Abstract
Posttranslationally modified proteins serve as autoimmunogenic targets in a wide spectrum of autoimmune diseases. Here, we identified a posttranslationally modified paraprotein target (paratargs) in monoclonal gammopathies of undetermined significance (MGUS), multiple myelomas (MM), and Waldenstrom’s macroglobulinemias (WM) using protein macroarrays that were sumoylated and screened for reactivity with paraproteins from MGUS, MM, and WM patients. We found that paraproteins from a proportion of European, African-American, and Japanese patients specifically reacted with the sumoylated heat-shock protein 90 β isoform-α (HSP90-SUMO1, where SUMO indicates small ubiquitin-like modifier), while no reactivity with HSP90-SUMO1 was detected in over 800 controls. HSP90-SUMO1 was present in blood cells from all patients with HSP90-SUMO1–binding paraproteins. We determined that the HSP90-SUMO1 carrier state is autosomal-dominantly inherited and caused by the inability of SUMO peptidase sentrin/SUMO-specific protease 2 (SENP2) to desumoylate HSP90-SUMO1. HSP90-SUMO1 was detected in a small percentage of healthy individuals from all backgrounds; however, only MGUS, MM, and WM patients who were HSP90-SUMO1 carriers produced HSP90-SUMO1–specific paraproteins, suggesting that sumoylated HSP90 promotes pathogenesis of these diseases through chronic antigenic stimulation. This study demonstrates that harboring HSP90-SUMO1 identifies healthy individuals at risk for plasma cell dyscrasias and that dominant inheritance of posttranslationally modified autoantigenic paratargs is one of the strongest molecular defined risk factors for MGUS, MM, and WM.
Introduction
We recently identified paraprotein target 7 (paratarg-7) as the target of up to 37% of paraproteins from monoclonal gammopathies of undetermined significance (MGUS)/multiple myeloma (MM) patients (1–3). All patients with paratarg-7–specific paraproteins were carriers of a hyperphosphorylated modification of paratarg-7 (pP-7) (4), and the pP-7 carrier state is inherited in an autosomal-dominant fashion (4). Besides paratarg-7, 10 additional paratargs were identified (5), for most of which hyperphosphorylation was shown to be a consistent finding and the most likely reason for chronic autoantigenic stimulation (6). In order not to miss posttranslationally modified autoantigenic targets of paraproteins that do not crossreact with the unmodified version, we posttranslationally modified membrane-bound array proteins in vitro. Here, we show that in vitro sumoylation of membrane-bound human protein macroarrays enabled the identification of an aberrantly sumoylated protein as a frequent and autosomal-dominantly inherited autoantigenic target of paraproteins.
Results
Identification of sumoylated paratargs.
Paraprotein-containing sera (6 IgA, 31 IgG, 3 IgM) diluted 1:107 were screened for reactivity with in vitro–sumoylated proteins represented on a fetal brain–derived macroarray. Two signals were obtained. These 2 immunoreactive clones were expressed in E. coli, and the corresponding His6-tagged proteins were sumoylated either in solution or after spotting onto polyvinylidene difluoride (PVDF) membranes. Screening of these 2 clones with 40 paraproteins from European patients resulted in the identification of 6/40 paraprotein-containing sera reacting with the sumoylated heat-shock protein 90 β isoform-α (HSP90-SUMO1, where SUMO indicates small ubiquitin-like modifier) (WT NP_031381), while the immunoreactivity of the other clone could not be confirmed. Patients’ sera showed no immunoreactivity against WT HSP90 or WT SUMO1 protein alone (represented by VCP-SUMO1), indicating an immune reaction specific for sumoylated HSP90 (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI76802DS1). No reaction was observed against HSP90-SUMO2 or HSP90-SUMO3, demonstrating that the signals were specific for the HSP90-SUMO1 isoform. In addition, absorption studies using HSP90-SUMO1 affinity columns proved that the entire monoclonal component targeted HSP90-SUMO1 (Supplemental Figure 2).
