Abstract
Since their introduction to biological imaging, quantum dots (QDs) have progressed from a little known, but attractive technology to one that has gained broad application in many areas of biology. The versatile properties of these fluorescent nanoparticles have allowed investigators to conduct biological studies with extended spatiotemporal capabilities that were previously not possible. In this review, we focus on QD applications that provide enhanced quantitative information on protein dynamics and localization, including single particle tracking (SPT) and immunohistochemistry (IHC), and finish by examining prospects of upcoming applications, such as correlative light and electron microscopy (CLEM) and super-resolution. Advances in single molecule imaging, including multi-color and 3D QD tracking, have provided new insights into the mechanisms of cell signaling and protein trafficking. New forms of QD tracking in vivo have allowed for observation of biological processes with molecular level resolution in the physiological context of the whole animal. Further methodological development of multiplexed QD-based immunohistochemistry assays are allowing more quantitative analysis of key proteins in tissue samples. These advances highlight the unique quantitative data sets that QDs can provide to further our understanding of biological and disease processes.
Keywords: QDs, single particle tracking, immunohistochemistry, fluorescence microscopy
In 1998, two papers appeared back-to-back in Science (Bruchez et al 1998; Chan and Nie 1998) describing the first applications of fluorescent semiconducting nanocrystals, or quantum dots (QDs), to biological imaging. The critical advance demonstrated in these papers was the development of water soluble QDs that could be conjugated to biomolecules for molecular targeting. The studies included the targeting of QDs to living cells via ligand coupling (Chan and Nie 1998) and multi-color labeling of structures in fixed cells (Bruchez et al 1998). Since these seminal papers, the application of QDs in bio-imaging has rapidly expanded to include many modalities that cover multiple time and length scales, from single molecule to in vivo imaging (Figure 1). The reason for their widespread use comes from several key advantages that QDs provide over conventional fluorophores (see Table 1). In particular, QDs have high photostability such that long-term imaging can be achieved without artifacts from photobleaching. Additionally, the broad absorption spectra and narrow emission spectra allow for simultaneous excitation of spectrally distinct QDs and easy spectral separation of emission for multiplex imaging. A number of excellent reviews contain detailed information on QD chemistry and photophysical properties (Michalet et al 2005; Giepmans et al 2006; Pons and Mattoussi 2009; Pinaud et al 2010; Petryayeva et al 2013). Here, we will highlight unique biological imaging applications that have been enabled by QDs, namely high-resolution imaging of protein behavior at the single molecule level and developments in multi-color, quantitative immunohistochemistry (IHC). Advances in bioconjugation techniques and applications in correlative light and electron microscopy (CLEM) and super-resolution are also discussed.
Figure 1. QDs are used in a range of biological imaging techniques.
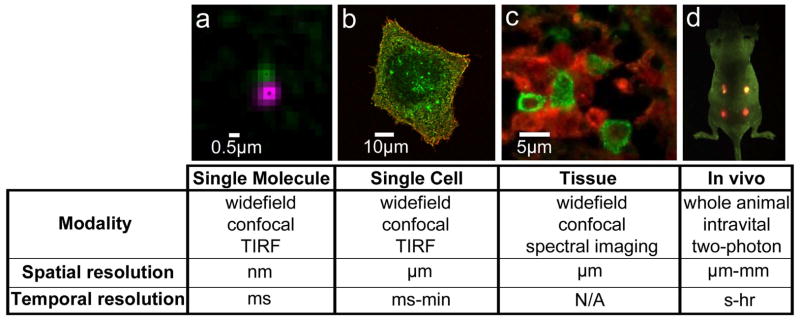
a) Single molecule detection provides high spatiotemporal resolution. Example of two-color QD tracking of QD-labeled EGF bound to EGFR (see also Low-Nam et al, 2011). b) Live cell imaging captures dynamics of cellular processes. Image shows QD-EGF (red) binding to EGFR (green) on the surface of an A431 cell (see also Lidke et al, 2004). c) QDs used in immunohistochemistry (IHC) assays allow for multiplex imaging. Example of two-color QD-IHC in human spleen tissue with QD-labeled antibodies against mast cell tryptase (green) and c-Kit (red) (image by E.W. Hatch, Lidke Lab). d) QDs can be visualized in in vivo imaging. Image shows simultaneous in vivo imaging of spectrally distinct QD-encoded microbeads. Image courtesy of X. Gao (see also Gao et al, 2004).
Table 1.
Relative comparison of QDs versus organic fluorophores.
| QDs | Organic Dyes | Fluorescent Proteins | |
|---|---|---|---|
| Size | 1 | ||
| Photostability | 2 | 2 | |
| Broad excitation | 3 | ||
| Multiplex imaging | 4 | ||
| Brightness | 5 | 6 | |
| Flexible conjugation | 7 | ||
| Genetically expressible | 8 | 8 | |
| Electron dense | 9 | 9 | |
| Sample fixation requirements | 10 | ||
| Continuous emission | 11 | ||
| Accessible time/length scales | 12 | 13 | 13 |
| In vivo imaging | 14 | 15 |
The color indicates relative performance level in each category: green is best, yellow intermediate and red poor.
QDs are larger than organic fluorophores (10–20 nm in diameter). This can lead to steric interference with protein function and must be carefully tested.
Photobleaching can be advantageous for techniques such as FRAP. Organic dye derivatives have improved photostability (Altman et al 2012).
QDs have a large Stoke’s shift and broad excitation into the UV, allowing for simultaneous excitation of distinct QD species.
QDs have narrow emission spectra that fit to a Gaussian profile, making spectral unmixing more straightforward.
While peak emission rate of QDs is less than that of organic fluorophores (QDs have a longer fluorescence lifetime), their high extinction coefficient (ε) and quantum yield (QY) result in higher brightness (ε*QY).
New fluorescent proteins demonstrate improved brightness (Shaner et al 2013).
QDs have a range of conjugation schemes; however it is difficult to achieve a truly monovalent coupling.
While fluorescent proteins are the only directly genetically expressible fluorophores, a number of small genetically expressible peptides can be used to target QDs and organic dyes (Regoes and Hehl 2005; Jacquier et al 2006; Szent-Gyorgyi et al 2008).
The QD core is inherently electron dense, but some organic fluorophores can provide contrast in EM (Shu et al 2011).
QDs have special requirements for fixation (PFA must be used, avoid MeOH or cold fixation) and mounting medium (some mounting media leads to QD signal degradation, nonpolar organic solvent based reagents are recommended, see Table 3).
QDs demonstrate intermittent fluorescence. Groups are working on the generation of non-blinking QDs (Ghosh et al 2012). Typically considered a disadvantage in single molecule imaging, though the blinking properties have been used in super-resolution (Lidke et al 2005; Lagerholm et al 2006; Dertinger et al 2009).
QD probes can be used in techniques that cover all spatiotemporal scales for biological imaging.
Photobleaching of organic fluorophores makes longer-term imaging difficult.
QDs are available in near IR wavelengths with a high two-photon cross-section and the potential for use as a theranostic, but there are concerns about toxicity.
Bright and stable near-infrared fluorescent proteins are being developed (Filonov et al 2011).
QD-enabled studies of single molecule behavior in living cells
Elucidating complex biological phenomena requires approaches that reveal the dynamic behaviors and organization of molecules in living systems. QD single particle tracking (QD-SPT) represents a powerful method for probing the dynamics of these individual proteins of interest in living cells with high spatial and temporal resolution. This capability is afforded by their high photostability and brightness that is superior to conventional fluorophores (fluorescent proteins and organic dyes). These advantageous properties overcome difficulties in photobleaching that limit fluorophore imaging time allowing for acquisition of biological events over long timescales and contribute to the QD’s utility as an ultrasensitive detection probe for SPT. Moreover, QD-SPT generates quantifiable dynamic information regarding diffusional properties, co-localization, and spatial and temporal heterogeneity of molecules inside living cells that conventional fluorescence and biochemical methods cannot capture (Courty et al 2006a; Cognet et al 2014; Breger et al 2014).
The method of QD-SPT proceeds through multiple steps. Briefly, the first step involves generating a QD probe targeting the molecule of interest. A number of strategies are available for targeting QDs to bio-molecules of interest in living cells (Medintz et al 2005; Petryayeva et al 2013). Second, once the QD probe is generated, a series of validations must be conducted to confirm that the QD probe binds with specificity to its cellular target and its function is not sterically hindered by QD size. Specificity of binding should be validated by comparing cellular labeling of QDs with and without components necessary for binding, e.g. streptavidin-QDs alone versus streptavidin-QDs coupled to the biotinylated targeting molecule. Methods for validating retention of biological function include comparing QD-labeled versus fluorescent dye-labeled (Cy3, Alexa dyes, fluorogen-activating proteins) targets and/or gold particle probes to ensure similar diffusion properties (Dahan et al 2003; Bannai et al 2006; Groc et al 2007; Schwartz et al 2014), measurement of protein signaling activity with QDs tagged to either ligand or protein (Lidke et al 2004; Andrews et al 2008) measurement of cellular activity such as cell outgrowth and survival with and without QDs (Cui et al 2007; Vermehren-Schmaedick et al 2014), and comparison of internalization kinetics in receptors pre-labeled with QDs to receptors post-labeled with QDs following fixation (Fichter et al 2010). Following labeling and validation of the QD probe, a sequence of fluorescence images are acquired in time lapse to capture the biological event. The temporal resolution achieved through this acquisition will usually be limited by the camera readout time rather than by the brightness of the probe (Courty et al 2006a). Finally, biological information is extracted from the recorded trajectories through single particle tracking to yield measurements such as diffusion coefficient and velocity that provide dynamic information about the molecule of interest (Bannai et al 2006). A variety of commercial (IDL - Research Systems, Boulder, CO), customized, and open source software (ImageJ plugins such as Particle Tracker, Manual Tracking) are available for conducting single QD tracking. Subsequent biophysical analyses of the data include computing molecular dynamic information of free and confined diffusive processes (Dahan et al 2003; Lidke et al 2005a; Bannai et al 2006; Crane et al 2008; Chang et al 2012), molecular state changes such as dimerization dynamics including dimer status, rates of dimerization, and dimer diffusivity (Chung et al 2010; Low-Nam et al 2011), and receptor protein trafficking dynamics (Pierobon et al 2009; Valentine et al 2012). Through the capabilities afforded by the sensitivity of QD-SPT, many groups have exploited the potential of QDs to unravel complex biological processes previously not possible. We highlight examples demonstrating recent work in the use of QD-SPT for tracking extracellular and intracellular protein targets.
