Abstract
CD4 T cells promote innate and adaptive immune responses, but how vaccine-elicited CD4 T cells contribute to immune protection remains unclear. Here we evaluated whether induction of virus-specific CD4 T cells by vaccination would protect mice against infection with chronic lymphocytic choriomeningitis virus (LCMV). Immunization with vaccines that selectively induced CD4 T cell responses resulted in catastrophic inflammation and mortality following challenge with a persistent strain of LCMV. Immunopathology required antigen-specific CD4 T cells and was associated with a cytokine storm, generalized inflammation, and multi-organ system failure. Virus-specific CD8 T cells or antibodies abrogated the pathology. These data demonstrate that vaccine-elicited CD4 T cells in the absence of effective antiviral immune responses can trigger lethal immunopathology.
CD4 T cells play an essential role in facilitating innate and adaptive immune responses. Absence of CD4 T cells at the time of priming results in impaired memory CD8 T cell responses(1–4) and severe CD8 T cell dysfunction with uncontrolled viral replication following persistent viral infections(5–8). Moreover, adoptive transfer of virus-specific CD4 T cells during chronic lymphocytic choriomeningitis virus (LCMV) infection has been shown to rescue cytotoxic and humoral responses, resulting in enhanced viral control(9). As a result, developing strategies that preferentially elicit CD4 T cell responses by candidate vaccines has been a research priority, and several CD4 T cell-based vaccines against smallpox and HIV are currently being tested(10–13). However, little is known about the role of vaccine-elicited CD4 T cells following viral challenge.
We explored whether a vaccine that elicited CD4 T cell responses would afford protective immunity against LCMV infection in mice. We first vaccinated C57BL/6 mice with a Listeria monocytogenes vector expressing the LCMV glycoprotein-specific I-Ab restricted CD4 T cell epitope GP61-80 (LM-GP61)(14). Vaccination elicited durable GP61-specific CD4 T cell responses (Fig. S1A) that peaked at day 8 and persisted for over 60 days following immunization (Fig. S1B).
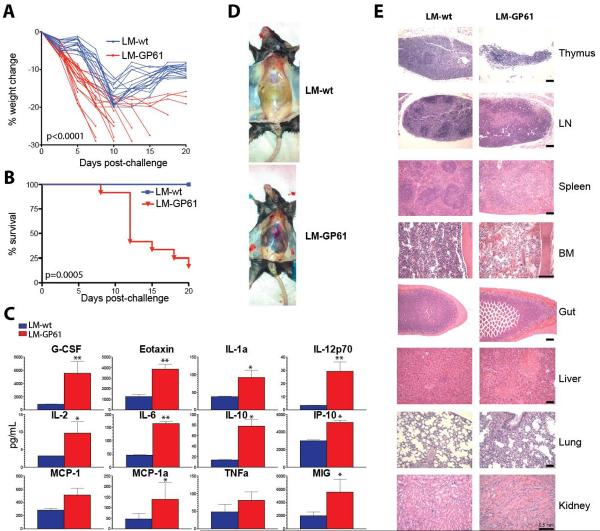
Vaccinated mice were then challenged with LCMV Clone-13 (Cl-13), which causes a systemic infection that lasts for 60–90 days(15). As expected, control mice (LM-wt) exhibited modest weight loss after challenge followed by recovery(16) (Fig. 1A). In contrast, LM-GP61 vaccinated mice exhibited immunopathology characterized by >20% weight loss (p<0.0001) (Fig. 1A) and 90% mortality by day 20 following challenge (P=0.0005) (Fig. 1B), which was associated with cytokine storm (Fig. 1C). Gross pathology of vaccinated mice following challenge showed widespread inflammation (Fig. 1D), and histopathology revealed involution of lymphoid tissues, impaired development of B cell follicles, and severe tissue destruction (Fig. 1E), consistent with multi-organ system failure.
Fig 1. CD4 T cell vaccines induce lethal immunopathology and systemic inflammation following LCMV Cl-13 challenge.