Frequency of HSP90-SUMO1–specific paraproteins and prevalence of HSP90-SUMO1 carriership in different ethnic groups.
Sera derived from European, Japanese, and African-American MM/MGUS patients were analyzed for immunoreactivity against HSP90-SUMO1 (Table 1 and Supplemental Table 1). At a dilution of 1:107, 27/226 (11.9%) sera from European, 9/80 (11.2%) from African-American, and 9/176 (5.1%) from Japanese patients reacted. No reactivity was detected in the 1:102 diluted sera from 550 European, 100 African-American, and 278 Japanese healthy controls (less diluted sera could not be tested due to background reactivity). Western blot and ELISA experiments revealed that whole blood from all MGUS/MM/Waldenstrom’s macroglobulinemias (WM) patients with an anti–HSP90-SUMO1 paraprotein expressed both sumoylated HSP90 and WT HSP90, while HSP90-SUMO1 was not seen in patients whose paraprotein did not bind to HSP90-SUMO1 (Figure 1 and data not shown). In contrast, 5/550 (0.9%) European, 2/100 (2%) African-American, and 2/278 (0.8%) Japanese controls carried HSP90-SUMO1, resulting in an odds ratio (OR) (as determined by univariate regression analysis in SPSS) of 14.8 for European, 6.2 for African-American, and 7.4 for Japanese healthy carriers for MGUS/MM/WM.
Table 1.
HSP90-SUMO1 immunoreactivity and carrier state in different ethnic groups
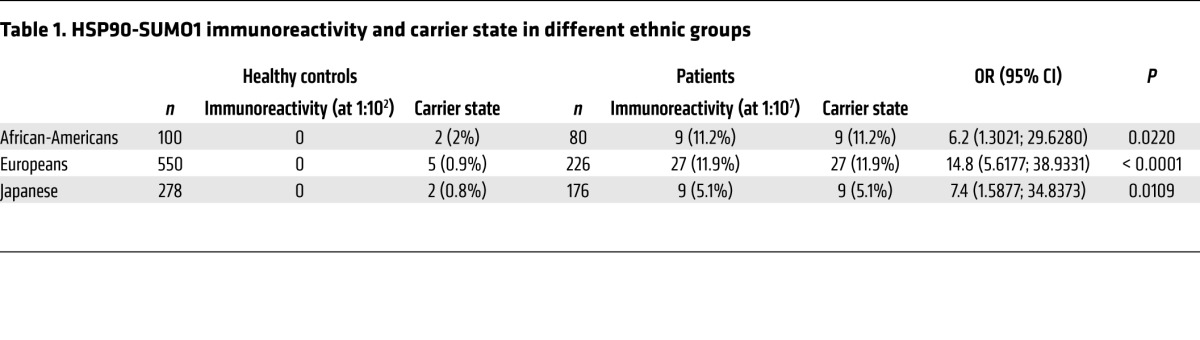
Figure 1. HSP90-SUMO1 in patients and in healthy donors.
Western blot (lanes 1–6) or immunoprecipitation (lanes 7–10) of blood cell lysates derived from patients and healthy donors. HSP90-SUMO1 was detected only in immunopositive patients. Lanes 1–3: 3 healthy donors; lane 4: patient with a paraprotein of other specificity; lanes 5 and 6: 2 patients with immunoreactivity against HSP90-SUMO1. Blood cell lysate from lane 7 represents a patient with a paraprotein of other specificity; lane 8 represents a patient with immunoreactivity versus HSP90-SUMO1. Both samples were immunoprecipitated with anti–HSP90-mAb, separated by SDS-PAGE, and detected with anti-HSP90. Lanes 9 and 10: same as in lanes 7 and 8, but detection with anti-SUMO1; lanes 11 and 12: same as in lanes 7 and 8, but detection with anti-SUMO2/3.
Antibody-binding epitope of HSP90-SUMO1.