QDs have served as a powerful tool for single particle tracking of membrane and cytoplasmic targets in a number of live cell studies (Figure 2, Table 2). The accessibility of membrane targets for QD labeling makes them ideal targets for QD tracking. Since the initial papers demonstrating QD tracking of membrane receptors (Dahan et al 2003; Lidke et al 2004), a wide variety of membrane proteins have been studied using live cell QD single particle tracking. These include dissecting the dynamic behavior of membrane bound targets such as receptors, channels, transporters, and membrane components. The first single QD tracking experiments (Dahan et al 2003) revealed the lateral diffusion of glycine receptors in living neurons for up to 20 minutes and showed that these diffusion behaviors varied in different synaptic domains. Here we describe recent examples of the use of QDs for understanding the dynamic properties of other membrane bound components such as membrane receptors, lipid raft constituents, and water channels (Crane et al 2008) in a variety of neuronal and epithelial cell systems (see Table 2). These include studies of the cystic fibrosis transmembrane conductance regulator (CFTR) that revealed the previously uncharacterized immobile behavior due to C-terminal PDZ interactions (Haggie et al 2006) and tracking of the aquaporin water channels that demonstrated non-anomalous diffusion (Crane and Verkman 2008). Chang et al studied the diffusion properties of lipid rafts by attaching QDs to the lipid raft constituent GM1 ganglioside. They found that lateral confinement persisted on similar time scales to signaling of raft-associated proteins, offering support for the role of lipid rafts as a possible signaling platform (Chang and Rosenthal 2012). In another study, the same group employed the use of antagonist-conjugated QDs for the study of serotonin transporter and found that populations of the transporter residing in cholesterol and ganglioside GM1-enriched microdomains displayed restricted mobility in comparison to the freely diffusing population of transporters not localized to these regions; these findings suggest a role for membrane microdomains in aspects of transporter regulation (Chang et al 2012). Altogether, these elegant studies serve to demonstrate the unique capability of QDs to explore the dynamic contributions of complex biological systems.
Figure 2. Single and multi-color QD Tracking of Extracellular and Intracellular Molecular Processes.
a) Single QD tracking reveals heterogeneous trafficking dynamics of BDNF-TrkB receptor transport in neurons. Left: receptor trajectory overlaid on wide field cell image (gray). Right: Magnified view of trajectory showing region of confinement. Images from Vermehren-Schmaedick et al. 2014; reprinted, with permission, from the investigators. b) Multicolor QD tracking of EGF receptors using hyperspectral microscopy allows for tracking of up to eight spectrally distinct QDs. Left: trajectories of selected QDs overlaid on hyperspectral image. Right: Particle trajectories plotted over time corresponding to the boxes region on the left. Color map (right) indicates QD emission peak. Images from Cutler et al. 2013; reprinted, with permission, from the investigators.
Table 2.
Example targets for QD-SPT
| SINGLE-COLOR | |||||
|---|---|---|---|---|---|
| Target | Novel Findings | Cell | Bioconjugation | Delivery | Location |
| CFTR (Haggie et al 2006) |
|
Epithelial (MDCK, COS7, HT29) | Anti-HA IgG, biotin-Fab, streptavidin-QD | NA | Extracellular |
| Ganglioside GM1 (lipid raft constitutent) (Chang and Rosenthal 2012) |
|
Serotonergic neurons (RN46A) | Biotin-cholera toxin B subunit (binds to GM1), streptavidin-QD | NA | Extracellular |
| AQP4 (Crane et al 2008) |
|
Primary astrocytes from mice Epithelial (MDCK, CHO-K1, COS-7) | Anti-c-myc IgG, IgG-QD | NA | Extracellular |
| EGFR (Chung et al 2010) |
|
Epithelial (CHO-K1) | EGFR-Fab-QD | NA | Extracellular |
| EGFR (Lidke et al 2004; Lidke et al 2005) |
|
Epithelial (CHO, A431, HeLa, MCF7) | Biotin-EGF, streptavidin-QD | Endocytosis (facilitated) | Extracellular and Intracellular |
| EGFR (Li et al 2012) |
|
Epithelial (A549) | Biotin-EGF, streptavidin-QD | Endocytosis (facilitated) | Extracellular and Intracellular |
| EGFR to observe endosomal trafficking (Zajac et al 2013) |
|
Epithelial (Arpe-19) | Biotin-EGF, streptavidin-QD | Endocytosis (facilitated) | Extracellular and Intracellular |
| WGA (Liu et al 2011) |
|
Epithelial (A549) | Biotin-WGA, streptavidin-QD | Endocytosis (facilitated) | Extracellular and Intracellular |
| Influenza Virus (Liu et al 2012) |
|
Epithelial (MDCK) | Biotin-virus, streptavidin-QD | Endocytosis (facilitated) | Extracellular and Intracellular |
| TrkA (Rajan et al 2008) |
|
Neuronal (PC12) | Biotin-NGF, streptavidin-QD | Endocytosis (facilitated) | Extracellular and Intracellular |
| TrkB (Vermehren-Schmaedick et al 2014) |
|
Primary nodose ganglion sensory neurons | Biotin-BDNF, streptavidin-QD | Endocytosis (facilitated) | Extracellular and Intracellular |
| IBB domain of snurportin-1 (to study transport across nuclear pore complex) (Lowe et al 2010) |
|
Epithelial (HeLa) | QDs functionalized to IBB z-domain through maleimide and sulphydryl reaction | Digitonin permeabilization (active) | Intracellular |
| Myosin and Kinesin (Nan et al 2005) |
|
Epithelial (A549) | Biotin-peptide, streptavidin-QD | Endocytosis (passive) | Intracellular |
| Cytosol (no specific target) (Keren et al 2009) |
|
Fibroblasts (keratocytes) | None | Electroporation (active) | Intracellular |
| MULTI-COLOR | ||||||
|---|---|---|---|---|---|---|
| Target | # Colors | Novel Findings | Cell | Bioconjugation | Delivery | Location |
| EGFR Liganded/unliganded (Low-Nam et al 2011) | 2 |
|
Epithelial (A431) | Biotin-EGF, streptavidin-QD Biotin-VHH, streptavidin-QD |
NA | Extracellular |
| GM1/CD59/EGFR (Clausen et al 2014) | Up to 4 |
|
Fibroblast (MEF) | QD-Cholera toxin to GM1 QD-CoA to ACP-tagged CD59 SAV-QD to biotynilated EGFR (BLAP-tag) |
NA | Extracellular |
| Fc RI/EGFR (Cutler et al 2013) | Up to 8 |
|
Mast cells (RBL) | Biotin-IgE, streptavidin-QD Biotin-EGF, streptavidin-QD |
NA | Extracellular |
New studies have also focused on tracking membrane protein trafficking, or the movement of specific proteins from the extracellular surface to intracellular environment in cells. Two areas of research include QD tracking of epidermal growth factor receptors (EGFR) and neuronal growth factor receptors. In neuronal systems, QDs have been most readily utilized in the study of binding and transport of neuronal receptor complexes such as nerve growth factor (NGF)-TrkA and brain-derived neurotrophic factor (BDNF)-TrkB (Figure 2a) (Cui et al 2007; Rajan et al 2008; Vermehren-Schmaedick et al 2014). In EGF receptor systems, Lidke et al demonstrated the first instance of the use of QDs for studying EGFR endosomal trafficking and found that EGFR is trafficked to the cell body along filopodia in a retrograde manner (Lidke et al 2004; Lidke et al 2005a). Since this study, a number of other groups have exploited the use of QDs for exploring other aspects of EGFR dynamic behavior including drug effects on EGFR endosomal trafficking duration (Li et al 2012), motility of early endosomes transporting EGFR as cargo (Zajac et al 2013), and dimerization activity of liganded and unliganded EGFR in different spatial contexts (Chung et al 2010). QD-enabled studies of NGF axonal transport using biotin-NGF and streptavidin-QDs has been able to uncover previously unrecognized “stop-and-go” motions of NGF during retrograde transport (Cui et al 2007), as well as to study the diffusive and active transport dynamics of NGF endosomal trafficking (Rajan et al 2008). Similar use of BDNF-QDs enabled study of the cytoplasmic trafficking dynamics of BDNF-TrkB endosomes and revealed that BDNF undergoes a combination of rapid and directed motions, interspersed with circuitous meanderings in the cell body that are characterized by mobile and immobile phases (Vermehren-Schmaedick et al 2014). The use of QDs for tracking biomolecules that move from the cell surface into the cell cytoplasm also includes non-receptor targets such as viral particles (Liu et al 2012), and lectins (Liu et al 2011a). The application of QDs will likely continue to further expand in the near future to other extracellular and extracellular/intracellular targets.
