. Biol. 2004;13:19–24. doi: 10.1111/j.1365-2583.2004.00454.x. [DOI] [PubMed] [Google Scholar]
- Hara S, Yamakawa M. Moricin, a novel type of antibacterial peptide isolated from the silkworm, Bombyx mori. J. Biol. Chem. 1995;270:29923–29927. doi: 10.1074/jbc.270.50.29923. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat. Immunol. 2002;3:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- Imler JL, Tauszig S, Jouanguy E, Forestier C, Hoffmann JA. LPS-induced immune response in Drosophila. J. Endotoxin Res. 2000;6:459–462. [PubMed] [Google Scholar]
- Inohara N, Nunez G. ML—a conserved domain involved in innate immunity and lipid metabolism. Trends Biochem. Sci. 2002;27:219–221. doi: 10.1016/s0968-0004(02)02084-4. [DOI] [PubMed] [Google Scholar]
- Jerala R. Structural biology of the LPS recognition. Int. J. Med. Microbiol. 2007;297:353–363. doi: 10.1016/j.ijmm.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Primary structure of ribosomal proteins S3 and S7 from Manduca sexta. Insect Mol. Biol. 1996;5:31–38. doi: 10.1111/j.1365-2583.1996.tb00038.x. [DOI] [PubMed] [Google Scholar]
- Kanaya T, Kobayashi J. Purification and characterization of an insect haemolymph protein promoting in vitro replication of the Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 2000;81:1135–1141. doi: 10.1099/0022-1317-81-4-1135. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Silverman N. Bacterial recognition and signalling by the Drosophila IMD pathway. Cell Microbiol. 2005;7:461–469. doi: 10.1111/j.1462-5822.2005.00504.x. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, Silverman N. Monomeric and polymeric Gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Katsuma S, Daimon T, Isono R, Omuro N, Mita K, Shimada T. Functional analysis of four Gloverin-like genes in the silkworm, Bombyx mori. Arch. Insect Biochem. Physiol. 2008;67:87–96. doi: 10.1002/arch.20223. [DOI] [PubMed] [Google Scholar]
- Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal structure of the TLR4–MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Lee E, Linder ME, Gilman AG. Expression of G-protein alpha subunits in Escherichia coli. Methods Enzymol. 1994;237:146–164. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc. Natl. Acad. Sci. U.S.A. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom A, Liu G, Kang D, Berzins K, Steiner H. Trichoplusia ni gloverin, an inducible immune gene encoding an antibacterial insect protein. Insect Biochem. Mol. Biol. 2002;32:795–801. doi: 10.1016/s0965-1748(01)00162-x. [DOI] [PubMed] [Google Scholar]
- Mackintosh JA, Gooley AA, Karuso PH, Beattie AJ, Jardine DR, Veal DA. A gloverin-like antibacterial protein is synthesized in Helicoverpa armigera following bacterial challenge. Dev. Comp. Immunol. 1998;22:387–399. doi: 10.1016/s0145-305x(98)00025-1. [DOI] [PubMed] [Google Scholar]
- Mancek-Keber M, Jerala R. Structural similarity between the hydrophobic fluorescent probe and lipid A as a ligand of MD-2. FASEB J. 2006;20:1836–1842. doi: 10.1096/fj.06-5862com. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S, Kitamura T, Kosugi A, Kimoto M, Miyake K. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat. Immunol. 2002;3:667–672. doi: 10.1038/ni809. [DOI] [PubMed] [Google Scholar]