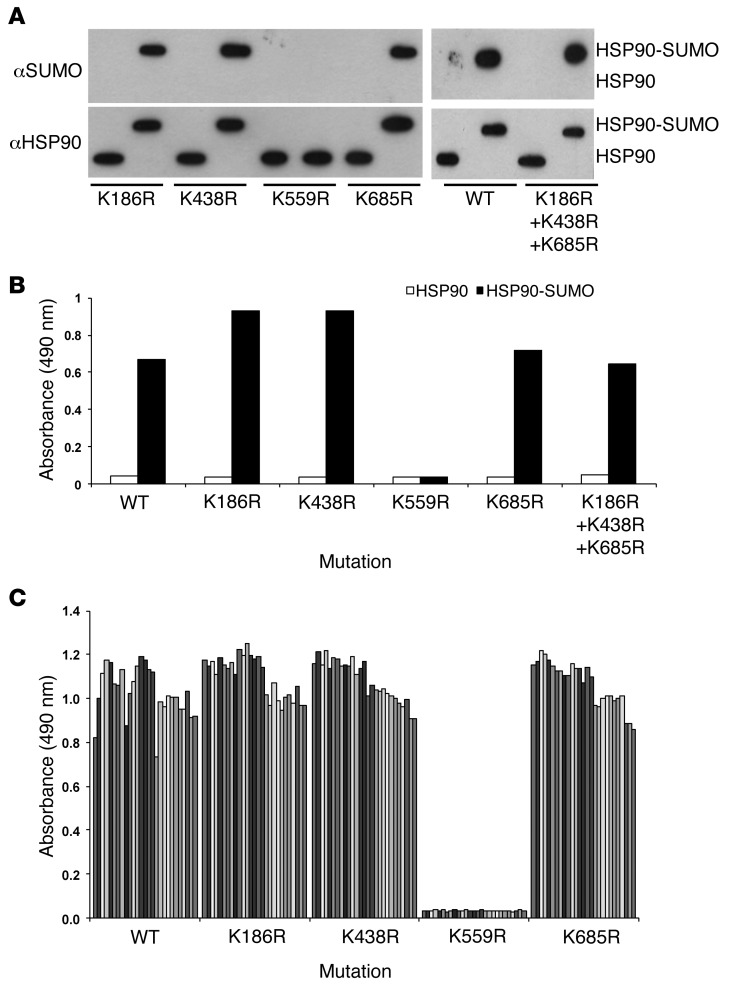
By in silico analysis, 17 possible sumoylation sites were predicted for HSP90 (SUMOplot Analysis Program; http://www.abgent.com/tools/), 5 of which have a high probability; 4 of these were located in the partial clone of HSP90-SUMO1 by which the HSP90-SUMO1–reacting paraproteins had been discovered. To identify the lysine and the epitope that, when sumoylated, bound the respective paraproteins, site-directed mutagenesis was performed. Only when HSP90 K559 was replaced by arginine (K559R) was no HSP90-SUMO1 serum reactivity detected, while replacement by K186R, K438R, and K685R (alone or in combination) had no effect on binding of the reactive paraproteins (Figure 2) or the B cell receptor–derived (BCR-derived) antigen-binding fragment (Fab, Supplemental Figure 3), indicating that the paraprotein immunoreactivity was specific for the HSP90-SUMO1 branch at K559 (HSP90-SUMO1K559). All HSP90-SUMO1–reactive paraproteins bound to the same HSP90-SUMO1K559 epitope.
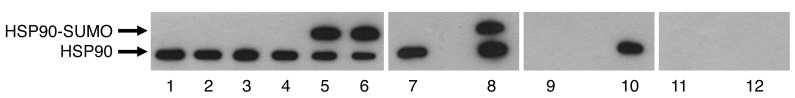
Figure 2. Identification of the immunogenic HSP90 sumoylation site.