While QDs have experienced a growth in their application for the study of extracellular targets, the use of QDs for the study of intracellular targets remains an area of slower growth due to the technical challenge of QD delivery into the cell interior. Despite these challenges, several groups have demonstrated successful targeting of QDs to intracellular targets. Studies have used non-specific membrane fusion to deliver endocytosed QDs into the cell interior. For example, in early work, Nan et al used tracked motor proteins transporting QD-endocytic cargoes to measure with high spatial resolution the 8 nm steps taken by microtubule motors in the plus- and minus-end directions (Nan et al 2005). Using a different technique to deliver QDs directly into the cytosol and bypass endocytic routes, Courty et al tracked QD-conjugated kinesin, enabling single molecule characterization of the dynamics of individual intracellular kinesin (Courty et al 2006b). QDs have also been used to probe exclusively intracellular events such as transport of QD-cargoes through the nuclear pore complexes (Lowe et al 2010), fluid flow in lamellipodia through non-specifically delivered QDs (Keren et al 2009), and diffusion of QD-mRNA in interchromatin regions (Ishihama and Funatsu 2009).
These and other studies have adopted a variety of strategies for introducing QDs into the cytosol, these include: (i) passive delivery, (ii) facilitated delivery, and (iii) active delivery of QDs into the cytosolic environment. Passive delivery of QDs occurs through induction of endocytic uptake through the inherent physical properties (surface coating and charge) of QDs (Nan et al 2005). Facilitated delivery of QDs occurs through association of the QD with a peptide or protein to promote intracellular uptake (Derfus et al 2004; Hild et al 2008; Delehanty et al 2009), or through use of methods such as pinocytosis (Courty et al 2006b), and transfection reagents (Xu et al 2013). Recent application of active delivery of QDs into the cellular environment include methods such as electroporation (Keren et al 2009), microinjection (Xu et al 2013), as well as novel methods such as photothermal nanoblade delivery (Xu et al 2012). While successful cytosolic delivery of QDs has been demonstrated, ensuring QD nonspecific binding in the absence of functional modification in these systems still remains a challenge. Attempts to demonstrate specific binding and functional integrity include comparing kinetics and behaviors of delivered QD-conjugates that are functionally versus non-functionally competent; for example, intact kinesin-QD conjugates versus denatured kinesin-QD conjugates (Courty et al 2006b) and QD conjugates with and without domains necessary for nuclear import (Lowe et al 2010). While strides have been made in strategies for cytosolic delivery of QDs, continued development of targeting strategies that improve delivery efficiency and specificity while retaining molecular function is necessary for exploring in greater detail the complex biological processes that occur inside cells.
Multi-color single QD tracking
Often in biological studies it is valuable to visualize protein behavior with respect to other proteins or their environment. The large Stokes shift of QDs makes them a good choice when imaging simultaneously blue shifted organic dyes or GFP. For example, using two-color TIRF microscopy, the motion of individual QD-labeled IgE receptors was tracked with respect to the landscape of the membrane proximal actin bundles, labeled by GFP-actin (Andrews et al 2008). Simultaneous imaging of the receptor and actin provided direct proof of the ability of actin to restrict membrane protein motion. Consistent with the actin corral hypothesis, the long QD tracks allowed for characterization of IgE receptor mobility when near actin and showed that the receptor is deflected by the actin boundary.
Due to their broad absorption spectra and narrow emission spectra, QDs are also ideally suited for multi-color imaging at the single molecule level. Two-color single QD tracking is relatively easy to achieve using a beam-splitter to separate the emission into separate spectral channels. Simultaneous imaging of distinctly tagged proteins allows for visualization of protein-protein interactions at the single molecule level. For example, homodimerization of the EGFR, erbB2 (HER2) and erbB3 (HER3) have been captured and quantified on live cells (Lidke et al 2005a; Low-Nam et al 2011; Steinkamp et al 2014). Labeling different protein species with spectrally distinct QDs also allows for the direct comparison of protein diffusion or capturing of heterodimer interactions (Low-Nam et al 2011; Steinkamp et al 2014). You et al have used pair correlation of dual-color imaging experiments to monitor QD-labeled IFNa2 binding to its receptor, IFNAR2, and the recruitment of STAT2 to IFNAR2 (You et al 2014). Torreno-Pina et al have used two-color single QD-tracking as part of a study to examine the role of glycans in micropatterning of the plasma membrane. By comparing mobility and protein interactions between DC-SIGN and a mutant that is not glycosylated, they found that glycan-mediated interactions did not lead to higher order clustering. However, glycosylated DC-SIGN exhibited more restricted mobility and these glycan-based interactions are important in regulating DC-SIGN interactions with clathrin (Torreno-Pina et al 2014).
Typically, experiments focus on two-color tracking due to technical limitations of using beam-splitters to separate the emission light into independent channels. Recently, several groups have developed methods for higher multiplexing capabilities. Lagerholm and colleagues have described a four-color beamsplitter approach that allows for the simultaneous tracking of four distinct QD species (Arnspang et al 2012). Recently, Clausen et al used this technology to simultaneously track three independent membrane components – CD59 (a GPI anchored protein), EGFR (a transmembrane protein) and GM1 (a lipid). Tracking of each species demonstrated that while each has a distinct diffusive behavior, the mobility of each is reduced in response to cholesterol depletion (Clausen et al 2014).
Keith Lidke and colleagues have developed a high-speed hyperspectral microscope that is not limited by filter-based detection, but acquires the full spectra of the sample in every pixel (Cutler et al 2013). This line-scanning confocal instrument allows for simultaneous single QD tracking of up to eight spectrally distinct QDs at 30 frames/sec (Figure 2b). The ability to increase the labeling up to eight colors allows for higher density labeling, increasing the probability of capturing protein-protein interactions and allowing high spatiotemporal resolution in diffusion maps (Cutler et al 2013). Since the confocal entrance slit to the spectrometer provides optical sectioning, tracking of membrane protein motion on the apical cell surface is easily achieved.
Tracking single molecule motion in 3D
The advances in multiplex imaging have opened new avenues for investigation of membrane component behavior. However, these experiments are still realistically limited to a two-dimensional focal plane. A number of groups have developed instrumentation, based on a range of approaches, that allows for single particle tracking in three dimensions (Kao and Verkman 1994; Schütz et al 2001; Prabhat et al 2007; Watanabe et al 2007; Lessard et al 2007; Wells et al 2009; Welsher and Yang 2014). In several cases, the brightness and photostability of QDs have been critical to the successful application of 3D tracking. Ober and colleagues have designed an instrument that allows for simultaneous imaging of multiple focal planes in the sample (Prabhat et al 2007). Using this microscope, they have monitored protein endocytosis, recycling and exocytosis in real-time (Prabhat et al 2007; Ram et al 2008). By following the endocytic trafficking of QD-labeled transferrin, they discovered the intercellular transfer of cargo between adjacent cells (Ram et al 2012). Izeddin et al used adaptive optics and point-spread function (PSF) engineering to track QD-labeled platelet-derived growth factor (PDGF) receptor motion in 3D. With this approach, they achieve 15 nm z-precision and captured the 3D landscape of the plasma membrane (Izeddin et al 2012). Werner and colleagues have developed a 3D tracking microscope that uses quad-APD detectors to monitor the emission of a single QD probe and track its motion in x, y, and z by moving the microscope stage to always keep the QD centered on the detector (Lessard et al 2007; Wells et al 2009). This instrument was used to capture endocytosis dynamics of the activated IgE receptor (Wells et al 2009), as the receptor underwent large (>1 m) changes in z position when trafficking from the plasma membrane to the cytosol. Recently, the group has collaborated with Hollingsworth group to bioconjugate their giant, non-blinking QDs (Ghosh et al 2012). The stable emission from these QDs allowed for much longer-term tracking of the IgE receptor motion in 3D on the cell surface (Keller et al 2014). Welsher et al have combined 3D tracking with two-photon microscopy, which allows for simultaneous imaging of cellular structure while tracking single molecules. With this approach, they were able to capture the real-time binding of individual Tat-coated QD nanoparticles to the cell surface (Welsher and Yang 2014).
Single QD tracking in whole animals
QDs have been used in in vivo animal models as imaging agents, enabling the high contrast visualization of the vasculature and lymph nodes, and as nanoparticle platforms to track the pharmaco/biodistribution of drugs and other biomolecules (Larson et al 2003; Kim et al 2004; Michalet et al 2005; Diagaradjane et al 2008; Jung et al 2011). An emerging area is the single particle tracking of QDs in in vivo animal models. Among the first studies of single QD tracking in vivo, Tada et al conducted a study that demonstrated the real-time tracking of single QDs in live in vivo animal preparations (Tada et al 2007). They imaged the movement of QDs that were conjugated to the antibody drug Hercep tin in mice containing tumors with overexpressed HER2 breast tumors. By using a confocal microscope with dorsal skinfold chamber, they observed the movement of QDs from the blood into and within the tumor, and were able to quantitate information such as velocity, direction and modes of transport. A study by Hamada et al has used the counting of single QD-VEGF probes in movies made in the blood vessels of ischemic mouse models undergoing angiogenesis (Hamada et al 2011) to examine the molecular distribution of vascular endothelial growth factor (VEGF) with high spatial resolution. Another study that exemplifies the use of QDs in high-resolution measurements of in vivo biological processes has been the use of QDs to track the motion of the protease-activated receptor 1 (PAR1) on the surface of tumor cells in order to study the membrane dynamics at high spatial resolution (~ 8 nm) during the process of extravasation (Gonda et al 2010). As well, QDs have been used to measure in real-time the length changes in the sarcomeres of myocytes in vivo by infusing QD solution over intact myocytes and allowing their internalization via membrane fusion (Serizawa et al 2011). Such studies, conducted in in vivo preparations are valuable in that one can look at biological processes that occur on the resolution scale of tens of nanometers in the physiologically relevant context of the larger scale biological system.