LM-wt or LM-GP61 immune C57BL/6 mice were challenged with 2×106 PFU of LCMV Cl-13. A) Weight loss. P-value on day 7 is indicated. B) Percent survival. Statistical analysis for survival plot was performed using the Mantel-Cox test. C) Average cytokine levels in serum by luminex assays at day 8. D) Gross pathology of inflammation and hemorrhage from representative mice at day 8. E) Hematoxylin & eosin (H&E) staining of lymphoid and non-lymphoid tissues at day 8 (20X, bone marrow and kidney; 10X, all other tissues). Black scale-bar represents 0.5 mm. Panels A and B present combined data from 4 experiments, N=3–5 mice/group per experiment. Panels C, D, E are representative data from 1 of 3 experiments, N=4 mice/group per experiment. *, P=0.05; **, P=0.02 (Mann-Whitney test). Error bars indicate SEM.
We next determined the generalizability of these observations. Immunization of C57BL/6 mice with dendritic cells (DCs) coated with various I-Ab restricted CD4 T cell epitopes (GP6, GP126, and NP309 with or without GP61) (Fig. S2A) resulted in mortality following LCMV Cl-13 challenge (Fig. S2B). Moreover, immunization of BALB/c mice with DCs pulsed with the I-Ad restricted NP116 epitope (Fig. S2C) similarly led to mortality following challenge (Fig. S2D). These data demonstrate that the CD4 T cell immunopathology observed with the LM-GP61 vaccine was not specific to the vaccine platform, target epitope, or host genetic background.
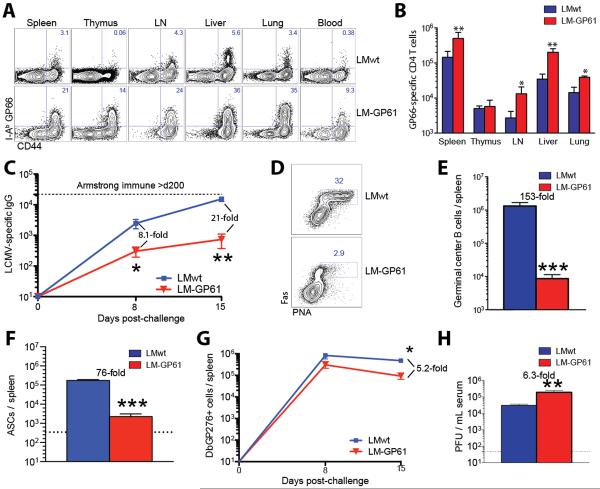
We next analyzed adaptive immune responses following challenge. Mice vaccinated with LM-GP61 and challenged with LCMV Cl-13 exhibited elevated GP61-specific CD4 T cell responses in tissues and blood at day 8 (25-fold greater than controls, p<0.0001) (Fig. 2A, 2B). By day 15, these vaccinated mice showed a 21-fold reduction in IgG responses (P=0.02) (Fig. 2C), a 153-fold reduction in the number of germinal center B cells (P=0.001) (Fig. 2D, 2E), a 76-fold reduction in the number of antibody-secreting cells (P=0.002) compared to controls (Fig. 2F). This decrease in humoral responses in mice that received the CD4 T cell vaccine paralleled the observations of our histological analyses, which showed absence of germinal centers in lymph nodes and spleen (Fig. 1F). Moreover, there was a 5.2-fold reduction in the number of GP276-specific CD8 T cells in the spleen (P=0.05) (Fig. 2G) (gating scheme shown in Fig. S3A–3C), which may have been due to a greater CD8 T cell deletion in the context of higher viral loads. Vaccinated mice also exhibited a 6.3-fold increase in viremia at day 8 (P=0.02) (Fig. 2H). Tissue viral loads were also increased (p≤0.05)(Fig. S4A), and the pattern of infected cells was similar between vaccinated and control mice at day 8 (Fig. S4B, S4C).
Fig 2. Uncontrolled anamnestic LCMV-specific CD4 T cells and impaired adaptive immunity following LCMV Cl-13 challenge.
A) Representative FACS plot showing I-Ab GP66-specific CD4 T cells in lymphoid and non-lymphoid tissues. B) Numbers of I-Ab GP66-specific CD4 T cells in lymphoid and non-lymphoid tissues. C) Longitudinal analysis of LCMV-specific IgG responses in sera. D) Representative FACS plot showing germinal center B cell responses in spleen. E) Number of germinal center B cells in spleen. F) Number of antibody secreting cells in spleen. G) Longitudinal analysis of LCMV-specific (DbGP276+) CD8 T cell responses in spleen. H) Viremia on day 8 following infection. Experiment was performed similarly to Fig. 1. Panels A, B are from day 8. Panels D, E, F are from day 15. Panels B–C and E–H are combined data from 5 experiments, n=3–4 mice/group per experiment. *, P=0.05; **, P=0.02; ***, P≤0.002 (Mann-Whitney test). Error bars indicate SEM.
