Abstract
Background
Gallbladder disease is highly related to inflammation, but the inflammatory processes are not well understood. Bile provides a direct substrate in assessing the local inflammatory response that develops in the gallbladder. To assess the reproducibility of measuring inflammatory markers in bile, we designed a methods study of 69 multiplexed immune-related markers measured in bile obtained from gallstone patients.
Methods
To evaluate assay performance, a total of 18 bile samples were tested twice within the same plate for each analyte, and the 18 bile samples were tested on two different days for each analyte. We used the following performance parameters: detectability, coefficient of variation (CV), intraclass correlation coefficient (ICC), and percent agreement (concordance among replicate measures above and below detection limit). Furthermore, we examined the association of analyte levels with gallstone characteristics such as type, numbers, and size.
Results
All but 3 analytes (Stem Cell Factor, SCF; Thrombopoietin, TPO; sIL-1RI) were detectable in bile. 52 of 69 (75.4%) analytes had detectable levels for at least 50% of the subjects tested. The within-plate CVs were ≤25% for 53 of 66 (80.3%) detectable analytes, and across-plate CVs were ≤25% for 32 of 66 (48.5%) detectable analytes. Moreover, 64 of 66 (97.0%) analytes had ICC values of at least 0.8. Lastly, the percent agreement was high between replicates for all of the analytes (median; within plate, 97.2%; across plate, 97.2%). In exploratory analyses, we assessed analyte levels by gallstone characteristics and found that levels for several analytes decreased with increasing size of the largest gallstone per patient.
Conclusions
Our data suggest that multiplex assays can be used to reliably measure cytokines and chemokines in bile. In addition, gallstone size was inversely related to the levels of select analytes, which may aid in identifying critical pathways and mechanisms associated with the pathogenesis of gallbladder diseases.
Keywords: Bile, Gallstones, Cytokines, Chemokines
1. Introduction
Gallbladder disease is the second highest cause of hospital admissions in the United States, with costs surpassing $2 billion [1]. Two key factors associated with gallbladder disease, gallstones and chronic inflammation, likely contribute to the hospital admissions. The chronic inflammation associated with gallstones in the gallbladder may be through the mechanical irritation of the gallstones rubbing against the gallbladder epithelial wall and gallstone-related cholestasis [2]. In addition, important risk factors for gallstones, such as obesity, serum lipids, and diabetes have also been linked to inflammation [3–5]. Furthermore, the use of nonsteroidal anti-inflammatory drugs (NSAIDs) has been associated with a reduction in gallbladder cancer [6], again suggesting that inflammation is an important mechanism in gallbladder pathogenesis.
Measurement of immune-related proteins in bile may help in characterizing the local inflammatory response that develops during gallbladder pathogenesis. However, these measurements can be challenging given the complex composition of bile, including bile salts, cholesterol, phospholipids, bilirubin, proteins, and inorganic ions (calcium, chloride, sodium, and bicarbonate) [7]. Previous studies measuring immune analytes in bile have largely been conducted in liver transplant patients where the bile was primarily collected from livers, T-tubes, or via endoscopic retrograde cholangiopancreaticography [8–16]. Also, all of these studies measured only a small number of analytes.
To facilitate future studies of inflammation and gallbladder disease, we evaluated the utility of multiplex assays to reliably measure multiple immune-related analytes in bile. Multiplex assays offer many advantages over singleplex assays such as ELISA, including 1) small sample volume requirement, 2) reduction in assay time, 3) reduced labor and material expenses, and 4) a larger range of quantification for each analyte. We have previously validated Luminex bead-based multiplex assays in serum and plasma [17], and applied these assays to identify inflammatory markers associated with risk of various types of cancer, including lung and non-Hodgkin lymphoma [18–20]. More recently, we established that it is feasible to perform these assays in cervical secretions [21], allowing future studies to evaluate the immunological associations with HPV-related cervical disease. In order to study the role of immune processes in gallbladder-related diseases, we developed methods to measure immune-related analytes in bile and examined the performance of multiplex cytokine assays in bile using samples from an NCI-sponsored study of Biliary Tract Cancers in Shanghai.
2. Material and Methods
2.1 Study Population
Biologic specimens and data were obtained from a population-based case-control study conducted in Shanghai, China [22]. All study participants provided written informed consent, and the Shanghai Cancer Institute and National Cancer Institute institutional review boards approved the study protocol and studies of inflammation, which includes the methods work described herein. For the assay optimization procedures, we randomly selected four gallstone patients with >1 mL of bile available, and for assessing assay reproducibility, we randomly selected 18 gallstone patients with >0.5 mL of bile available.
2.2 Optimization of Immune-related Analyte Assays
To determine whether Luminex bead-based assays could be used to measure cytokines in bile accurately, we tested bile from four gallstone patients. The Luminex 200 instrument can detect 100 unique bead sets through a unique internal fluorescent dye signature, so the readout is highly dependent on the integrity of the fluidics system and beads. Due to the viscosity (mucins and lipids) and dark pigmentation of bile, we first evaluated whether bile would affect bead aggregation and classification. Bead aggregation ranged from 20% to 40%; however, the beads were gated to exclude doublets during acquisition.