QDs for multiplex IHC
In addition to advancing single molecule and single cell imaging, QDs are being used to improve the multiplexing capabilities and sensitivity of immunohistochemistry (IHC). IHC is a well-established method of diagnosis in surgical pathology that allows for in situ identification of characteristic antigens indicative of cell type and origin as well as disease state. Typically, IHC of pathological samples involves the labeling of paraformaldehyde fixed, paraffin embedded tissue with antibodies to specific targets. The extent of antibody labeling is traditionally detected by enzyme-substrate based chromogenic reporters using a transmission light microscope. Fluorescence is becoming an attractive alternative to chromogenic detection in IHC. Specifically, fluorescence has the advantage when it comes to quantification since the amount of labeling scales linearly with fluorescence intensity, whereas the enzyme-based deposition is dependent on parameters such as time of incubation, temperature, and concentration of the substrate. Additionally, analysis of chromogenic substrates is only semi-quantitative, typically utilizing an H-score or other qualitative analysis (Barrow et al 2011; Gonda et al 2012).
In the early 2000’s, QD-based IHC (QD-IHC) protocols were developed and have since been applied to a range of tissues (Sun et al 2001; Zahavy et al 2005; True and Gao 2007; Byers and Hitchman 2011). The unique properties of QDs (Table 1), provide enhance capabilities for sensitivity, multiplexing and quantification. These allow for more detailed analysis of tissue structure, cellular localization, relative amounts of antigens, and colocalization as well as allowing for sparing use of limited tissue samples, such as in biopsy tissue (Akhtar et al 2007; Caldwell et al 2008; Yu et al 2013).
The QD-IHC protocol has come a long way from its debut in 2001 and many publications have examined the special considerations for fluorescence and QD label usage (Table 3). In addition to the considerations for traditional IHC, such as sample care and antigen retrieval, Xing et al and Monton et al provide a particularly rigorous analysis of reagents that may alter fluorescent signals as well as recommended conjugation and multiplexing methods (Xing et al 2007; Montón et al 2012). Additionally, key reviews have chronicled the progress of QD-IHC methodologies (Byers and Hitchman 2011; Chen et al 2012; Fang et al 2012; Kairdolf et al 2013).
Table 3.
Key differences in QD-immunohistochemistry protocol.
| IHC Workflow | QD-IHC Specific |
|---|---|
| Sample Preparation | Works well with fresh, frozen or formalin-fixed, paraffin embedded samples |
| Deparaffinization/Rehydration | Toluene recommended (replacing xylene*) |
| Decloaking of antigens | Same as traditional temperature, pressure, pH and time of cycles is crucial to retrieval of epitopes |
| Blocking and Antibody incubation | Hydrophilic barrier pens such as ImmEdge by Vector* (Xing et al 2007) |
| Dehydration and mounting | Toluene recommended (replacing xylene*) - both for rinses and mounting media |
| Microscopy | Spectral camera and software recommended (e.g. Nuance) |
| Analysis | Spectral unmixing; generation of spectral fingerprints or comparisons of label intensity |
An important underlying theme in the difference between traditional and QD-IHC protocols is to utilize solvents such as toluene, chloroform and hexane to prevent interaction with QD surface chemistry that may lead to quenching.
Sensitivity is an important characteristic of IHC for its traditional applications in diagnostic medicine, such as for detection of receptor targets before initiation of therapies including those targeting the estrogen receptor (ER) (Hammond et al 2010; Gonda et al 2012). There have been many studies validating detection by QDs in IHC (Chen et al 2009; Xu et al 2012; Tabatabaei-Panah et al 2013; Yu et al 2013). The recent key studies that have demonstrated QD-IHC’s statistical and functional advantages over other forms of IHC include work in which QD-IHC was found to have 5% better sensitivity and 10% better specificity over traditional IHC when examining the Tn antigen in breast cancer tissues (Au et al 2014) and demonstrations that QD-IHC detection of the proliferation marker Ki67 was able to more accurately predict 5-year disease free survival in breast cancer patients (Sun et al 2014). Additionally, QD-IHC is amenable to coupling with signal amplifying techniques such as tyramide signal amplification (TSA) to further enhance detection of subtle antigenic signal (Akhtar et al 2007).
Multiplex QD-IHC has allowed for new insights into spatial organization and biomarker relationships in disease processes. Since the advent of multi-color QD-IHC in 2005 (Zahavy et al 2005), studies have examined colocalization of biomarkers (Storch et al 2007), tissue and microenvironment heterogeneity and co-evolution (Chen et al 2010; Liu et al 2010a; Liu et al 2010b; Faratian et al 2011), and structural changes, such as invasion (Xing et al 2007; Peng et al 2011; Liu et al 2011b; Fang et al 2012). An example of multiplex QD-IHC in tissue is shown in Figure 3. Liu et al targeted multiple antigens implicated in prostate cancer to create a signal map of patient tissues that cumulatively indicated architectural abrogation and demonstrated the potential of QD-IHC to reconstruct malignant transformation of a heterogeneous tissue (Liu et al 2010a). Peng et al demonstrated patterns of spatial and temporal coevolution of gastric and breast cancer cells and their surrounding stroma by following markers of basement membrane integrity or breakdown, angiogenesis, and macrophage invasion (Peng et al 2011). Additionally, QD-IHC has been combined with other techniques such as QD in situ hybridization (QD-ISH) to examine RNA transcripts and provide additional spatial information (Matsuno et al 2005; Matsuno et al 2006).
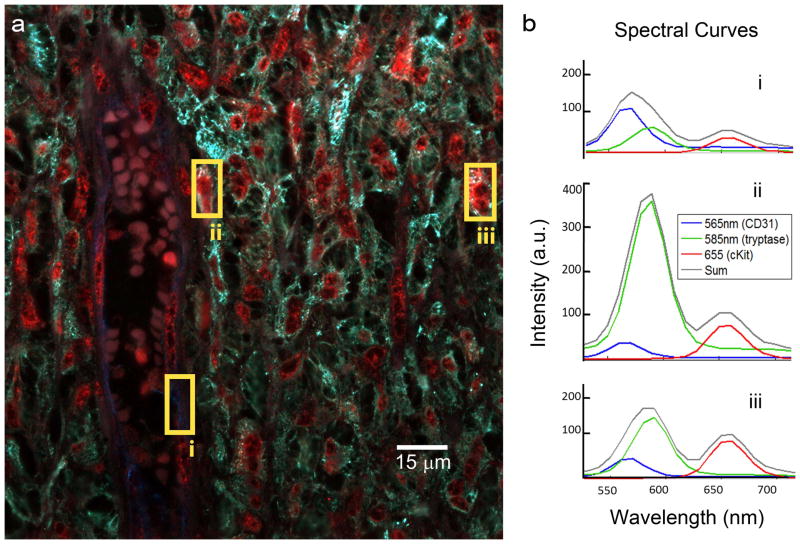
Figure 3. Multiplex QD-immunohistochemistry.
a) QD-IHC performed on human spleen tissue of a patient with aggressive systemic mastocytosis (tissue courtesy of Tracy George, MD). Staining includes anti-CD31 (QD565, blue), anti-tryptase (QD585, green) and anti-cKit (QD655, red), with autofluorescence represented as white. Visible in the image is a CD31+ splenic capillary (blue) filled with red blood cells; surrounding it is an aggregate of mast cells which are positive for both tryptase and cKit biomarkers. b) Spectral curves of the corresponding boxes in A. The splenic capillary is high in QD565 signal that labels CD31 (i) and mast cells show high labeling of QD585-tryptase and QD655-cKit (ii). Images acquired by E.W. Hatch using Nuance spectral camera, spectral unmixing performed using custom software.
Despite the advantages of QD-IHC for achieving quantitative information, to date, only relative quantification has been implemented in practice through various means such as spectral image acquisition, ROI assignment and signal discretization, and the subsequent quantification of intensity values. Hardware and software are major components dictating the quality of data collected from QD-IHC. While hardware is beyond the scope of this review it is worth mentioning the state of software options. Many packages are available to separate fluorescence signatures in spectral images; the most published is the commercially available Nuance software (PerkinElmer, MA, USA) and spectral capabilities of ZEN software (Carl Zeiss Microimaging GmbH). Many groups additionally are writing their own unmixing protocols, which are similarly based on spectral separation, such as via a Gaussian mixture model, defining and thresholding a masked region of interest, and quantifying intensity based on maxima and total area. Examples include WuDa Image Analysis System (Wuhan University) (Lv et al 2013), inForm (Caliper Life Sciences, Hopkinton, MA) and the open source wares FARSIGHT and Q-IHC (www.miblab.org) (Xing et al 2007; Yu et al 2013). Analysis may expand on this relative quantification by providing a comparison between signals, where one common housekeeping antigen may be used as an internal control to draw conclusions about relative quantity of biomarkers. Common housekeeping targets include nuclear stains and proteins intimately involved in normal cellular processes, such as elongation factor 1 alpha (EF1α) (Xing et al 2007). Quantitative multiplex QD-IHC is an area of open opportunity by which to exploit the unique properties of QD photostability, multiplexed emission, and bright intensity to enable improved tissue diagnostics.