Highly predicted sumoylation sites of HSP90 were single or triple mutated as indicated and tested for immunoreactivity with patients’ sera (dilution 1:107) by ELISA. (A) Verification of the mutations by Western blot analysis using anti-HSP90 or anti-SUMO mAbs. (B) ELISA: As an example, immunoreactivity of one patient’s serum versus mutated and sumoylated HSP90 is shown. Patient’s serum did not react with WT HSP90 or HSP90-SUMO1 (K559R), while all other mutations did not affect the immunoreactions. (C) ELISA: all sera derived from all 27 HSP90-SUMO1–immunopositive patients showed no immunoreactivity when K559 was replaced by arginine (K559R), which lacks the respective sumoylation site. All other mutations had no effect on immunoreactivity. Each column represents an individual HSP90-SUMO1–positive patient.
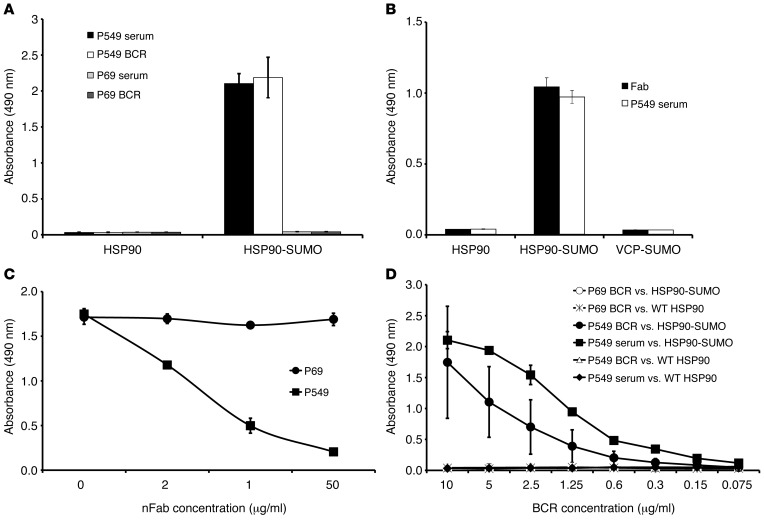
Demonstration of the monoclonal BCR-derived origin of HSP90-SUMO1 paraproteins.
The paraprotein-mediated reaction against HSP90-SUMO1 was demonstrated by cloning the BCR from a bone marrow smear (plasma cell infiltration approximately 80%) of a patient with an anti–HSP90-SUMO1–specific paraprotein. The Ig genes (VH, Vκ, Vλ) were characterized by PCR and cloned into a phagemid vector to produce Fabs. Both the BCR-derived Fab and the paraprotein recognized HSP90-SUMO1 as target (Figure 3), and the patient’s paraprotein displaced the BCR-derived recombinant Fab in a dose-dependent manner (Figure 3C) in a competition assay, demonstrating that both the recombinant Fab and the paraprotein bind to the same epitope of HSP90-SUMO1.
Figure 3. Specificity of plasma cell antigen receptor–derived (BCR-derived) Fab and HSP90-SUMO1–specific paraproteins by ELISA.
P549 represents a HSP90-SUMO1 reactive paraprotein, while P69 is a paraprotein with specificity for paratarg-7. (A) Reactivity. (B) Specificity. Sumoylated valosin-containing protein (VCP) (CDC48) as a control for WT SUMO. (C) Competition ELISA. (D) Titer determination of paraprotein and recombinant BCR.
Expression of HSP90-SUMO1 in peripheral blood cells.
While no HSP90-SUMO1 was detected in any magnetic cell separation–isolated subpopulation of PBMCs from carriers of WT HSP90, white blood cells from HSP90-SUMO1 carriers expressed both forms of HSP90. B cells from HSP90-SUMO1 carriers expressed more HSP90-SUMO1 than WT HSP90, while WT HSP90 was the predominant form in all other blood cell subpopulations. In addition, in mouth swabs from HSP90-SUMO1 carriers, only WT HSP90 was detected, whereas HSP90-SUMO1 was absent. EBV-transformed lymphoblastoid cell lines (LCLs) from HSP90-SUMO1 carriers expressed both HSP90 variants throughout their life spans in culture (tested from passages 1 up to 50; Supplemental Figure 4), while only WT HSP90 was expressed in LCLs derived from WT HSP90 carriers.