We evaluated three methods to improve measurement reliability and bead aggregation: filtration, delipidation, and dilution. Serial filtration was performed using 1.2, 0.44, and 0.22 micron filters. Samples clogged the filters at each pore size, which minimized the utility of filtration. For delipidation, bile samples were spiked with known concentrations of several cytokines, incubated with Cleanascite™ (1:2), and treated according to the manufacture’s recommendations. The removal of lipids provided minimal improvement in bead aggregation, and only three of the 19 spiked cytokines were recovered at an acceptable range (70%–130%) using the Cleanascite™ reagent (Supplementary Table 1). The bile samples were serially diluted in assay buffer from the respective kit being tested, and the set of dilutions for each assay are described in Supplementary Table 1. Typically, we noticed a marked improvement in bead aggregation with diluting the bile; however, the overall improvement in bead aggregation varied with subjects. Consequently, there was a decrease in detectability at the highest dilution as compared to preceding dilutions for each multiplex panel examined, which was taken into consideration on selecting the appropriate dilution for each assay.
Furthermore, serial dilutions of bile were used to evaluate the linearity and recovery of each spiked analyte (Supplementary Table 1). We considered analytes with more than a 2-fold change between dilutions for at least half the subjects to be acceptable, except for the Milliplex soluble receptor panel, which was evaluated starting at a 1.2-fold change between dilutions. At least two concentrations of standards from each assay were spiked into the bile samples to assess recovery. A recovery of 70%–130% of the spiked analyte for at least half the subjects was considered to be acceptable for bile.
2.3 Additional Assays
Three analytes (C-reactive protein, CRP; Serum Amyloid A, SAA; and Serum Amyloid P, SAP) in the Luminex-based Milliplex assay (Cardiovascular Disease Panel 2) had poor spike recovery and dilution patterns for each analyte even though the linearity passed our criteria (Supplementary Table 1). Given the increasing interest in these markers, particularly CRP, in cancer, we evaluated the Vascular Injury Panel 2 (Meso Scale Discovery, MSD; Rockville, MD), which contains CRP, SAA, Intercellular Adhesion Molecule-1 (ICAM-1), and Vascular Cell Adhesion Molecule-1 (VCAM-1). Overall, the performance of the MSD Panel was better than the Milliplex panel as indicated by the acceptable linearity and spike recovery for each analyte (Supplementary Table 1), so the MSD Vas cular Injury Panel was utilized for the reproducibility evaluations.
Because the concentration of bile can vary from person to person, we evaluated two different proteins that could be used as an overall measurement of bile concentration against which measurements of immune-related markers could be normalized. We examined the reproducibility of total protein (BCA, Thermo Fisher Scientific Inc., Rockford, IL), which has been used to normalize the levels of immune markers in cervical secretions [23], and albumin (Bethyl Laboratories, Inc., Montgomery, TX). The bile samples were diluted 1:100 for total protein, and 1:75000 for albumin. The overall CV of BCA and albumin was 14.7%, and 7.9%, respectively. Additionally, hemoglobin (Bethyl Laboratories, Inc., Montgomery, TX), was used as a surrogate for contamination of bile with peripheral blood. The overall CV of hemoglobin (diluted 1:10000) was 19.2%.
Samples undetectable at the indicated dilutions above were repeated at a different dilution for BCA (n=3; 1:2), albumin (n=7; 1:5000), and hemoglobin (n=6; 1:20 and 1:200) in order to interpolate a detectable level based on a 5-parameter logistic curve using SoftMax Pro 6.1 (Molecular Devices, LLC., Sunnyvale, CA).
2.4 Samples for Reproducibility Assessment
The reproducibility of each analyte was assessed from the bile of 18 randomly selected gallstone patients. We created four child aliquots for each panel of analytes; two child aliquots were tested within the same plate on day 1, and the other two child aliquots were tested within the same plate on day 2. Each sample aliquot had a unique ID, so the technician testing the samples was blinded to the replicate aliquots (n=2) examined in each plate. The Milliplex panels and MSD panel were tested according to the manufactures’ recommendations. As determined by the optimization procedures described above, the bile samples were diluted 1:10 in assay buffer for all of the Milliplex panels (Cytokine Panel 1, 23-plex; Cytokine Panel 2, 17-plex; Cytokine Panel 3, 7-plex) except for Soluble Receptor Panel (13-plex, samples diluted 1:40) and Adipokine Panel 1 (5-plex, samples diluted 1:100). All samples in the MSD panel (Vascular Injury Panel 2, 4-plex) were diluted 1:100. The levels of each marker within the Milliplex panels were evaluated on either a 5- or 4-parameter logistic curve using BioPlex Manager 6.1 software (BioRad, Hercules, CA). The markers in the MSD panel were evaluated with a 5-parameter logistic curve using Discovery Workbench 4.0 Software (MSD, Rockville, MD).
2.5 Statistical Analysis
For each marker, we assessed the reproducibility based on the crude measurements, as well as the measurements normalized by total protein (BCA) [(pg/mL of analyte)/(ug/mL of total protein)] and albumin [(pg/mL of analyte)/(ng of albumin)]. We used the following four measures: 1) percent detectability, which was assessed at subject level, where a subject was considered as having a detectable analyte level if all four samples per subject were above the minimum detectable standard, 2) within-plate and across-plate coefficient of variation (CVs), which were based on analyte levels above minimum detectable standard, 3) the intraclass correlation coefficient (ICC), which was based on analyte levels above minimum detectable standard, and 4) the percent agreement (i.e., the concordance among replicate measures above and below detection limit). CVs and ICCs were calculated based on untransformed analyte values. The within-plate CV, across-plate CV, and ICC were calculated based on least squares estimates obtained from models fitted with “Proc GLM” (SAS 9.1.3). When evaluating the performance criteria as a whole, we classified the analytes into four groups: excellent (detectability of samples ≥50%, within-plate and across-plate CVs ≤25%, and ICC ≥0.8); good (detectability of samples ≥25%, within-plate and across-plate CVs ≤40%, and ICC ≥0.8); fair (detectability of samples ≥25%, within-plate and across-plate CVs ≤75%, and ICC ≥0.8); and poor (all remaining analytes). The percent agreement was calculated as the percentage of paired samples with analyte levels both above or both below the minimum detectable standard. Furthermore, we assessed the association of analyte levels with tertiles of hemoglobin levels detected in the bile samples using the Kruskal-Wallis test.