Progress in QD bioconjugation paradigms
QDs must be bioconjugated with specific biomolecules (e.g. proteins, DNA, ligands, drugs) to conduct intended biological applications such as imaging, delivery, and biosensing (Medintz et al 2005; Petryayeva et al 2013). Typically, QD bioconjugation entails (Medintz et al 2005; Petryayeva et al 2013). QD bioconjugation entails the attachment of biomolecules to amphiphilic polymers that are assembled at the QD surface and serve to render the QD water-soluble (Medintz et al 2005). QD bioconjugation methodologies that are currently used to generate functionalized, targeted probes have been derived from standard protein labeling chemistries and have been described in recent reviews (Petryayeva et al 2013; Blanco-Canosa et al 2014). Covalent chemistries are the preferred method of choice for use in cellular studies due to the more stable nature of these reactions. Covalent chemistries include carboiimide chemistries, which offer cheap and fast covalent binding to amine and carboxyl groups of the biomolecule of interest (Hermanson 2013). A disadvantage of these chemistries, however, is a lack of precise control over orientation and stoichiometry of cross-linked biomolecules at the QD surface. Furthermore, aggregation is often a concern. Non-covalent biotin-streptavidin interactions are likely the most widely used for cellular-QD applications. This is due to the versatility of biotin-streptavidin bonds, which offer high affinity, wide pH and salt stability (Hermanson 2013), as well as practical ease of use and some quantitative control of biomolecular valency at the QD surface. Biotinylation kits and biotinylated biomolecules are available from a wide range of suppliers, and streptavidin-QDs are commercially available and can be easily paired with a protein of interest. Other chemistries successfully used include self-assembly by histidine-metal affinity; this involves a His-appended biomolecule that interacts with the inorganic ZnS shell of the QD (Blanco-Canosa et al 2014).
Despite the availability of methods in use, new bioconjugation chemistries will still be needed to continue to generate QD bioconjugates with improved precise control and specific functionality necessary to achieve specific biological tasks (Zrazhevskiy et al 2010). Alternative bioconjugation schemes that produce repeatable product, control the valency of the biomolecule on the QD surface, as well as produce a desired orientation of the biomolecule on the QD surface and do not compromise the function of the biomolecule still await further developments which could provide a greater impetus for the expansion and establishment of QD applications in biology and medicine. Along these lines, recent efforts include the work in improving QD probes for single protein tracking by producing monovalent QD bioconjugates by using peptide surface coatings and demonstration of new bioconjugation schemes employing hydrazide, aldehyde, and thiol-based linkages (You et al 2010; Clarke et al 2010; Iyer et al 2011). The development of new bioconjugation schemes will be powerful and necessary for achieving multifunctional tasks such as combined imaging, targeting and delivery (Zrazhevskiy et al 2010). Development of the use of specific types of QD bioconjugates along with their integration into novel methodologies will also be important for achieving multifunctional imaging, targeting and delivery tasks (Zrazhevskiy et al 2010). Recent work along these lines, include flexible, adaptable procedures designed to tap the full potential of multicolor QDs for tagging multiple targets in the same biological sample (Zrazhevskiy and Gao 2013; Zrazhevskiy et al 2013). Another emerging development is the use of ‘click’ chemistries which are appealing due to their rapidity, high yield, and ease of use at room temperature. Click chemistries originally employed copper(I)-catalyzed azide-alkyene reactions which are well-suited for high-selectivity because these groups do not interact with native biological functional groups (Kolb et al 2001). Click chemistries have been used to bioconjugate biomolecules to magnetic, gold, and other colloidal nanoparticles. Because copper may alter the luminescent properties of QDs, recently available copper-free bio-orthogonal approaches (Bernardin et al 2010; Han et al 2010; Schieber et al 2012) have made possible the use of click chemistries for bioconjugation of QDs which in the near future are likely to grow in application.
Potential areas for further development of QDs
A potential future opportunity for the application of QDs is in the exciting and growing area of correlated microscopy. Correlated electron and fluorescence microscopy (CLEM) offers the opportunity to visualize fluorescently-tagged proteins in their surrounding context at high-resolution in EM. The dual QD fluorescence and electron-dense properties make QDs advantageous for CLEM (Giepmans et al 2006; Sjollema et al 2012). QDs can be distinctly discriminated at the EM level, and the simultaneous labeling of multiple endogenous proteins has been demonstrated for CLEM in cells and tissue as well as the postsection labeling of nuclear proteins (Nisman et al 2004; Deerinck et al 2007). 3D EM reconstruction of QDs in corresponding fluorescence optical sections have been demonstrated to elucidate details of neurofibrillary tangles in Alzheimer’s disease in the study by Uematsu et al, who also employed energy-dispersive X-ray chemical analysis of the Cd and Se to confirm the presence of QDs in appropriate locations in the EM image (Uematsu et al 2012). A unique attribute of QDs for use in CLEM is the capability to distinguish in multiplexed fashion multiple QD probes in EM based on their differing size and shape and fluorescent color (Sosinsky et al 2007). As yet, the increased use of QDs in CLEM remains to be fully exploited. Another powerful use of QD correlative microscopy on the horizon is the combination of QD with other super-resolution fluorescence probes to bridge dynamic information of individual proteins in relation to their spatial organization in the cell. For example, information on the diffusive motion of glycine receptors using QD-SPT have been studied in the context of the glycine receptor subsynaptic distribution using PALM (Specht et al 2013).
QDs also possess properties that make them uniquely suited for super-resolution imaging. To achieve resolution beyond the diffraction limit, a number of super-resolution techniques rely on the ability to switch the fluorescence emission from emitters on and off. Lidke et al were the first to propose and demonstrate that blinking fluorophores could be used for super-resolution (Lidke et al 2005b). The authors used the natural blinking of QDs to independently localize individual emitters with separations less than the diffraction limit. In a similar manner, Lagerholm et al used blinking to localize QDs attached to the ends of double-stranded DNA that were separated by 42 nm (Lagerholm et al 2006). Hoyer et al have recently extended this idea to 3D super-resolution (Wang et al 2013). In 2009, Dertinger et al applied QDs for SOFI (Super-resolution Optical Fluctuating Imaging) where statistical analysis of the fluorescence fluctuations throughout a time series can achieve a super-resolution image (Dertinger et al 2009; Dertinger et al 2013). While photostability is one of the greatest advantages of QDs, it is possible to induce a blue-shift in their emission spectra (“blueing”) in the QD emission in response to high intensity illumination. Hoyer et al used this property to achieve resolution below the diffraction limit using a simple webcam for detection (Hoyer et al 2011).
Conclusions
From single molecules to single cells to tissue, QDs have provided unique quantitative data sets for better understanding of biological and disease processes. The ability of QDs to enhance bio-imaging has been enabled by the development of instrumentation that can take advantage of QD’s photophysical properties and sophisticated analysis routines to extract biological parameters from single molecule and spectral imaging data. Improvements in QD properties, such as reduced size, constant emission and monovalent conjugation will further increase their utility. Therefore, while QDs have already had a strong impact on biological imaging, it is likely that we have yet to realize the full potential of QDs for quantitative imaging.
Acknowledgments
This work was supported by NIH 1RO1NS071116, NIH 1R21NS073113 to TQV and the OHSU Neuroscience Imaging Center (P30-NS061800); NIH 1R01GM100114 and NSF MCB-0845062 to DSL, and the NM Spatiotemporal Modeling Center (NIH P50GM085273). EWH was supported by the NM Cancer Nanotechnology Training Grant. We thank Dr. Tracy George for collaboration in generation of the QD-IHC samples in Figures 1 and 3.