Despite the massive expansion of GP61-specific CD4 T cells, the lethal immunopathology was associated with a 2.7-fold reduction in the total number of CD4 T cells (P=0.05) (Fig. S5A, S5B), suggesting impaired maintenance of CD4 T cells. Mice that received the LM-GP61 vaccine also showed a 3.6-fold reduction in the frequencies of Tregs (P=0.03) (Fig. S5A, S5C), a 15.4-fold increase in the effector to Treg ratio (P=0.003) (Fig. S5D), and were moderately lymphopenic (Fig. S5E) as compared to controls following LCMV Cl-13 challenge. It is unlikely that partial Treg collapse alone caused the observed mortality, since complete Treg ablation typically induces immunopathology after 2–3 weeks(17, 18), and the mortality reported here was fulminant, consistent with cytokine storm rather than autoimmunity.
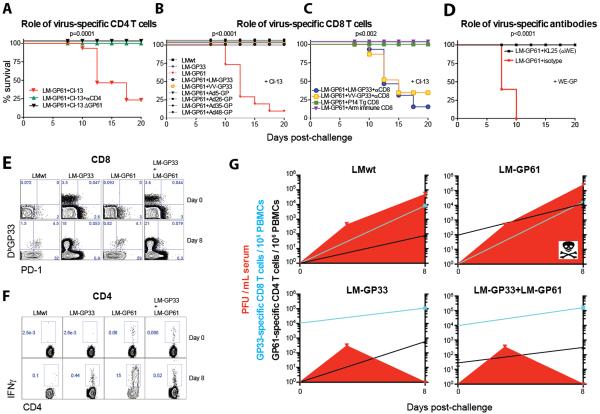
We next explored the mechanism of the observed lethal immunopathology. First, LM-GP61-vaccinated mice that were challenged with a mutant LCMV Cl-13 virus strain that specifically lacked the GP61-80 epitope (rCl-13/WE-GP ΔGP61) demonstrated no mortality, and depletion of CD4 T cells prior to LCMV Cl-13 challenge abrogated the immunopathology (Fig. 3A). These data suggest that virus-specific CD4 T cells are required for the observed immunopathology. Second, we assessed whether the immunopathology could be recapitulated simply by increasing the precursor frequency of virus-specific CD4 T cells. We challenged mice with LCMV Cl-13 one day after adoptive transfer with between 103 and 105 naïve SMARTA cells (TCR-transgenic CD4 T cells specific for the LCMV GP66-77 epitope) (Fig. S6A). Transfer of 105 SMARTA cells resulted in significant mortality (Fig. S6B) and impaired antiviral immunity (Fig. S6C–S6D), similar to what we observed after vaccination with LM-GP61 or peptide-pulsed DCs. These data suggest that the lethal immunopathology could be recapitulated by increasing the precursor frequency of CD4 T cells.
Fig 3. Prevention of CD4 T cell mediated pathology by antiviral CD8 T cells and antibodies.
A) Percent survival in LM-GP61 vaccinated mice following LCMV Cl-13 challenge with or without CD4 T cell depletion or following rCl-13/WE-GP ΔGP61 challenge. B) Percent survival following co-immunization with LM-GP61 and Listeria, poxvirus, or adenovirus based vaccines that elicited CD8 T cell responses prior to challenge. C) Percent survival in co-immunized mice following CD8 T cell depletion or in LM-GP61 immunized mice following adoptive transfer of P14 transgenic CD8 T cells or LCMV-specific CD8 T cells from LCMV Armstrong immune mice prior to chronic viral challenge. D) Percent survival following infection with rCl-13/WE-GP expressing the glycoprotein of the WE strain, with or without infusion with the WE-GP-specific mAb KL25. E) Representative FACS plots showing the kinetics of GP33-specific CD8 T cell responses before and after chronic viral challenge. F) Representative FACS plots showing the kinetics of GP61-specific CD4 T cell responses before and after chronic viral challenge. G) Summary of CD8 T cell responses, CD4 T cell responses and viral loads following chronic viral infection. Statistical analyses for survival plots were performed using the Mantel-Cox test. Experiment was performed similarly to Fig. 1. Panels A–D and G present combined data from 2 experiments, n=5–6 mice/group per experiment. Error bars indicate SEM.