We also explored whether any of the multiplex analyte levels differed by the type of gallstones (cholesterol and mixed), number of gallstones (1–3, 4–8, and >8 gallstones), or size of the largest gallstone (<10 mm, 10–14 mm, and ≥15 mm) recovered from the gallbladder following surgery. Analysis of gallstone characteristics were evaluated using either the Mann-Whitney test for comparisons between two groups or the Kruskal-Wallis test for comparisons between three groups. For all analyses, a p-value <0.05 was considered significant. All statistics were calculated with SAS 9.1.3 (SAS Institute Inc., Cary, NC) or JMP 10.0.2 (SAS Institute Inc., Cary, NC).
3. Results
The gallstone patients included 8 males and 10 females, with a median age of 65 (range: 37–73) (Table 1).
Table 1.
Characteristics of 18 gallstone patients utilized in reproducibility analysis.
| N (%) | ||
|---|---|---|
| Gender | ||
| Male | 8 (44%) | |
| Female | 10 (56%) | |
| Age, y | ||
| ≤ 55 | 4 (22%) | |
| 55–65 | 6 (33%) | |
| ≥ 66 | 8 (44%) | |
| Type of Gallstones/patient | ||
| Cholesterol | 5 (28%) | |
| Pigment | 2 (11%) | |
| Mixed | 9 (50%) | |
| Undetermined | 2 (11%) | |
| Number of Gallstones/patient | ||
| 1–3 | 7 (39%) | |
| 4–8 | 7 (39%) | |
| >8 | 4 (22%) | |
| Size of Largest Gallstone | ||
| <10 mm | 6 (33%) | |
| 10–14 mm | 6 (33%) | |
| ≥15 mm | 5(28%) | |
| Undetermined | 1 (6%) | |
| Smoking status | ||
| Never | 13 (72%) | |
| Former | 1 (6%) | |
| Current | 4 (22%) | |
These patients largely had cholesterol (5/18, 27.8%) or mixed cholesterol (9/18, 50%) gallstones, followed by pigmented gallstones (2/18, 11.1%) and stones of undetermined type (2/18, 11.1%). The number of gallstones collected after surgery ranged from 1 to 30, and the size of the stones ranged from 2 mm to 30 mm. The majority of patients were never smokers (13/18, 72.2%), followed by current smokers (4/18, 22.2%) and former smokers (1/18, 5.6%).
We were concerned that the chemical composition of bile would affect the Luminex assay, so we performed a set of optimization studies to first assess the effects of bile on the beads and analyte concentrations. We noticed that the amount of bead aggregation after incubating bile with the Luminex beads varied by sample; the aggregation increased with samples that were more viscous than others. The most effective solution in reducing bead aggregation was diluting the sample at least 10-fold with assay buffer. Diluting the bile had minimal effects on detectability since nearly 75% of the analytes tested were detectable in at least 50% of the bile samples. Step-wise filtration was problematic because the bile clogged the filter membrane, and removing the lipids using Cleanascite™ resulted in only minor improvement in bead aggregation.
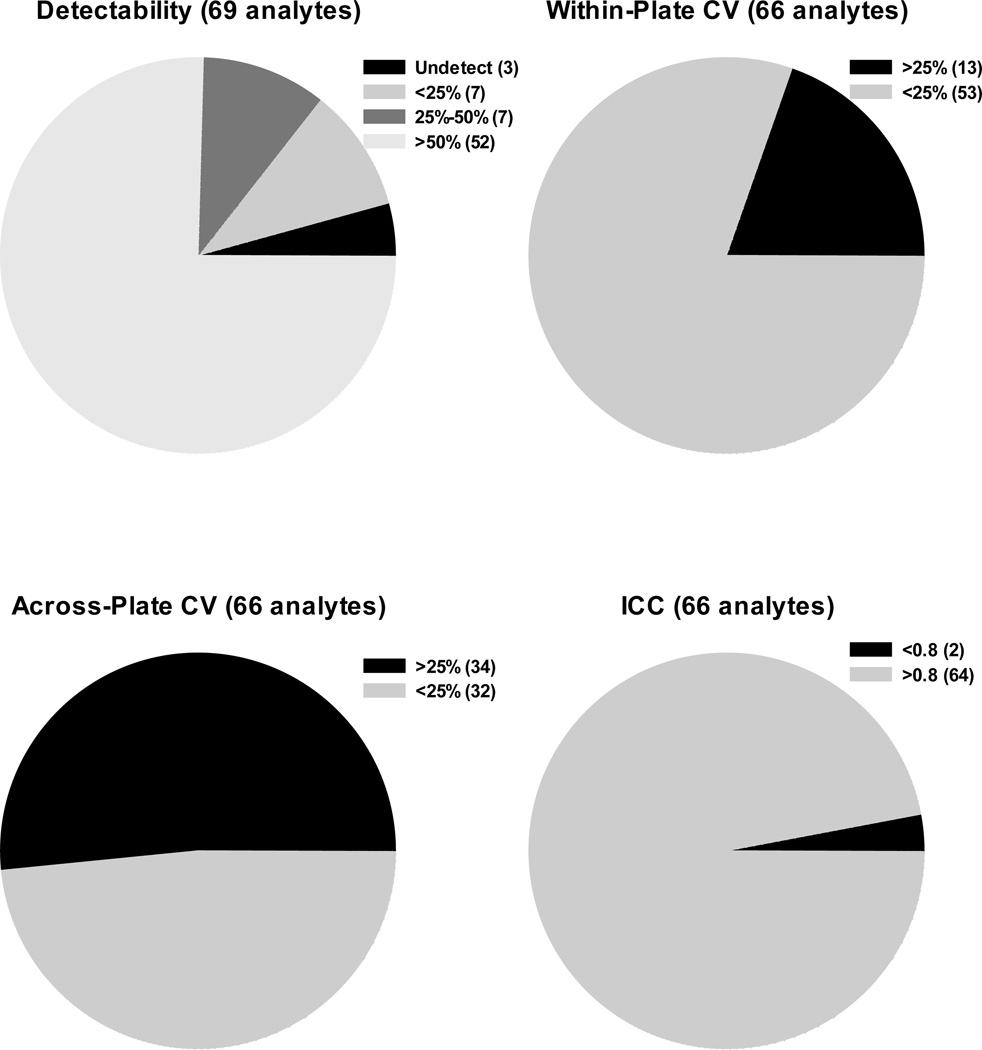
Sixty-nine analytes were evaluated for percent detectability. Seven analytes were detected in less than 25% of the subjects; seven analytes were detectable in 25% to 50% of the subjects; and 52 analytes were detected in at least 50% of the subjects tested (Figure 1).