- Ohto U, Fukase K, Miyake K, Satow Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science. 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- Oizumi Y, Hemmi H, Minami M, Asaoka A, Yamakawa M. Isolation, gene expression and solution structure of a novel moricin analogue, antibacterial peptide from a lepidopteran insect, Spodoptera litura. Biochim. Biophys. Acta. 2005;1752:83–92. doi: 10.1016/j.bbapap.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Pinner RW, Teutsch SM, Simonsen L, Klug LA, Graber JM, Clarke MJ, Berkelman RL. Trends in infectious diseases mortality in the United States. J. Am. Med. Assoc. 1996;275:189–193. [PubMed] [Google Scholar]
- Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Akashi S, Yamada T, Tanimura N, Kobayashi M, Konno K, Matsumoto F, Fukase K, Kusumoto S, Nagai Y, Kusumoto Y, Kosugi A, Miyake K. Lipid A antagonist, lipid IVa, is distinct from lipid A in interaction with Toll-like receptor 4 (TLR4)-MD-2 and ligand-induced TLR4 oligomerization. Int. Immunol. 2004;16:961–969. doi: 10.1093/intimm/dxh097. [DOI] [PubMed] [Google Scholar]
- Schromm AB, Lien E, Henneke P, Chow JC, Yoshimura A, Heine H, Latz E, Monks BG, Schwartz DA, Miyake K, Golenbock DT. Molecular genetic analysis of an endotoxin nonresponder mutant cell line: a point mutation in a conserved region of MD-2 abolishes endotoxin-induced signaling. J. Exp. Med. 2001;194:79–88. doi: 10.1084/jem.194.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenbak CR, Ryu JH, Leulier F, Pili-Floury S, Parquet C, Herve M, Chaput C, Boneca IG, Lee WJ, Lemaitre B, Mengin-Lecreulx D. Peptidoglycan molecular requirements allowing detection by the Drosophila immune deficiency pathway. J. Immunol. 2004;173:7339–7348. doi: 10.4049/jimmunol.173.12.7339. [DOI] [PubMed] [Google Scholar]
- Suderman RJ, Andersen SO, Hopkins TL, Kanost MR, Kramer KJ. Characterization and cDNA cloning of three major proteins from pharate pupal cuticle of Manduca sexta. Insect Biochem. Mol. Biol. 2003;33:331–343. doi: 10.1016/s0965-1748(02)00247-3. [DOI] [PubMed] [Google Scholar]
- Viriyakosol S, Tobias PS, Kitchens RL, Kirkland TN. MD-2 binds to bacterial lipopolysaccharide. J. Biol. Chem. 2001;276:38044–38051. doi: 10.1074/jbc.M105228200. [DOI] [PubMed] [Google Scholar]
- Visintin A, Mazzoni A, Spitzer JA, Segal DM. Secreted MD-2 is a large polymeric protein that efficiently confers lipopolysaccharide sensitivity to Toll-like receptor 4. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12156–12161. doi: 10.1073/pnas.211445098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu. Rev. Cell Dev. Biol. 2006;22:409–437. doi: 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Kanost MR. Developmental expression of Manduca sexta hemolin. Arch. Insect Biochem. Physiol. 1999;42:198–212. doi: 10.1002/(SICI)1520-6327(199911)42:3<198::AID-ARCH4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Kanost MR. Immulectin-2, a lipopolysaccharide-specific lectin from an insect, Manduca sexta, is induced in response to Gram-negative bacteria. J. Biol. Chem. 2000;275:37373–37381. doi: 10.1074/jbc.M003021200. [DOI] [PubMed] [Google Scholar]
- Yu B, Hailman E, Wright SD. Lipopolysaccharide binding protein and soluble CD14 catalyze exchange of phospholipids. J. Clin. Invest. 1997;99:315–324. doi: 10.1172/JCI119160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XQ, Gan H, Kanost MR. Immulectin, an inducible C-type lectin from an insect, Manduca sexta, stimulates activation of plasma prophenol oxidase. Insect Biochem. Mol. Biol. 1999;29:585–597. doi: 10.1016/s0965-1748(99)00036-3. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Tracy ME, Ling E, Scholz FR, Trenczek T. Anovel C-type immulectin-3 from Manduca sexta is translocated from hemolymph into the cytoplasm of hemocytes. Insect Biochem. Mol. Biol. 2005;35:285–295. doi: 10.1016/j.ibmb.2005.01.004. [DOI] [PubMed] [Google Scholar]