To explore the mechanism further, we assessed whether suppressing viral replication with LCMV-specific CD8 T cells or antibodies would abrogate this pathology. All mice that were co-immunized with LM-GP61 and various vaccines that expressed the CD8 epitope GP33 or the full-length LCMV glycoprotein survived the LCMV Cl-13 challenge (Fig. 3B). The abrogation of the immunopathology was specifically due to vaccine-elicited CD8 T cells, since depletion of CD8 T cells in co-immunized mice before LCMV Cl-13 challenge recapitulated the observed mortality (Fig. 3C). In addition, adoptive transfer of 106 P14 CD8 T cells (TCR transgenic CD8 T cells specific for LCMV GP33), or purified CD8 T cells from mice that cleared LCMV Armstrong (which is a strain that is acutely cleared and induces functional responses) prevented the immunopathology (Fig. 3C). In order to test the role of antibodies at preventing the observed immunopathology, we challenged vaccinated mice with a recombinant LCMV Cl-13 strain expressing LCMV WE GP (LCMV Cl-13/WE-GP), which can be neutralized by administering the monoclonal antibody KL25(19). Similarly, administration of this neutralizing antibody, but not an isotype-matched control antibody, abrogated the lethal pathology in LM-GP61 vaccinated mice following challenge with neutralization sensitive LCMV Cl-13/WE-GP (Fig. 3D).
Mice that were co-immunized with vaccines that induce both CD4 and CD8 T cell responses demonstrated more robust CD8 T cell recall responses (Fig. 3E), which was associated with a 40.3-fold reduction in CD4 T cell responses (P=0.04) (Fig. 3F–3G) and complete virological control (P=0.007) by day 8 post-challenge (Fig. 3G). These data support the proposed model of antigen-driven hyperstimulation of vaccine-elicited CD4 T cells. Sufficient antiviral CD8 T cells or antibodies limit viral replication and thereby reduce the antigen-dependent activation of memory CD4 T cells, thus abrogating the observed immunopathology. The absence of mortality in LM-wt immunized mice was likely due to the low numbers of virus-specific CD4 T cell responses relative to LM-GP61 immunized mice.
Many CD4 T cell epitopes incorporate smaller CD8 T cell epitopes(20–24). For example, within the LCMV GP61-80 CD4 epitope lies an embedded H-2Kb-restricted GP70-77 CD8 T cell epitope(25) (Fig. S7). However, this CD8 T cell response was too subdominant to control viral replication; H-2Kb deficient mice (which cannot generate GP70-specific CD8 T cells) and wild type mice vaccinated with LM-GP61 similarly succumbed following LCMV Cl-13 challenge (Fig. S8).
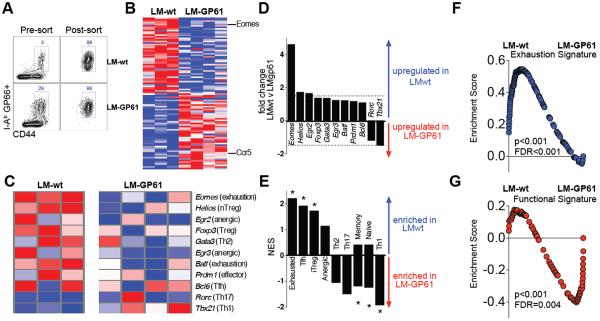
Finally, we assessed differences in the transcriptional profiles of CD4 T cells on day 8 following chronic viral challenge by gene expression profiling. Transcriptional analysis of purified GP66-specific CD4 T cells (Fig. 4A) identified numerous differentially expressed genes (Fig. 4B–4C and Table S1–S3). Following LCMV Cl-13 challenge, LCMV-specific CD4 T cells from both experimental groups remained FoxP3- (Fig. S9A), and some exhibited T follicular helper (Tfh) differentiation (Fig. S9B), as expected(26). GP66-specific CD4 T cells from LM-GP61 vaccinated mice showed lower levels of Eomes (which is a transcription factor associated with expression of inhibitory receptors and exhaustion) (27), and expressed higher levels of CCR5 (the HIV coreceptor) relative to controls (Fig. 4B, 4C, S9C, S9D).