Figure 1.
Detectability, Within- and Across-Plate CVs, and ICC for Multiplex Analytes. The pie charts represent each performance characteristic evaluated for all analytes tested, and the figure legend for each pie chart presents the criteria and absolute number of analytes that belong to the set criteria.
The results were similar when based on the number of samples rather than the number of subjects (Supplementary Table 2).
Three of 69 (4.3%) analytes had undetectable values for all subjects tested (Figure 1): Stem Cell Factor (SCF), Thrombopoietin (TPO), and sIL-1RI. These three analytes were subsequently excluded from coefficient of variation (CV), intraclass correlation coefficient (ICC), and percent agreement calculations, so the total number of analytes evaluated was 66.
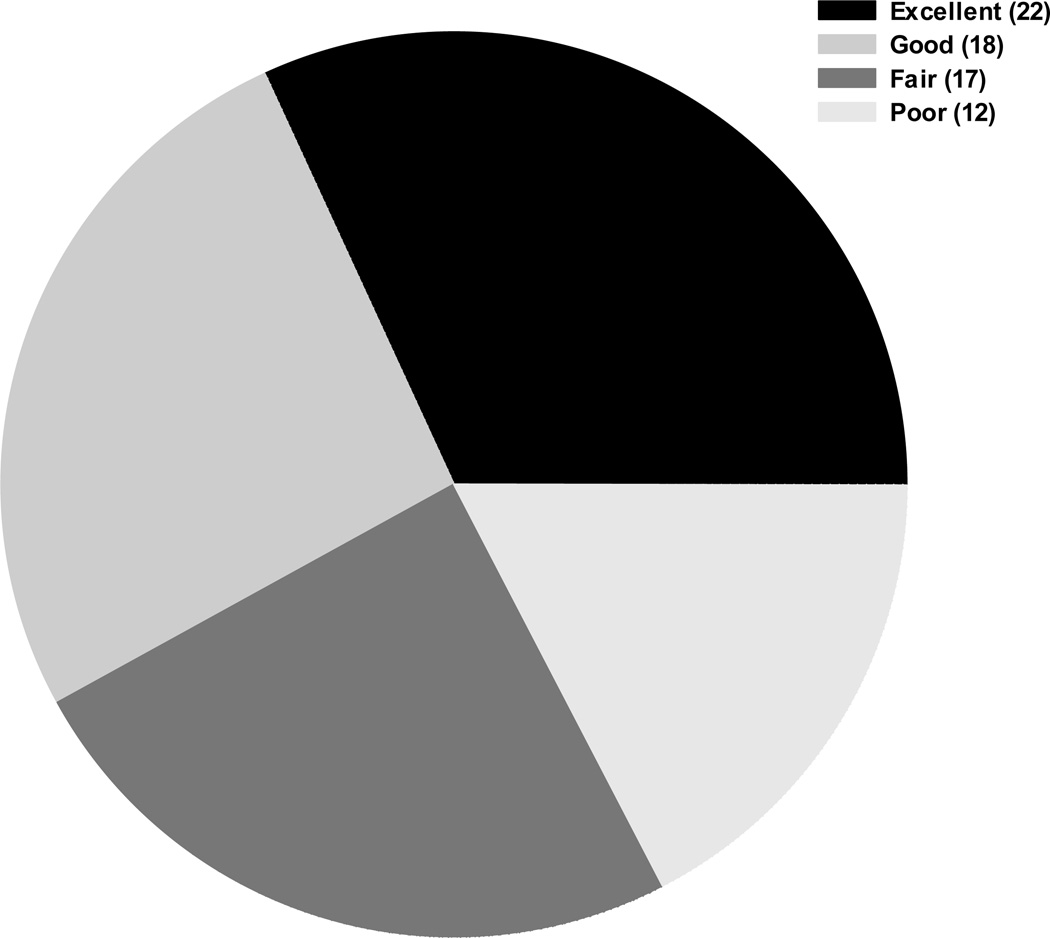
The CVs for each detectable analyte were separated into two categories: within-plate CV and across-plate CV. Fifty-three of 66 (80.3%) analytes had CVs ≤25% within-plate, and 32 of 66 (48.5%) analytes had across-plate CVs ≤25%. The median agreement of within-plate replicates was 97.2% (range: 77.8%–100%), and the median agreement of across-plate replicates was 97.2% (range: 72.2%–100%). Sixty-four of 66 (97.0%) analytes had an ICC value of at least 0.8 (Figure 1). Only two analytes had an ICC below 0.8: sVEGFR3 (0.79) and IL-33 (0.72). As seen in Figure 2, a majority of the analytes fall within the excellent and good classification (n=40). Of note, two analytes (Macrophage Inflammatory Protein-1α [MIP-1a] and stromal cell-derived factor-1α+β [SDF-1a+b]) were classified as having good performance and one analyte (6Ckine) was classified as having fair performance, yet these analytes had poor linearity and spike recovery as noted in Supplementary Table 1.