HSP90-SUMO1 in other malignancies and autoimmune diseases.
In all European patients with other cancer types and autoimmune diseases, we found a prevalence of HSP90-SUMO1 similar to that in healthy controls (Supplemental Tables 2 and 3), indicating that the HSP90-SUMO1 carrier state and immunoreactivity against this posttranslational modification are increased only in patients with MM/MGUS/WM.
Inheritance of HSP90-SUMO1.
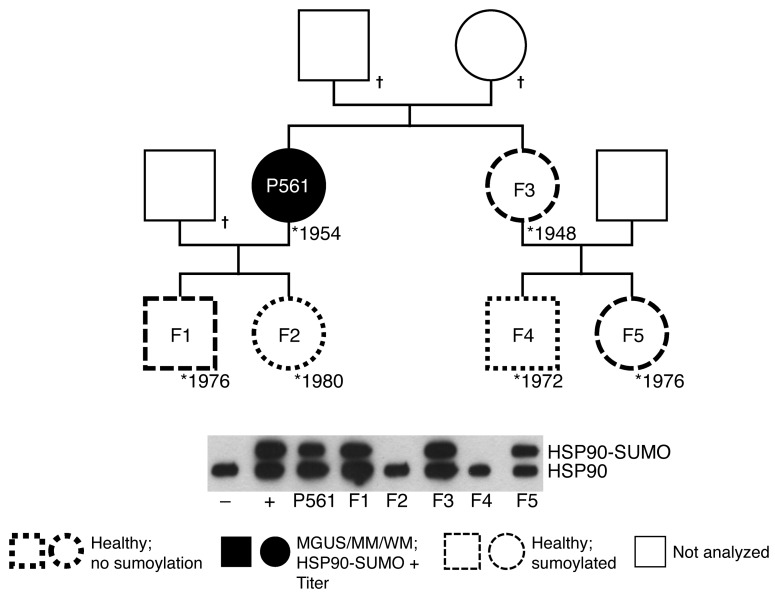
The members of 3 families with patients who were carriers of and had paraproteins specific for HSP90-SUMO1 in their serum gave written consent to be studied for the HSP90-SUMO1 carrier state. In all these families, the analysis revealed that the sumoylated carrier state of HSP90 is inherited in an autosomal-dominant fashion (Figure 4 and Supplemental Figure 5).
Figure 4. Pedigree of a family with a patient with MM and HSP90-SUMO1 as the paratarg.
Shown is the pedigree of the family and a Western blot for the HSP90 state. Sumoylated HSP90 was detected in the female patient having MM and in her healthy sister. Each of the 2 sisters had 2 children, one of whom was a carrier of HSP90-SUMO1, while the other was not. All children were healthy.
Regulation of HSP90-SUMO1 in patients.