Fig 4. Vaccine-elicited CD4 T cells are highly functional and bypass typical functional exhaustion following LCMV Cl-13 challenge.
Microarray analysis was performed on sorted GP66-specific CD4 T cells at day 8. A) Cell purity following FACS-sorting of I-Ab restricted GP66-specific CD44+ CD4 T cells. B) Heat map of the most differentially expressed genes. C) Heat map comparing the expression of transcription factors involved in CD4 T cell differentiation. D) Fold change in the expression of transcription factors involved in CD4 T cell differentiation. Dashed line represents the cut-off for significance (≥1.5-fold change). E) Enrichment score for different CD4 T cell subsets by gene set enrichment analysis (GSEA). Asterisks represent significant values (p value & FDR q value <0.01). F) GSEA demonstrating enrichment for a CD4 T cell exhaustion signature in chronically infected mice that had received the control vaccine. G) GSEA demonstrating enrichment for a functional Th1 CD4 T cell response signature in chronically infected mice that had received the CD4 T cell vaccine. Experiment was performed similarly to Fig. 1. Presented data are from LM-wt (n=3) and LM-GP61 (n=4) vaccinated mice at day 8 following challenge with LCMV Cl-13.
GP61-specific CD4 T cells from controls exhibited the expected CD4 T cell exhaustion signature characterized by high Eomes expression (Fig. 4B–4D). In contrast, GP61-specific CD4 T cells from LM-GP61 vaccinated mice showed marked enrichment of genes and expression of cytokines associated with highly functional, effector T helper 1 (Th1) responses (Fig. 4C–4E, S10). Moreover, by gene set enrichment analysis, immunopathologic CD4 T cells displayed an activated Th1 CD4 T cell signature, suggesting that these cells failed to undergo normal physiologic exhaustion (Fig. 4E–4G). Furthermore, the gene expression profiles in LM-GP61 vaccinated mice following challenge showed enrichment for cellular processes involved in T cell activation and lymphocyte activation (Fig. S11, Table S1–S3). Moreover, the lethal immunopathology required ongoing viral replication, as LM-GP61 vaccinated mice challenged with LCMV Armstrong showed no mortality, with a modestly enhanced memory CD8 T cell differentiation and viral control (Fig. S12). Taken together, these data are consistent with a model in which uncontrolled viral replication resulted in overstimulation of vaccine-elicited Th1 CD4 cells, leading to generalized inflammation and multi-organ system failure (Fig. S13).
Our data demonstrate that a vaccine that elicits primarily CD4 T cells can result in lethal immunopathology following challenge with a persistently replicating virus by a mechanism that involves hyperstimulation of vaccine-elicited CD4 T cells by uncontrolled viral replication. Importantly, both antiviral CD8 T cells and antibodies that limit viral replication abrogate this pathology. These data show that vaccine-elicited CD4 T cells can trigger immunopathology and death in certain settings.
Although the extent to which this phenomenon may occur in humans has not yet been determined, this mechanism is potentially generalizable to other vaccines that primarily induce CD4 T cells in the absence of other effective antiviral immune responses. A previous study reported that a vaccine that encoded an SIV CD4 T cell epitope led to higher viral loads and accelerated AIDS progression as compared with controls following SIV challenge in rhesus monkeys(28), although SIV-specific CD4 T cell responses were not directly measured in this prior study. Moreover, since activated CD4 T cells also can serve directly as targets for HIV, vaccine-elicited CD4 T cells could theoretically have multifactorial negative effects(29). These findings warrant a thorough re-evaluation of CD4 T cell responses, especially in the context of chronic infection.