Figure 2.
Overall Performance Characteristics o f 69 Analytes. The pie chart represents the overall performance characteristics evaluated for all analytes tested, and the figure legend defines the classification given to each analyte. Excellent (detectability of samples ≥50%, within-plate and across-plate CVs ≤25%, and ICC ≥0.8); Good (detectability of samples ≥25%, within-plate and across-plate CVs ≤40%, and ICC ≥0.8); Fair (detectability of samples ≥25%, within-plate and across-plate CVs ≤75%, and ICC ≥0.8); and Poor (all remaining analytes).
Normalization using either BCA or albumin resulted in an increase in CVs, while the ICCs remained relatively unchanged (Supplementary Table 3). For example, the range of within-plate CVs for the 66 detectable analytes was 0.5%–48.5% without normalization, 3.8%–102.3% with BCA-based normalization, and 5.4%–170.6% with albumin-based normalization. Similarly, the range of ICC values for the 66 detectable analytes was 0.72–1.0 without normalization, 0.76–1.0 with BCA-based normalization, and 0.6–1.0 with albumin-based normalization.
Using hemoglobin as a surrogate for contamination of bile with peripheral blood, we assessed the association of analyte levels with tertiles of hemoglobin levels detected in the bile samples. Several analytes were significantly associated with hemoglobin (Supplementary Table 2). For example, the levels of the five analytes in the Adipokine Panel (Adiponectin, Adipsin, Lipocalin-2, Resistin, and PAI-1) were positively associated with hemoglobin. Furthermore, albumin was significantly associated with hemoglobin, while BCA was not significantly associated with hemoglobin, which corresponds to the increase in analytes significantly associated with hemoglobin after normalization to albumin (Supplementary Table 3).
After evaluating whether marker levels differed by gallstone characteristics (Supplementary Table 4), we found significantly different marker levels for only one marker [epithelial-derived neutrophil-activating peptide 78 (ENA-78)] by gallstone type (p=0.05) and one marker [C-reactive protein (CRP)] by number of gallstones (p=0.04). Interestingly, nine analytes showed significant changes in levels that tended to decrease as the size of the largest gallstone increased from <10 mm to 10–14 mm to ≥15 mm: epidermal growth factor (EGF), macrophage inflammatory protein-1 beta (MIP-1b), Eotaxin-2, monocyte chemoattractant protein-2 (MCP-2), macrophage inflammatory protein-1 delta (MIP-1d), sIL-1RII, TNF-related apoptosis-inducing ligand (TRAIL), soluble tumor necrosis factor receptor I (sTNFRI), and sTNFRII.
4. Discussion
Given our success in using the Luminex bead-based platform to conduct immune profiling studies using serum, plasma, and cervical secretions [17, 19–21], we assessed feasibility of expanding the use of this platform for measuring multiple immune-related analytes in bile. After an initial series of optimization experiments, we also tested four analytes (CRP, SAA, ICAM-1, and VCAM-1) using the MSD platform. Of the 69 analytes tested, 66 (95.7%) were detectable in bile. Of the 66 detectable analytes, 52 had detectable levels for at least 50% of the subjects tested, and only 7 had poor detectability (≤25%). In addition, 53 of the detectable analytes (80.3%) had within-plate CVs ≤25%, 32 (48.5%) had across-plate CVs ≤25%, and 64 (97.0%) had ICC values of ≥0.8. Of note, 22 analytes had excellent performance characteristics (detection in >50% subjects, ICC ≥0.8, CV≤25%) and 18 analytes had good performance characteristics (detection in >25% subjects and ICC >0.8) despite having somewhat elevated variability (CV between 25%–40%). Taken together, our data suggest that multiplex assays can be used to reliably measure cytokines and chemokines in bile, which may aid in identifying critical markers and pathways associated with gallbladder pathogenesis.
Gallbladder pathogenesis is strongly associated with gallstones and inflammation; however, the lithogenic mechanisms, particularly with respect to the role of inflammation in gallstone formation, are still unclear. Gallstones form during a hypersaturated state of cholesterol in the bile, which leads to crystal formation [24]. One suggested contributing factor to gallstone formation is the interaction of gel-forming mucins with the cholesterol crystals, which is further supported by the detectability of mucins within gallstones [25]. The mechanism associated with the overproduction of mucins, especially MUC5AC, has been linked to EGFR and TNFa [26]. In vitro studies have shown that TNFa in combination with EGFR ligands (TGFa) triggers significant production of MUC5AC mRNA and protein.