References
- Akhtar RS, Latham CB, Siniscalco D, et al. Immunohistochemical detection with quantum dots. Methods Mol Biol. 2007;374:11–28. doi: 10.1385/1-59745-369-2:11. [DOI] [PubMed] [Google Scholar]
- Altman RB, Terry DS, Zhou Z, et al. Cyanine fluorophore derivatives with enhanced photostability. Nat Methods. 2012;9:68–71. doi: 10.1038/nmeth.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NL, Lidke KA, Pfeiffer JR, et al. Actin restricts FcepsilonRI diffusion and facilitates antigen-induced receptor immobilization. Nat Cell Biol. 2008;10:955–63. doi: 10.1038/ncb1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnspang EC, Brewer JR, Lagerholm BC. Multi-color single particle tracking with quantum dots. PLoS One. 2012;7:e48521. doi: 10.1371/journal.pone.0048521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au GHT, Mejias L, Swami VK, et al. Quantitative assessment of Tn antigen in breast tissue micro-arrays using CdSe aqueous quantum dots. Biomaterials. 2014;35:2971–80. doi: 10.1016/j.biomaterials.2013.12.034. [DOI] [PubMed] [Google Scholar]
- Bannai H, Lévi S, Schweizer C, et al. Imaging the lateral diffusion of membrane molecules with quantum dots. Nat Protoc. 2006;1:2628–34. doi: 10.1038/nprot.2006.429. [DOI] [PubMed] [Google Scholar]
- Barrow E, Evans DG, McMahon R, et al. A comparative study of quantitative immunohistochemistry and quantum dot immunohistochemistry for mutation carrier identification in Lynch syndrome. J Clin Pathol. 2011;64:208–14. doi: 10.1136/jcp.2010.084418. [DOI] [PubMed] [Google Scholar]
- Bernardin A, Cazet A, Guyon L, et al. Copper-free click chemistry for highly luminescent quantum dot conjugates: application to in vivo metabolic imaging. Bioconjug Chem. 2010;21:583–8. doi: 10.1021/bc900564w. [DOI] [PubMed] [Google Scholar]
- Blanco-Canosa JB, Wu M, Susumu K, et al. Recent progress in the bioconjugation of quantum dots. Coord Chem Rev. 2014;263–264:101–137. doi: 10.1016/j.ccr.2013.08.030. [DOI] [Google Scholar]
- Breger J, Delehanty JB, Medintz IL. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2014. Continuing progress toward controlled intracellular delivery of semiconductor quantum dots. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchez M, Moronne M, Gin P, et al. Semiconductor Nanocrystals as Fluorescent Biological Labels. Science (80-) 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Byers RJ, Hitchman ER. Quantum dots brighten biological imaging. Prog Histochem Cytochem. 2011;45:201–37. doi: 10.1016/j.proghi.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Caldwell ML, Moffitt RA, Liu J, et al. Simple quantification of multiplexed quantum dot staining in clinical tissue samples. Conf Proc Annu Int Conf IEEE Eng Med Biol Soc IEEE Eng Med Biol Soc Annu Conf. 2008;2008:1907–10. doi: 10.1109/IEMBS.2008.4649559. [DOI] [PubMed] [Google Scholar]
- Chan W, Nie S. Quantum Dot Bioconjugates for Ultrasensitive Nonisotopic Detection. Science (80-) 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- Chang JC, Rosenthal SJ. Visualization of lipid raft membrane compartmentalization in living RN46A neuronal cells using single quantum dot tracking. ACS Chem Neurosci. 2012;3:737–43. doi: 10.1021/cn3000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, Tomlinson ID, Warnement MR, et al. Single molecule analysis of serotonin transporter regulation using antagonist-conjugated quantum dots reveals restricted, p38 MAPK-dependent mobilization underlying uptake activation. J Neurosci. 2012;32:8919–29. doi: 10.1523/JNEUROSCI.0048-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Peng J, Sun S-R, et al. Tapping the potential of quantum dots for personalized oncology: current status and future perspectives. Nanomedicine (Lond) 2012;7:411–28. doi: 10.2217/nnm.12.9. [DOI] [PubMed] [Google Scholar]
- Chen C, Peng J, Xia H, et al. Quantum-dot-based immunofluorescent imaging of HER2 and ER provides new insights into breast cancer heterogeneity. Nanotechnology. 2010;21:095101. doi: 10.1088/0957-4484/21/9/095101. [DOI] [PubMed] [Google Scholar]
- Chen C, Peng J, Xia H-S, et al. Quantum dots-based immunofluorescence technology for the quantitative determination of HER2 expression in breast cancer. Biomaterials. 2009;30:2912–8. doi: 10.1016/j.biomaterials.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Chung I, Akita R, Vandlen R, et al. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–7. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- Clarke S, Pinaud F, Beutel O, et al. Covalent monofunctionalization of peptide-coated quantum dots for single-molecule assays. Nano Lett. 2010;10:2147–54. doi: 10.1021/nl100825n. [DOI] [PubMed] [Google Scholar]
- Clausen MP, Arnspang EC, Ballou B, et al. Simultaneous multi-species tracking in live cells with quantum dot conjugates. PLoS One. 2014;9:e97671. doi: 10.1371/journal.pone.0097671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognet L, Leduc C, Lounis B. Advances in live-cell single-particle tracking and dynamic super-resolution imaging. Curr Opin Chem Biol. 2014;20C:78–85. doi: 10.1016/j.cbpa.2014.04.015. [DOI] [PubMed] [Google Scholar]
- Courty S, Bouzigues C, Luccardini C, et al. Tracking individual proteins in living cells using single quantum dot imaging. Methods Enzymol. 2006a;414:211–28. doi: 10.1016/S0076-6879(06)14012-4. [DOI] [PubMed] [Google Scholar]
- Courty S, Luccardini C, Bellaiche Y, et al. Tracking Individual Kinesin Motors in Living Cells Using Single Quantum-Dot Imaging. Nano Lett. 2006b;6:1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- Crane JM, Van Hoek AN, Skach WR, Verkman AS. Aquaporin-4 dynamics in orthogonal arrays in live cells visualized by quantum dot single particle tracking. Mol Biol Cell. 2008;19:3369–78. doi: 10.1091/mbc.E08-03-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane JM, Verkman AS. Long-range nonanomalous diffusion of quantum dot-labeled aquaporin-1 water channels in the cell plasma membrane. Biophys J. 2008;94:702–13. doi: 10.1529/biophysj.107.115121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Wu C, Chen L, et al. One at a time, live tracking of NGF axonal transport using quantum dots. Proc Natl Acad Sci U S A. 2007;104:13666–71. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler PJ, Malik MD, Liu S, et al. Multi-color quantum dot tracking using a high-speed hyperspectral line-scanning microscope. PLoS One. 2013;8:e64320. doi: 10.1371/journal.pone.0064320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan M, Lévi S, Luccardini C, et al. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302:442–5. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- Deerinck TJ, Giepmans BNG, Smarr BL, et al. Light and electron microscopic localization of multiple proteins using quantum dots. Methods Mol Biol. 2007;374:43–53. doi: 10.1385/1-59745-369-2:43. [DOI] [PubMed] [Google Scholar]
- Delehanty JB, Mattoussi H, Medintz IL. Delivering quantum dots into cells: strategies, progress and remaining issues. Anal Bioanal Chem. 2009;393:1091–105. doi: 10.1007/s00216-008-2410-4. [DOI] [PubMed] [Google Scholar]
- Derfus AM, Chan WCW, Bhatia SN. Intracellular Delivery of Quantum Dots for Live Cell Labeling and Organelle Tracking. Adv Mater. 2004;16:961–966. doi: 10.1002/adma.200306111. [DOI] [Google Scholar]
- Dertinger T, Colyer R, Iyer G, et al. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI) Proc Natl Acad Sci U S A. 2009;106:22287–92. doi: 10.1073/pnas.0907866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dertinger T, Pallaoro A, Braun G, et al. Advances in superresolution optical fluctuation imaging (SOFI) Q Rev Biophys. 2013;46:210–21. doi: 10.1017/S0033583513000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diagaradjane P, Orenstein-Cardona JM, Colón-Casasnovas NE, et al. Imaging epidermal growth factor receptor expression in vivo: pharmacokinetic and biodistribution characterization of a bioconjugated quantum dot nanoprobe. Clin Cancer Res. 2008;14:731–41. doi: 10.1158/1078-0432.CCR-07-1958. [DOI] [PubMed] [Google Scholar]
- Fang M, Peng C-W, Pang D-W, Li Y. Quantum dots for cancer research: current status, remaining issues, and future perspectives. Cancer Biol Med. 2012;9:151–63. doi: 10.7497/j.issn.2095-3941.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faratian D, Christiansen J, Gustavson M, et al. Heterogeneity mapping of protein expression in tumors using quantitative immunofluorescence. J Vis Exp. 2011:e3334. doi: 10.3791/3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichter KM, Flajolet M, Greengard P, Vu TQ. Kinetics of G-protein-coupled receptor endosomal trafficking pathways revealed by single quantum dots. Proc Natl Acad Sci U S A. 2010;107:18658–63. doi: 10.1073/pnas.1013763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov GS, Piatkevich KD, Ting L-M, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–61. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Cui Y, Levenson RM, et al. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–76. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- Ghosh Y, Mangum BD, Casson JL, et al. New insights into the complexities of shell growth and the strong influence of particle volume in nonblinking “giant” core/shell nanocrystal quantum dots. J Am Chem Soc. 2012;134:9634–43. doi: 10.1021/ja212032q. [DOI] [PubMed] [Google Scholar]
- Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–24. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Gonda K, Miyashita M, Watanabe M, et al. Development of a quantitative diagnostic method of estrogen receptor expression levels by immunohistochemistry using organic fluorescent material-assembled nanoparticles. Biochem Biophys Res Commun. 2012;426:409–14. doi: 10.1016/j.bbrc.2012.08.105. [DOI] [PubMed] [Google Scholar]