Supplementary Material
Acknowledgements
The authors thank A. Wieland, M. Rasheed, A. Kamphorst, K. Araki, S. Crotty, B. Walker, Christine Bricault, P. Abbink, and F. Ball for generous advice, assistance, and reagents. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. This work was supported by grants from the NIH (AI007245 / AI07387 P.P.M.; AI078526 / AI096040 D.H.B.; AI030048 R.A.), the Bill and Melinda Gates Foundation (OPP1033091 D.H.B.), the Swiss National Science Foundation (310030_149340/1 D.D.P.), the European Research Council (D.D.P.), and the Ragon Institute of MGH, MIT, and Harvard (D.H.B.). D.L.B. was supported by the Intramural Research Program of NIAID/NIH. Gene expression data have been uploaded to GEO (#GSE63825).
Footnotes
The authors declare no financial conflicts of interest.
References
- 1.Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–342. doi: 10.1126/science.1083317. published online EpubApr 11 (10.1126/science.1083317 300/5617/339 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–339. doi: 10.1126/science.1082305. published online EpubApr 11 (10.1126/science.1082305) [DOI] [PubMed] [Google Scholar]
- 3.Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–856. doi: 10.1038/nature01441. published online EpubFeb 20 (10.1038/nature01441) [DOI] [PubMed] [Google Scholar]
- 4.Janssen EM, Droin NM, Lemmens EE, Pinkoski MJ, Bensinger SJ, Ehst BD, Griffith TS, Green DR, Schoenberger SP. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature. 2005;434:88–93. doi: 10.1038/nature03337. published online EpubMar 3 (10.1038/nature03337) [DOI] [PubMed] [Google Scholar]
- 5.Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. Journal of virology. 1994;68:8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. published online EpubDec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battegay M, Moskophidis D, Rahemtulla A, Hengartner H, Mak TW, Zinkernagel RM. Enhanced establishment of a virus carrier state in adult CD4+ T-cell-deficient mice. Journal of virology. 1994;68:4700–4704. doi: 10.1128/jvi.68.7.4700-4704.1994. published online EpubJul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardin RD, Brooks JW, Sarawar SR, Doherty PC. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. The Journal of experimental medicine. 1996;184:863–871. doi: 10.1084/jem.184.3.863. published online EpubSep 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, Murthy KK, Rice CM, Walker CM. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302:659–662. doi: 10.1126/science.1088774. published online EpubOct 24 (10.1126/science.1088774) [DOI] [PubMed] [Google Scholar]
- 9.Aubert RD, Kamphorst AO, Sarkar S, Vezys V, Ha SJ, Barber DL, Ye L, Sharpe AH, Freeman GJ, Ahmed R. Antigen-specific CD4 T-cell help rescues exhausted CD8 T cells during chronic viral infection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21182–21187. doi: 10.1073/pnas.1118450109. published online EpubDec 27 (10.1073/pnas.1118450109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lichterfeld M, Gandhi RT, Simmons RP, Flynn T, Sbrolla A, Yu XG, Basgoz N, Mui S, Williams K, Streeck H, Burgett-Yandow N, Roy G, Janssens M, Pedneault L, Vandepapeliere P, Koutsoukos M, Demoitie MA, Bourguignon P, McNally L, Voss G, Altfeld M. Induction of strong HIV-1-specific CD4+ T-cell responses using an HIV-1 gp120/NefTat vaccine adjuvanted with AS02A in antiretroviral-treated HIV-1-infected individuals. J Acquir Immune Defic Syndr. 2012;59:1–9. doi: 10.1097/QAI.0b013e3182373b77. published online EpubJan 1 (10.1097/QAI.0b013e3182373b77) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amara RR, Nigam P, Sharma S, Liu J, Bostik V. Long-lived poxvirus immunity, robust CD4 help, and better persistence of CD4 than CD8 T cells. Journal of virology. 2004;78:3811–3816. doi: 10.1128/JVI.78.8.3811-3816.2004. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez F, Harkins S, Redwine JM, de Pereda JM, Whitton JL. CD4(+) T cells induced by a DNA vaccine: immunological consequences of epitope-specific lysosomal targeting. Journal of virology. 2001;75:10421–10430. doi: 10.1128/JVI.75.21.10421-10430.2001. published online EpubNov (10.1128/JVI.75.21.10421-10430.