In evaluating analyte levels with gallstone characteristics, we found nine markers with significant differences that tended to decrease across categories of gallstone size. For type of gallstone and number of gallstones, only one marker (ENA-78 and CRP, respectively) differed significantly, likely reflecting chance given multiple comparisons. Nine markers, however, is more than twice as many than would be expected by chance at α=0.05. The consistent trend of decreasing levels across categories of gallstone size for the nine markers, especially between the 10–14 mm and ≥15 mm category, suggests that there is a biologic relationship between immune-related markers and gallstone size. Many of the analytes are chemokines (MIP-1b, MIP-1d, Eotaxin-2, and MCP-2), which suggests a role for the recruitment and localization of immune infiltrates into the gallbladder epithelium during the early stages of gallstone formation. The decrease in the level of these analytes as the gallstone size increases may represent a suppression in the immune response at some point in gallbladder pathogenesis. Moreover, this may provide further evidence for the mechanism of gallstone formation and associations of increased gallstone size with increased risk of gallbladder carcinogenesis [27, 28]. While we did not have bile from healthy donors to formally evaluate the level of these markers in bile from gallstones patients compared to healthy controls, the trends we observed by gallstone characteristics among gallstone patients suggest that we can use the multiplex analyte assay to gain a better perspective on the local soluble analyte response and its role in gallbladder pathogenesis. Hence, the multiplex assays serve as a unique tool to capture the level of many cytokines and chemokines, which represent a multitude of pathways (anti-viral, apoptotic, proinflammatory, proliferation, and chemotactic). Measurement of these analytes can be used to gain a better understanding of the local milieu and how that may play an active role in lithogenesis and potentially gallbladder carcinogenesis. Moreover, the local cytokine and chemokine milieu in the bile may allude to specific immune cells present in the gallbladder. Future studies are needed to evaluate the role these analytes across the full spectrum of gallbladder pathogenesis.
Not only is bile a good biospecimen f or assessing the local environment during gallbladder diseases, but bile also has great utility in monitoring liver complications following transplant. For example, a previous study found that the levels of sIL-2R and sICAM-1 in bile were higher in patients with acute liver graft rejection compared to patients with complications due to infection and stable liver graft transplants [29, 30]. Furthermore, in a study that measured five cytokines (TNF-a, IL-4, IL-5, IL-6, and IL-13) in bile, IL-4 and IL-5 levels were significantly higher in patients with IgG4-related cholangitis compared to patients with primary sclerosing cholangitis, bacterial cholangitis, and benign strictures of the extrahepatic bile ducts not due to cholangitis [16]. In vitro, theses cytokines were important in modulating the function of the tight junctions of biliary epithelial cells. Both of these studies examined only a few analytes by ELISA; however, the utilization of multiplex assays may help to capture a larger group of analytes capable of distinguishing between acute liver graft rejection and infection with better sensitivity and specificity, which may help guide clinical management, as well as identifying additional markers involved in pathogenesis.
Bile is a particularly challenging medium to work with, since it is a complex matrix of bile acids, lipids/cholesterol, pigments, and mucins [7]. In our optimization experiments, we evaluated several methods to reduce bead aggregation. While step-wise filtration and removing lipids with Cleanascite™ did not substantively improve bead aggregation, diluting the sample at least 10-fold with assay buffer was effective and did not impair detectability. We also assessed the recovery and linearity of each analyte. While most markers exhibited good recovery and linearity, we found strange patterns for a few markers. For example, the calculated recovery of EGF with three different spiked concentrations of EGF was between 70% and 130% at a 1:10 sample dilution, indicating good recovery. However, the calculated concentration increased 10-fold with each subsequent dilution from neat (pure bile) to 1:10 and 1:100. This increase in the observed amount of EGF with increasing dilutions may suggest that the EGF antibodies were non-specifically binding to factors in pure bile and lower-level dilutions. Even so, the reproducibility characteristics of EGF were acceptable (within-plate CV 5.9% and across-plate CV 20.3%, ICC=0.93), despite the irregularities in the concentration of EGF observed at different dilutions. These findings highlight the importance of performing methodological studies on an assay prior to reporting the results so the results can be interpreted accurately. In addition, when applying these assays to epidemiological studies it is important to confirm significant results by replicating with an independent group of samples and a different assay.
Although most of the analytes tested in this study were based on the Luminex assay, we evaluated CRP, SAA, ICAM-1, and VCAM-1 in the MSD planar multiplex assay system because CRP and SAA (the only two of these four acute phase analytes included on the Luminex panel) did not perform well in bile during our optimization study with the Luminex assay. While the detectability, CV, ICC, and agreement among replicates were acceptable for CRP, SAA, ICAM-1, and VCAM-1 with the MSD assay, the Luminex assay is preferable in biospecimens where it performs well since the MSD system is limited in its multiplexing capacity and currently can only detect up to 10 different analytes in a given well.
5. Conclusions
In summary, we optimized multiplexed assays for use in b ile, providing new access to efficient exploratory tools for screening markers in this medium. A majority of analytes could be reliably measured from bile and used in epidemiologic studies, suggesting that we can efficiently use multiplex technology to examine more comprehensively the immunological milieu present in bile. Furthermore, we found an inverse association with several analytes and gallstone size. These assays could be applied to assist with the monitoring of liver transplant patients for graft rejection and other complications, may help inform clinical management, and could be used to investigate underlying pathogenic mechanisms. In particular, epidemiologic studies using these assays would provide insightful clues to the factors reflective of the host and cancer microenvironment and potentially associated with the progression from gallstones to gallbladder cancer.
Supplementary Material
We evaluated the performance of 69 analytes in bile with multiplex assays.
Most analytes can be reliably measured from bile and used in epidemiologic studies.