- Gonda K, Watanabe TM, Ohuchi N, Higuchi H. In vivo nano-imaging of membrane dynamics in metastatic tumor cells using quantum dots. J Biol Chem. 2010;285:2750–7. doi: 10.1074/jbc.M109.075374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groc L, Lafourcade M, Heine M, et al. Surface trafficking of neurotransmitter receptor: comparison between single-molecule/quantum dot strategies. J Neurosci. 2007;27:12433–7. doi: 10.1523/JNEUROSCI.3349-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggie PM, Kim JK, Lukacs GL, Verkman AS. Tracking of quantum dot-labeled CFTR shows near immobilization by C-terminal PDZ interactions. Mol Biol Cell. 2006;17:4937–45. doi: 10.1091/mbc.E06-08-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada Y, Gonda K, Takeda M, et al. In vivo imaging of the molecular distribution of the VEGF receptor during angiogenesis in a mouse model of ischemia. Blood. 2011;118:e93–e100. doi: 10.1182/blood-2010-12-322842. [DOI] [PubMed] [Google Scholar]
- Hammond MEH, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer (Unabridged Version) Arch Pathol Lab Med. 2010;134:e48–e72. doi: 10.5858/134.7.e48. [DOI] [PubMed] [Google Scholar]
- Han H-S, Devaraj NK, Lee J, et al. Development of a bioorthogonal and highly efficient conjugation method for quantum dots using tetrazine-norbornene cycloaddition. J Am Chem Soc. 2010;132:7838–9. doi: 10.1021/ja101677r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson GT. Bioconjugate Techniques. Academic Press; Amsterdam: 2013. [Google Scholar]
- Hild WA, Breunig M, Goepferich A. Quantum dots - nano-sized probes for the exploration of cellular and intracellular targeting. Eur J Pharm Biopharm. 2008;68:153–68. doi: 10.1016/j.ejpb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Hoyer P, Staudt T, Engelhardt J, Hell SW. Quantum dot blueing and blinking enables fluorescence nanoscopy. Nano Lett. 2011;11:245–50. doi: 10.1021/nl103639f. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Funatsu T. Single molecule tracking of quantum dot-labeled mRNAs in a cell nucleus. Biochem Biophys Res Commun. 2009;381:33–8. doi: 10.1016/j.bbrc.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Iyer G, Pinaud F, Xu J, et al. Aromatic aldehyde and hydrazine activated peptide coated quantum dots for easy bioconjugation and live cell imaging. Bioconjug Chem. 2011;22:1006–11. doi: 10.1021/bc100593m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeddin I, El Beheiry M, Andilla J, et al. PSF shaping using adaptive optics for three-dimensional single-molecule super-resolution imaging and tracking. Opt Express. 2012;20:4957–67. doi: 10.1364/OE.20.004957. [DOI] [PubMed] [Google Scholar]
- Jacquier V, Prummer M, Segura J-M, et al. Visualizing odorant receptor trafficking in living cells down to the single-molecule level. Proc Natl Acad Sci U S A. 2006;103:14325–30. doi: 10.1073/pnas.0603942103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K-H, Choe YS, Paik J-Y, Lee K-H. 99mTc-Hydrazinonicotinamide epidermal growth factor-polyethylene glycol-quantum dot imaging allows quantification of breast cancer epidermal growth factor receptor expression and monitors receptor downregulation in response to cetuximab therapy. J Nucl Med. 2011;52:1457–64. doi: 10.2967/jnumed.111.087619. [DOI] [PubMed] [Google Scholar]
- Kairdolf BA, Smith AM, Stokes TH, et al. Semiconductor quantum dots for bioimaging and biodiagnostic applications. Annu Rev Anal Chem (Palo Alto Calif) 2013;6:143–62. doi: 10.1146/annurev-anchem-060908-155136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HP, Verkman AS. Tracking of single fluorescent particles in three dimensions: use of cylindrical optics to encode particle position. Biophys J. 1994;67:1291–300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller AM, Ghosh Y, DeVore MS, et al. Live Cell Imaging: 3-Dimensional Tracking of Non-blinking “Giant” Quantum Dots in Live Cells (Adv. Funct. Mater. 30/2014) Adv Funct Mater. 2014;24:4795–4795. doi: 10.1002/adfm.201470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren K, Yam PT, Kinkhabwala A, et al. Intracellular fluid flow in rapidly moving cells. Nat Cell Biol. 2009;11:1219–24. doi: 10.1038/ncb1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lim YT, Soltesz EG, et al. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nat Biotechnol. 2004;22:93–7. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Lagerholm BC, Averett L, Weinreb GE, et al. Analysis method for measuring submicroscopic distances with blinking quantum dots. Biophys J. 2006;91:3050–60. doi: 10.1529/biophysj.105.079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zipfel WR, Williams RM, et al. Water-soluble quantum dots for multiphoton fluorescence imaging in vivo. Science (80-) 2003;300:1434–6. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- Lessard GA, Goodwin PM, Werner JH. Three-dimensional tracking of individual quantum dots. Appl Phys Lett. 2007;91:224106. doi: 10.1063/1.2819074. [DOI] [Google Scholar]
- Li H, Duan Z-W, Xie P, et al. Effects of paclitaxel on EGFR endocytic trafficking revealed using quantum dot tracking in single cells. PLoS One. 2012;7:e45465. doi: 10.1371/journal.pone.0045465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke DS, Lidke KA, Rieger B, et al. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J Cell Biol. 2005a;170:619–26. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke DS, Nagy P, Heintzmann R, et al. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- Lidke KA, Rieger B, Jovin TM, Heintzmann R. Superresolution by localization of quantum dots using blinking statistics. Opt Express. 2005b;13:7052. doi: 10.1364/OPEX.13.007052. [DOI] [PubMed] [Google Scholar]
- Liu J, Lau SK, Varma VA, et al. Molecular mapping of tumor heterogeneity on clinical tissue specimens with multiplexed quantum dots. ACS Nano. 2010a;4:2755–65. doi: 10.1021/nn100213v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lau SK, Varma VA, et al. Multiplexed detection and characterization of rare tumor cells in Hodgkin’s lymphoma with multicolor quantum dots. Anal Chem. 2010b;82:6237–43. doi: 10.1021/ac101065b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S-L, Zhang Z-L, Sun E-Z, et al. Visualizing the endocytic and exocytic processes of wheat germ agglutinin by quantum dot-based single-particle tracking. Biomaterials. 2011a;32:7616–7624. doi: 10.1016/j.biomaterials.2011.06.046. [DOI] [PubMed] [Google Scholar]
- Liu S-L, Zhang Z-L, Tian Z-Q, et al. Effectively and Efficiently Dissecting the Infection of Influenza Virus by Quantum-Dot-Based Single-Particle Tracking. ACS Nano. 2012;6:141–150. doi: 10.1021/nn2031353. [DOI] [PubMed] [Google Scholar]
- Liu X-L, Peng C-W, Chen C, et al. Quantum dots-based double-color imaging of HER2 positive breast cancer invasion. Biochem Biophys Res Commun. 2011b;409:577–82. doi: 10.1016/j.bbrc.2011.05.052. [DOI] [PubMed] [Google Scholar]
- Lowe AR, Siegel JJ, Kalab P, et al. Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking. Nature. 2010;467:600–3. doi: 10.1038/nature09285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Nam ST, Lidke KA, Cutler PJ, et al. ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat Struct Mol Biol. 2011;18:1244–9. doi: 10.1038/nsmb.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, Lei X, Ji M, et al. Clinical significance of EBP50 overexpression assessed by quantum dot analysis in gastric cancer. Oncol Lett. 2013;5:1844–1848. doi: 10.3892/ol.2013.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno A, Itoh J, Takekoshi S, et al. Three-dimensional imaging of the intracellular localization of growth hormone and prolactin and their mRNA using nanocrystal (Quantum dot) and confocal laser scanning microscopy techniques. J Histochem Cytochem. 2005;53:833–8. doi: 10.1369/jhc.4A6577.2005. [DOI] [PubMed] [Google Scholar]
- Matsuno A, Mizutani A, Takekoshi S, et al. Analyses of the mechanism of intracellular transport and secretion of pituitary hormone, with an insight of the subcellular localization of pituitary hormone and its mRNA. Brain Tumor Pathol. 2006;23:1–5. doi: 10.1007/s10014-005-0189-y. [DOI] [PubMed] [Google Scholar]
- Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4:435–46. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- Michalet X, Pinaud FF, Bentolila LA, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science (80-) 2005;307:538–44. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montón H, Roldán M, Merkoçi A, et al. The use of quantum dots for immunochemistry applications. Methods Mol Biol. 2012;906:185–92. doi: 10.1007/978-1-61779-953-2_13. [DOI] [PubMed] [Google Scholar]
- Nan X, Sims PA, Chen P, Xie XS. Observation of Individual Microtubule Motor Steps in Living Cells with Endocytosed Quantum Dots. J Phys Chem B. 2005;109:24220–24224. doi: 10.1021/jp056360w. [DOI] [PubMed] [Google Scholar]
- Nisman R, Dellaire G, Ren Y, et al. Application of Quantum Dots as Probes for Correlative Fluorescence, Conventional, and Energy-filtered Transmission Electron Microscopy. J Histochem Cytochem. 2004;52:13–18. doi: 10.1177/002215540405200102. [DOI] [PubMed] [Google Scholar]
- Peng C-W, Liu X-L, Chen C, et al. Patterns of cancer invasion revealed by QDs-based quantitative multiplexed imaging of tumor microenvironment. Biomaterials. 2011;32:2907–17. doi: 10.1016/j.biomaterials.2010.12.053. [DOI] [PubMed] [Google Scholar]
- Petryayeva E, Algar WR, Medintz IL. Quantum Dots in Bioanalysis: A Review of Applications Across Various Platforms for Fluorescence Spectroscopy and Imaging. Appl Spectrosc. 2013;67:215–252. doi: 10.1366/12-06948. [DOI] [PubMed] [Google Scholar]