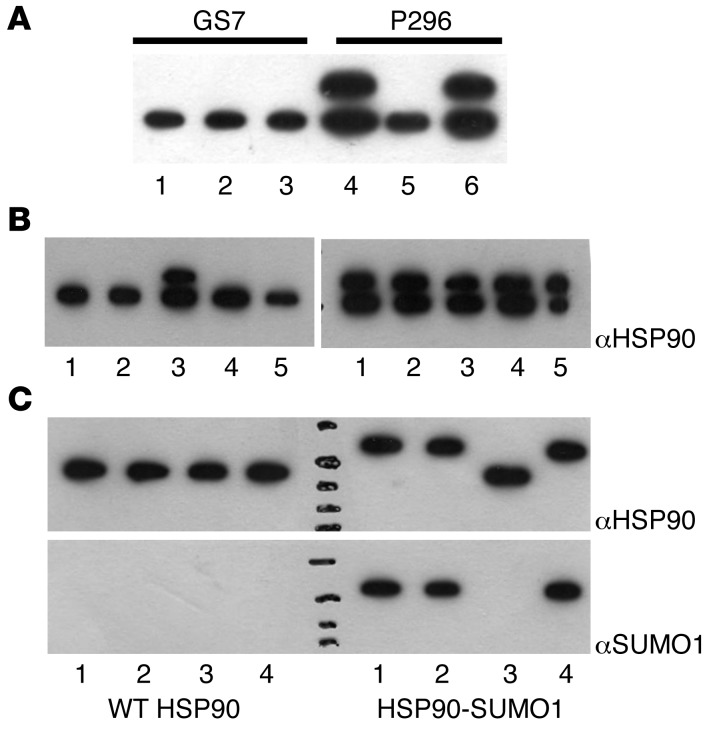
The observation that HSP90-SUMO1 was only detected in patients, but not in healthy persons, raised the question of the enzymes responsible for sumoylation and desumoylation of HSP90. Complementation experiments using native enzyme extracts derived from the blood of patients and healthy persons showed that desumoylation, but not sumoylation, of HSP90-SUMO1 was (partially) inactive in patients (Figure 5A). For knockdown experiments, LCLs were used because LCLs maintain the carrier state of the donor from which they are established. Knockdown experiments using sentrin/SUMO-specific protease–specific (SENP-specific) shRNAs in LCLs derived from healthy people identified SENP2 as responsible for desumoylation of HSP90-SUMO1 in healthy people, while inhibition of other SENPs had no influence on HSP90-SUMO1. This result was confirmed by incubation of recombinant HSP90-SUMO1 together with recombinant SENPs: only SENP2, but none of the other SENPs, cleaved HSP90-SUMO1, confirming that the (specific) inability of SENP2 to desumoylate HSP90-SUMO1 was responsible for HSP90-SUMO1 carriership (Figure 5, B and C). Neither DNA modifications nor differences in SENP2 expression at the mRNA and protein levels were detected (Supplemental Figure 6); similarly, no up- or downregulation of other sumoylating or desumoylating enzymes was found (Supplemental Figure 7). To test patient-derived cells for SENP2 activity, SENP2 enzymes from patients’ PBMCs and from healthy controls were enriched by immunoaffinity chromatography and tested for their capability to cleave the indicator substrate RanGAP. RanGAP was cleaved by SENP2 derived from patients and healthy donors (Figure 6), indicating that patients had no general defect of SENP2 enzyme activity.
Figure 5. Regulation of HSP90-SUMO1 in patients.
(A) Complementation assay. Native cell extracts derived from fresh blood of a HSP90-SUMO1–carrying patient (P296) and of a healthy donor (GS7) were prepared, used for complementation, and analyzed by Western blot and immunodetection using anti-HSP90 mAb. The upper band represents HSP90-SUMO1, the lower HSP90. HSP90-SUMO1 was cleaved by native enzyme extract derived from the healthy donor, while the native enzymes derived from the patient did not cleave. Lanes 1–3: PBMC cell extract of healthy donor blood; lanes 4–6: PBMC cell extract of patient blood. Lanes 1 and 4: extract plus PBS as control; lanes 2 and 5: extract plus healthy native enzyme extract; lanes 3 and 6: extract plus patients’ native enzyme extract. (B) Desumoylation of HSP90-SUMO1. LCLs derived from a healthy donor (left) or a patient (right) and transformed with shRNAs specific for individual SENPs were checked for HSP90 by Western blot and immunodetection using anti-HSP90 mAb. HSP90-SUMO1 was detectable only when SENP2 was inhibited. Lane 1: incubation with scrambled shRNA; lanes 2–5: incubation with shRNA versus SENP1 (lane 2), SENP2 (lane 3), SENP3 (lane 4), and SENP5 (lane 5). (C) Incubation of purified recombinant WT HSP90 (left) or HSP90-SUMO1 (right) with purified recombinant SENPs. Analysis was done by Western blot and immunodetection using anti-HSP90 or anti-SUMO1. Only SENP2 cleaved HSP90-SUMO1, while the other SENPs did not. Lane 1: no SENP; lane 2: SENP1; lane 3: SENP2; lane 4: SENP3.