Both bead-based and planar multiplex assays were utilized for select analytes.
Technology may aid in identifying critical markers associated with gallbladder cancer.
Acknowledgements
We thank the surgeons and pathologists in Shanghai for assistance in patient recruitment and pathology review; Chia-Rong Cheng and Kai Wu of the Shanghai Cancer Institute for data collection; Shelley Niwa of West at for data management; Michael Curry of IMS, Inc. for data analysis; and Marcus Williams and Yuanji Pan for technical assistance in the HPV Immunology Laboratory.
Financial support: This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Furthermore, The Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics, USA provided funds for the current study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: All authors declare no competing interest.
Authors’ contributions: TJK drafted the manuscript and participated in the design of the study, data analysis, and interpretation; FAC participated in its design, data analysis, interpretation, and helped to draft the manuscript; AH participated in its design, interpretation, and helped to draft the manuscript; YG participated in the design and conduct of the Population-based case–control study and helped to draft the manuscript; LN participated in its design and helped to draft the manuscript; BW participated in the design and conduct of the Population-based case–control study; LS participated in the design and conduct of the Population-based case–control study; GS carried out the immunoassays and helped to draft the manuscript; RMP participated in its design, helped with statistical analysis, and helped to draft the manuscript; AWH participated in the design and conduct of the Population-based case–control study and helped to draft the manuscript; JK participated in its design, data analysis, interpretation, coordination, and helped to draft the manuscript; and LAP participated in its design, data analysis, interpretation, coordination, and helped to draft the manuscript.
Contributor Information
Troy J. Kemp, Email: kemptj@mail.nih.gov.
Felipe A. Castro, Email: felipe.castro@nih.gov.
Yu-Tang Gao, Email: ytgao@vip.sina.com.
Allan Hildesheim, Email: hildesha@exchange.nih.gov.
Leticia Nogueira, Email: leticia.nogueira@nih.gov.
Bingsheng Wang, Email: wang.bingsheng@zs-hospital.sh.cn.
Lu Sun, Email: sunlush@126.com.
Gloriana Shelton, Email: Gloriana.Shelton@nih.gov.
Ruth M. Pfeiffer, Email: pfeiffer@mail.nih.gov.
Ann W. Hsing, Email: Ann.Hsing@cpic.org.
Ligia A Pinto, Email: pintol@mail.nih.gov.
Jill Koshiol, Email: koshiolj@mail.nih.gov.
References
- 1.Peery AF, Dellon ES, Lund J, Crockett SD, McGowan CE, Bulsiewicz WJ, et al. Burden of gastrointestinal disease in the United States: 2012 update. Gastroenterology. 2012;143:1179–1187. e1–e3. doi: 10.1053/j.gastro.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wistuba II, Gazdar AF. Gallbladder cancer: lessons from a rare tumour. Nat Rev Cancer. 2004;4:695–706. doi: 10.1038/nrc1429. [DOI] [PubMed] [Google Scholar]
- 3.Shoelson SE, Herrero L, Naaz A. Obesity, inflammation, and insulin resistance. Gastroenterology. 2007;132:2169–2180. doi: 10.1053/j.gastro.2007.03.059. [DOI] [PubMed] [Google Scholar]
- 4.Maury E, Brichard SM. Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Mol Cell Endocrinol. 2010;314:1–16. doi: 10.1016/j.mce.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Hernandez H, Simental-Mendia LE, Rodriguez-Ramirez G, Reyes-Romero MA. Obesity and inflammation: epidemiology, risk factors, and markers of inflammation. Int J Endocrinol. 2013;2013:678159. doi: 10.1155/2013/678159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu E, Sakoda LC, Gao YT, Rashid A, Shen MC, Wang BS, et al. Aspirin use and risk of biliary tract cancer: a population-based study in Shanghai, China. Cancer Epidemiol Biomarkers Prev. 2005;14:1315–1318. doi: 10.1158/1055-9965.EPI-05-0032. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann AF. Bile composition. In: Johnson LR, editor. Encyclopedia of Gastroenterology. Amsterdam: Academic Press; 2004. pp. 176–184. [Google Scholar]
- 8.Belov L, Meher-Homji V, Putaswamy V, Miller R. Western blot analysis of bile or intestinal fluid from patients with septic shock or systemic inflammatory response syndrome, using antibodies to TNF-alpha, IL-1 alpha and IL-1 beta. Immunol Cell Biol. 1999;77:34–40. doi: 10.1046/j.1440-1711.1999.00796.x. [DOI] [PubMed] [Google Scholar]
- 9.Kondera-Anasz Z, Michalski A, Gil D, Gil B, Starzewski J, Gonciarz Z. Accuracy of t-PA, u-PA, PAI-1 and PAI-2 estimation in human bile by ELISA kits. Med Sci Monit. 2000;6:616–617. [PubMed] [Google Scholar]
- 10.Koopmann J, Thuluvath PJ, Zahurak ML, Kristiansen TZ, Pandey A, Schulick R, et al. Mac-2-binding protein is a diagnostic marker for biliary tract carcinoma. Cancer. 2004;101:1609–1615. doi: 10.1002/cncr.20469. [DOI] [PubMed] [Google Scholar]
- 11.Okada N, Ishida H, Murata N, Hashimoto D, Seyama Y, Kubota S. Matrix metalloproteinase-2 and-9 in bile as a marker of liver metastasis in colorectal cancer. Biochem Biophys Res Commun. 2001;288:212–216. doi: 10.1006/bbrc.2001.5741. [DOI] [PubMed] [Google Scholar]