- Pierobon P, Achouri S, Courty S, et al. Velocity, processivity, and individual steps of single myosin V molecules in live cells. Biophys J. 2009;96:4268–75. doi: 10.1016/j.bpj.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud F, Clarke S, Sittner A, Dahan M. Probing cellular events, one quantum dot at a time. Nat Methods. 2010;7:275–85. doi: 10.1038/nmeth.1444. [DOI] [PubMed] [Google Scholar]
- Pons T, Mattoussi H. Investigating biological processes at the single molecule level using luminescent quantum dots. Ann Biomed Eng. 2009;37:1934–59. doi: 10.1007/s10439-009-9715-0. [DOI] [PubMed] [Google Scholar]
- Prabhat P, Gan Z, Chao J, et al. Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proc Natl Acad Sci U S A. 2007;104:5889–94. doi: 10.1073/pnas.0700337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan SS, Liu HY, Vu TQ. Ligand-bound quantum dot probes for studying the molecular scale dynamics of receptor endocytic trafficking in live cells. ACS Nano. 2008;2:1153–66. doi: 10.1021/nn700399e. [DOI] [PubMed] [Google Scholar]
- Ram S, Kim D, Ober RJ, Ward ES. 3D single molecule tracking with multifocal plane microscopy reveals rapid intercellular transferrin transport at epithelial cell barriers. Biophys J. 2012;103:1594–603. doi: 10.1016/j.bpj.2012.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram S, Prabhat P, Chao J, et al. High accuracy 3D quantum dot tracking with multifocal plane microscopy for the study of fast intracellular dynamics in live cells. Biophys J. 2008;95:6025–43. doi: 10.1529/biophysj.108.140392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regoes A, Hehl AB. SNAP-tag mediated live cell labeling as an alternative to GFP in anaerobic organisms. Biotechniques. 2005;39:809–10. 812. doi: 10.2144/000112054. [DOI] [PubMed] [Google Scholar]
- Schieber C, Bestetti A, Lim JP, et al. Conjugation of transferrin to azide-modified CdSe/ZnS core-shell quantum dots using cyclooctyne click chemistry. Angew Chem Int Ed Engl. 2012;51:10523–7. doi: 10.1002/anie.201202876. [DOI] [PubMed] [Google Scholar]
- Schütz GJ, Axmann M, Schindler H. Imaging Single Molecules in Three Dimensions. Single Mol. 2001;2:69–74. doi: 10.1002/1438-5171(200107)2:2<69::AID-SIMO69>3.0.CO;2-N. [DOI] [Google Scholar]
- Schwartz SL, Yan Q, Telmer CA, et al. Fluorogen Activating Proteins Provide Tunable Labeling Densities for Tracking FcεRI Independent of IgE. ACS Chem Biol. 2014 doi: 10.1021/cb5005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa T, Terui T, Kagemoto T, et al. Real-time measurement of the length of a single sarcomere in rat ventricular myocytes: a novel analysis with quantum dots. Am J Physiol Cell Physiol. 2011;301:C1116–27. doi: 10.1152/ajpcell.00161.2011. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013;10:407–9. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Lev-Ram V, Deerinck TJ, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjollema KA, Schnell U, Kuipers J, et al. Methods Mol Biol. Academic Press; 2012. Correlated Light Microscopy and Electron Microscopy; pp. 157–173. [DOI] [PubMed] [Google Scholar]
- Sosinsky GE, Giepmans BNG, Deerinck TJ, et al. Markers for correlated light and electron microscopy. Methods Cell Biol. 2007;79:575–91. doi: 10.1016/S0091-679X(06)79023-9. [DOI] [PubMed] [Google Scholar]
- Specht CG, Izeddin I, Rodriguez PC, et al. Quantitative nanoscopy of inhibitory synapses: counting gephyrin molecules and receptor binding sites. Neuron. 2013;79:308–21. doi: 10.1016/j.neuron.2013.05.013. [DOI] [PubMed] [Google Scholar]
- Steinkamp MP, Low-Nam ST, Yang S, et al. erbB3 is an active tyrosine kinase capable of homo- and heterointeractions. Mol Cell Biol. 2014;34:965–77. doi: 10.1128/MCB.01605-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KN, Taatjes DJ, Bouffard NA, et al. Alpha smooth muscle actin distribution in cytoplasm and nuclear invaginations of connective tissue fibroblasts. Histochem Cell Biol. 2007;127:523–30. doi: 10.1007/s00418-007-0275-9. [DOI] [PubMed] [Google Scholar]
- Sun B, Xie W, Yi G, et al. Microminiaturized immunoassays using quantum dots as fluorescent label by laser confocal scanning fluorescence detection. J Immunol Methods. 2001;249:85–89. doi: 10.1016/S0022-1759(00)00331-8. [DOI] [PubMed] [Google Scholar]
- Sun J-Z, Chen C, Jiang G, et al. Quantum dot-based immunofluorescent imaging of Ki67 and identification of prognostic value in HER2-positive (non-luminal) breast cancer. Int J Nanomedicine. 2014;9:1339–46. doi: 10.2147/IJN.S58881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgyi C, Schmidt BF, Schmidt BA, et al. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat Biotechnol. 2008;26:235–40. doi: 10.1038/nbt1368. [DOI] [PubMed] [Google Scholar]
- Tabatabaei-Panah A-S, Jeddi-Tehrani M, Ghods R, et al. Accurate sensitivity of quantum dots for detection of HER2 expression in breast cancer cells and tissues. J Fluoresc. 2013;23:293–302. doi: 10.1007/s10895-012-1147-9. [DOI] [PubMed] [Google Scholar]
- Tada H, Higuchi H, Wanatabe TM, Ohuchi N. In vivo real-time tracking of single quantum dots conjugated with monoclonal anti-HER2 antibody in tumors of mice. Cancer Research. 2007;67(3):1138–44. doi: 10.1158/0008-5472.CAN-06-1185. [DOI] [PubMed] [Google Scholar]
- Torreno-Pina JA, Castro BM, Manzo C, et al. Enhanced receptor-clathrin interactions induced by N-glycan-mediated membrane micropatterning. Proc Natl Acad Sci U S A. 2014;111:11037–42. doi: 10.1073/pnas.1402041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True LD, Gao X. Quantum dots for molecular pathology: their time has arrived. J Mol Diagn. 2007;9:7–11. doi: 10.2353/jmoldx.2007.060186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu M, Adachi E, Nakamura A, et al. Atomic identification of fluorescent Q-dots on tau-positive fibrils in 3D-reconstructed pick bodies. Am J Pathol. 2012;180:1394–7. doi: 10.1016/j.ajpath.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Valentine CD, Verkman AS, Haggie PM. Protein trafficking rates assessed by quantum dot quenching with bromocresol green. Traffic. 2012;13:25–9. doi: 10.1111/j.1600-0854.2011.01287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermehren-Schmaedick A, Krueger W, Jacob T, et al. Heterogeneous intracellular trafficking dynamics of brain-derived neurotrophic factor complexes in the neuronal soma revealed by single quantum dot tracking. PLoS One. 2014;9:e95113. doi: 10.1371/journal.pone.0095113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fruhwirth G, Cai E, et al. 3D super-resolution imaging with blinking quantum dots. Nano Lett. 2013;13:5233–41. doi: 10.1021/nl4026665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe TM, Sato T, Gonda K, Higuchi H. Three-dimensional nanometry of vesicle transport in living cells using dual-focus imaging optics. Biochem Biophys Res Commun. 2007;359:1–7. doi: 10.1016/j.bbrc.2007.04.168. [DOI] [PubMed] [Google Scholar]
- Wells NP, Lessard GA, Phipps ME, et al. Going beyond 2D: following membrane diffusion and topography in the IgE-Fc[epsilon]RI system using 3-dimensional tracking microscopy. Proc SPIE. 2009;7185:71850Z1–71850Z13. doi: 10.1117/12.809412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsher K, Yang H. Multi-resolution 3D visualization of the early stages of cellular uptake of peptide-coated nanoparticles. Nat Nanotechnol. 2014;9:198–203. doi: 10.1038/nnano.2014.12. [DOI] [PubMed] [Google Scholar]
- Xing Y, Chaudry Q, Shen C, et al. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nat Protoc. 2007;2:1152–65. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- Xu J, Chang J, Yan Q, et al. Labeling Cytosolic Targets in Live Cells with Blinking Probes. J Phys Chem Lett. 2013;4:2138–2146. doi: 10.1021/jz400682m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Teslaa T, Wu T-H, et al. Nanoblade delivery and incorporation of quantum dot conjugates into tubulin networks in live cells. Nano Lett. 2012;12:5669–72. doi: 10.1021/nl302821g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C, Richter CP, Löchte S, et al. Dynamic submicroscopic signaling zones revealed by pair correlation tracking and localization microscopy. Anal Chem. 2014;86:8593–602. doi: 10.1021/ac501127r. [DOI] [PubMed] [Google Scholar]
- You C, Wilmes S, Beutel O, et al. Self-controlled monofunctionalization of quantum dots for multiplexed protein tracking in live cells. Angew Chem Int Ed Engl. 2010;49:4108–12. doi: 10.1002/anie.200907032. [DOI] [PubMed] [Google Scholar]
- Yu J, Monaco SE, Onisko A, et al. A validation study of quantum dot multispectral imaging to evaluate hormone receptor status in ductal carcinoma in situ of the breast. Hum Pathol. 2013;44:394–401. doi: 10.1016/j.humpath.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Zahavy E, Freeman E, Lustig S, et al. Double labeling and simultaneous detection of B- and T cells using fluorescent nano-crystal (q-dots) in paraffin-embedded tissues. J Fluoresc. 2005;15:661–5. doi: 10.1007/s10895-005-2972-x. [DOI] [PubMed] [Google Scholar]
- Zajac AL, Goldman YE, Holzbaur ELF, Ostap EM. Local cytoskeletal and organelle interactions impact molecular-motor- driven early endosomal trafficking. Curr Biol. 2013;23:1173–80. doi: 10.1016/j.cub.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrazhevskiy P, Gao X. Quantum dot imaging platform for single-cell molecular profiling. Nat Commun. 2013;4:1619. doi: 10.1038/ncomms2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrazhevskiy P, Sena M, Gao X. Designing multifunctional quantum dots for bioimaging, detection, and drug delivery. Chem Soc Rev. 2010;39:4326–54. doi: 10.1039/b915139g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrazhevskiy P, True LD, Gao X. Multicolor multicycle molecular profiling with quantum dots for single-cell analysis. Nat Protoc. 2013;8:1852–69. doi: 10.1038/nprot.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]