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Streeck H, D'Souza MP, Littman DR, Crotty S. Harnessing CD4(+) T cell responses in HIV vaccine development. Nature medicine. 2013;19:143–149. doi: 10.1038/nm.3054. published online EpubFeb (10.1038/nm.3054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams MA, Ravkov EV, Bevan MJ. Rapid culling of the CD4+ T cell repertoire in the transition from effector to memory. Immunity. 2008;28:533–545. doi: 10.1016/j.immuni.2008.02.014. published online EpubApr (10.1016/j.immuni.2008.02.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. Journal of virology. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waggoner SN, Cornberg M, Selin LK, Welsh RM. Natural killer cells act as rheostats modulating antiviral T cells. Nature. 2012;481:394–398. doi: 10.1038/nature10624. published online EpubJan 19 (10.1038/nature10624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penaloza-MacMaster P, Kamphorst AO, Wieland A, Araki K, Iyer SS, West EE, O'Mara L, Yang S, Konieczny BT, Sharpe AH, Freeman GJ, Rudensky AY, Ahmed R. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. The Journal of experimental medicine. 2014;211:1905–1918. doi: 10.1084/jem.20132577. published online EpubAug 25 (10.1084/jem.20132577) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nature immunology. 2007;8:191–197. doi: 10.1038/ni1428. published online EpubFeb (ni1428 [pii] 10.1038/ni1428) [DOI] [PubMed] [Google Scholar]
- 19.Bruns M, Cihak J, Muller G, Lehmann-Grube F. Lymphocytic choriomeningitis virus. VI. Isolation of a glycoprotein mediating neutralization. Virology. 1983;130:247–251. doi: 10.1016/0042-6822(83)90135-6. published online EpubOct 15. [DOI] [PubMed] [Google Scholar]
- 20.Dao T, Korontsvit T, Zakhaleva V, Haro K, Packin J, Scheinberg DA. Identification of a human cyclin D1-derived peptide that induces human cytotoxic CD4 T cells. PloS one. 2009;4:e6730. doi: 10.1371/journal.pone.0006730. 10.1371/journal.pone.0006730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dow C, Oseroff C, Peters B, Nance-Sotelo C, Sidney J, Buchmeier M, Sette A, Mothe BR. Lymphocytic choriomeningitis virus infection yields overlapping CD4+ and CD8+ T-cell responses. Journal of virology. 2008;82:11734–11741. doi: 10.1128/JVI.00435-08. published online EpubDec (10.1128/JVI.00435-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May RJ, Dao T, Pinilla-Ibarz J, Korontsvit T, Zakhaleva V, Zhang RH, Maslak P, Scheinberg DA. Peptide epitopes from the Wilms' tumor 1 oncoprotein stimulate CD4+ and CD8+ T cells that recognize and kill human malignant mesothelioma tumor cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:4547–4555. doi: 10.1158/1078-0432.CCR-07-0708. published online EpubAug 1 (10.1158/1078-0432.CCR-07-0708) [DOI] [PubMed] [Google Scholar]
- 23.Ou D, Mitchell LA, Decarie D, Gillam S, Tingle AJ. Characterization of an overlapping CD8+ and CD4+ T-cell epitope on rubella capsid protein. Virology. 1997;235:286–292. doi: 10.1006/viro.1997.8704. published online EpubSep 1 (10.1006/viro.1997.8704) [DOI] [PubMed] [Google Scholar]
- 24.Abrams SI, Stanziale SF, Lunin SD, Zaremba S, Schlom J. Identification of overlapping epitopes in mutant ras oncogene peptides that activate CD4+ and CD8+ T cell responses. European journal of immunology. 1996;26:435–443. doi: 10.1002/eji.1830260225. published online EpubFeb (10.1002/eji.1830260225) [DOI] [PubMed] [Google Scholar]
- 25.Homann D, Lewicki H, Brooks D, Eberlein J, Mallet-Designe V, Teyton L, Oldstone MB. Mapping and restriction of a dominant viral CD4+ T cell core epitope by both MHC class I and MHC class II. Virology. 2007;363:113–123. doi: 10.1016/j.virol.2006.12.025. published online EpubJun 20 (10.1016/j.virol.2006.12.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fahey LM, Wilson EB, Elsaesser H, Fistonich CD, McGavern DB, Brooks DG. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. The Journal of experimental medicine. 2011;208:987–999. doi: 10.1084/jem.20101773. published online EpubMay 9 (10.1084/jem.20101773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crawford A, Angelosanto JM, Kao C, Doering TA, Odorizzi PM, Barnett BE, Wherry EJ. Molecular and Transcriptional Basis of CD4(+) T Cell Dysfunction during Chronic Infection. Immunity. 