- 12.Rosen HR, Winkle PJ, Kendall BJ, Diehl DL. Biliary interleukin-6 and tumor necrosis factor-alpha in patients undergoing endoscopic retrograde cholangiopancreatography. Dig Dis Sci. 1997;42:1290–1294. doi: 10.1023/a:1018822628096. [DOI] [PubMed] [Google Scholar]
- 13.Vaishnavi C, Singh S, Kochhar R, Singh G, Singh K. C-reactive protein in patients with gallbladder and biliary tract diseases. Trop Gastroenterol. 2004;25:73–75. [PubMed] [Google Scholar]
- 14.Warle MC, Metselaar HJ, Hop WC, Gyssens IC, Kap M, Kwekkeboom J, et al. Early differentiation between rejection and infection in liver transplant patients by serum and biliary cytokine patterns. Transplantation. 2003;75:146–151. doi: 10.1097/00007890-200301150-00026. [DOI] [PubMed] [Google Scholar]
- 15.Watt JK, Hawkins K, Zhang M, Lipschitz J, Sandha G, Gong Y, et al. Transforming growth factor-beta (TGF-beta) protein levels are not elevated in the blood or bile of patients with primary sclerosing cholangitis: a pilot study. Dig Dis Sci. 2004;49:5–8. doi: 10.1023/b:ddas.0000011594.94294.12. [DOI] [PubMed] [Google Scholar]
- 16.Muller T, Beutler C, Pico AH, Otten M, Durr A, Al-Abadi H, et al. Increased T-helper 2 cytokines in bile from patients with IgG4-related cholangitis disrupt the tight junction-associated biliary epithelial cell barrier. Gastroenterology. 2013;144:1116–1128. doi: 10.1053/j.gastro.2013.01.055. [DOI] [PubMed] [Google Scholar]
- 17.Chaturvedi AK, Kemp TJ, Pfeiffer RM, Biancotto A, Williams M, Munuo S, et al. Evaluation of multiplexed cytokine and inflammation marker measurements: a methodologic study. Cancer Epidemiol Biomarkers Prev. 2011;20:1902–1911. doi: 10.1158/1055-9965.EPI-11-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodelon C, Polley MY, Kemp TJ, Pesatori AC, McShane LM, Caporaso NE, et al. Circulating levels of immune and inflammatory markers and long versus short survival in early-stage lung cancer. Ann Oncol. 2013;24:2073–2079. doi: 10.1093/annonc/mdt175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purdue MP, Hofmann JN, Kemp TJ, Chaturvedi AK, Lan Q, Park JH, et al. A prospective study of 67 serum immune and inflammation markers and risk of non-Hodgkin lymphoma. Blood. 2013;122:951–957. doi: 10.1182/blood-2013-01-481077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiels MS, Pfeiffer RM, Hildesheim A, Engels EA, Kemp TJ, Park JH, et al. Circulating Inflammation Markers and Prospective Risk of Lung Cancer. J Natl Cancer Inst. 2013 doi: 10.1093/jnci/djt309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshiol J, Sklavos M, Wentzensen N, Kemp T, Schiffman M, Dunn ST, et al. Evaluation of a multiplex panel of immune-related markers in cervical secretions: A methodologic study. Int J Cancer. 2013 doi: 10.1002/ijc.28354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsing AW, Gao YT, Han TQ, Rashid A, Sakoda LC, Wang BS, et al. Gallstones and the risk of biliary tract cancer: a population-based study in China. Br J Cancer. 2007;97:1577–1582. doi: 10.1038/sj.bjc.6604047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marks MA, Eby Y, Howard R, Gravitt PE. Comparison of normalization methods for measuring immune markers in cervical secretion specimens. J Immunol Methods. 2012;382:211–215. doi: 10.1016/j.jim.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carey MC. Pathogenesis of gallstones. Recenti Prog Med. 1992;83:379–391. [PubMed] [Google Scholar]
- 25.Smith BF, LaMont JT. Identification of gallbladder mucin-bilirubin complex in human cholesterol gallstone matrix. Effects of reducing agents on in vitro dissolution of matrix and intact gallstones. J Clin Invest. 1985;76:439–445. doi: 10.1172/JCI111991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finzi L, Barbu V, Burgel PR, Mergey M, Kirkwood KS, Wick EC, et al. MUC5AC, a gel-forming mucin accumulating in gallstone disease, is overproduced via an epidermal growth factor receptor pathway in the human gallbladder. Am J Pathol. 2006;169:2031–2041. doi: 10.2353/ajpath.2006.060146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Csendes A, Becerra M, Rojas J, Medina E. Number and size of stones in patients with asymptomatic and symptomatic gallstones and gallbladder carcinoma: a prospective study of 592 cases. J Gastrointest Surg. 2000;4:481–485. doi: 10.1016/s1091-255x(00)80090-6. [DOI] [PubMed] [Google Scholar]
- 28.Diehl AK. Gallstone size and the risk of gallbladder cancer. JAMA. 1983;250:2323–2326. [PubMed] [Google Scholar]
- 29.Adams DH, Wang L, Hubscher SG, Elias E, Neuberger JM. Soluble interleukin-2 receptors in serum and bile of liver transplant recipients. Lancet. 1989;1:469–471. doi: 10.1016/s0140-6736(89)91368-8. [DOI] [PubMed] [Google Scholar]
- 30.Adams DH, Mainolfi E, Elias E, Neuberger JM, Rothlein R. Detection of circulating intercellular adhesion molecule-1 after liver transplantation--evidence of local release within the liver during graft rejection. Transplantation. 1993;55:83–87. doi: 10.1097/00007890-199301000-00016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.