2014;40:289–302. doi: 10.1016/j.immuni.2014.01.005. published online EpubFeb 20 (10.1016/j.immuni.2014.01.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staprans SI, Barry AP, Silvestri G, Safrit JT, Kozyr N, Sumpter B, Nguyen H, McClure H, Montefiori D, Cohen JI, Feinberg MB. Enhanced SIV replication and accelerated progression to AIDS in macaques primed to mount a CD4 T cell response to the SIV envelope protein. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13026–13031. doi: 10.1073/pnas.0404739101. published online EpubAug 31 (10.1073/pnas.04047391010404739101 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fauci AS, Marovich MA, Dieffenbach CW, Hunter E, Buchbinder SP. Immunology. Immune activation with HIV vaccines. Science. 2014;344:49–51. doi: 10.1126/science.1250672. published online EpubApr 4 (10.1126/science.1250672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed R, Oldstone MB. Organ-specific selection of viral variants during chronic infection. The Journal of experimental medicine. 1988;167:1719–1724. doi: 10.1084/jem.167.5.1719. published online EpubMay 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recher M, Lang KS, Hunziker L, Freigang S, Eschli B, Harris NL, Navarini A, Senn BM, Fink K, Lotscher M, Hangartner L, Zellweger R, Hersberger M, Theocharides A, Hengartner H, Zinkernagel RM. Deliberate removal of T cell help improves virus-neutralizing antibody production. Nature immunology. 2004;5:934–942. doi: 10.1038/ni1102. published online EpubSep (10.1038/ni1102) [DOI] [PubMed] [Google Scholar]
- 32.Flatz L, Bergthaler A, de la Torre JC, Pinschewer DD. Recovery of an arenavirus entirely from RNA polymerase I/II-driven cDNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4663–4668. doi: 10.1073/pnas.0600652103. published online EpubMar 21 (10.1073/pnas.0600652103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Badovinac VP, Messingham KA, Jabbari A, Haring JS, Harty JT. Accelerated CD8+ T-cell memory and prime-boost response after dendritic-cell vaccination. Nature medicine. 2005;11:748–756. doi: 10.1038/nm1257. published online EpubJul (10.1038/nm1257) [DOI] [PubMed] [Google Scholar]
- 34.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. The Journal of experimental medicine. 1984;160:521–540. doi: 10.1084/jem.160.2.521. published online EpubAug 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. published online EpubMar 23 (10.1126/science.10588671058867 [pii]) [DOI] [PubMed] [Google Scholar]
- 36.Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, Russell K, Toth I, Piechocka-Trocha A, Dolfi D, Angelosanto J, Crawford A, Shin H, Kwon DS, Zupkosky J, Francisco L, Freeman GJ, Wherry EJ, Kaufmann DE, Walker BD, Ebert B, Haining WN. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nature medicine. 2010;16:1147–1151. doi: 10.1038/nm.2232. published online EpubOct (10.1038/nm.2232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnitz RA, Imam S, Yates K, Haining WN. Isolation of RNA and the synthesis and amplification of cDNA from antigen-specific T cells for genome-wide expression analysis. Methods Mol Biol. 2013;979:161–173. doi: 10.1007/978-1-62703-290-2_13. 10.1007/978-1-62703-290-2_13) [DOI] [PubMed] [Google Scholar]
- 38.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O'Shea JJ, Zhao K. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. published online EpubJan 16 (10.1016/j.immuni.2008.12.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. published online EpubJul 1 (10.4049/jimmunol.0903505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Safford M, Collins S, Lutz MA, Allen A, Huang CT, Kowalski J, Blackford A, Horton MR, Drake C, Schwartz RH, Powell JD. Egr-2 and Egr-3 are negative regulators of T cell activation. Nature immunology. 2005;6:472–480. doi: 10.1038/ni1193. published online EpubMay (10.1038/ni1193) [DOI] [PubMed] [Google Scholar]
- 41.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. 10.1186/1471-2105-10-48) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS computational biology. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. published online EpubMar 23 (10.1371/journal.pcbi.0030039) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.