Figure 6. Activity of SENP2 in patient-derived PBMCs.
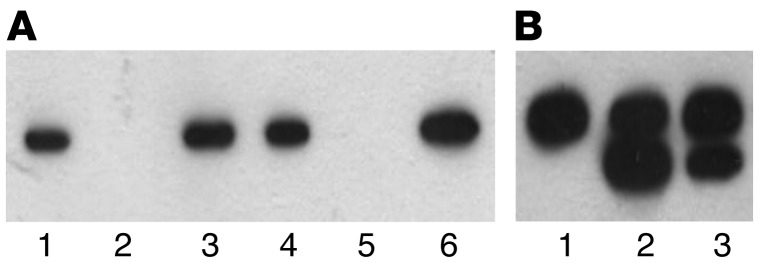
SENP2 enzymes derived from PBMCs of a patient and a healthy donor were purified by affinity chromatography and incubated with GST-RanGAP-SUMO1. Analysis was done by Western blotting and immunodetection using GST mAb. Desumoylation was detected for both preparations, indicating an active SENP2 enzyme in patients and healthy controls. (A) SENP2 isolation from PBMCs. Detection was done by Western blot using anti-SENP2. Lanes 1–3: healthy donor; lanes 4–6: patient. Lanes 1 and 4: lysate; lanes 2 and 5: flow through; lanes 3 and 6: eluate. (B) Test for SENP2 activity. Detection was done by Western blot using anti-GST. Lane 1: GST-RanGAP as control; lane 2: GST-RanGAP plus patient-derived SENP2; lane 3: GST-RanGAP plus SENP2 from healthy donors. The upper band represents GST-RanGAP-SUMO1, the lower GST-RanGAP.
Discussion
Posttranslationally modified proteins have been identified as autoimmunogenic targets in a wide spectrum of autoimmune diseases, e.g., nuclear antigens in systemic lupus erythematosus (SLE), citrullinated vimentin in rheumatoid arthritis (7), and myelin-associated glycoprotein in chronic inflammatory demyelinating polyneuropathy (8, 9); posttranslationally modified autoantigenic targets of the BCR in B cell malignancies have been described for chronic lymphatic leukemia (CLL) (oxidized LDL; ref. 10) and MGUS/MM/WM (hyperphosphorylated paratargs) (2, 4, 6).
The hyperphosphorylated paratargs were detected using unmodified protein macroarrays because the respective paraproteins reacted with both the native protein on the macroarrays and the hyperphosphorylated variant. In order not to miss posttranslationally modified targets of paraproteins that do not crossreact with the native or WT protein, we subjected the commercially available protein macroarrays to sumoylation in vitro, thus succeeding in identifying HSP90-SUMO1 as a frequent target of paraproteins from MGUS/MM/MW patients. The fact that the reaction of the respective paraproteins was specific for sumoylated HSP90, while WT HSP90 or SUMO1 showed no reaction, suggested that the branch region of the HSP90 protein that combines HSP90 and SUMO1 might be the epitope binding the respective paraproteins. Performing site-directed mutagenesis, we identified lysine at position 559 of HSP90 (K559) as the site of sumoylation and part of the paraprotein-binding epitope.
Having shown that HSP90 sumoylation at lysine 559 is inherited in an autosomal-dominant fashion, it was of interest to check for the prevalence of MGUS/MM patients with HSP90-SUMO1–specific paraproteins and the prevalence of healthy HSP90-SUMO1 carriers in different ethnic groups. Since the HSP90-SUMO1 carrier state is rare in healthy controls from all 3 ethnic groups studied here, carriers of the variant are at a significantly increased risk for MGUS/MM/WM, with ORs ranging from 6.2 in African-Americans and 7.4 in Japanese to 14.8 in European carriers of the sumoylated variant, making HSP90-SUMO1 carriership the second strongest risk factor after hyperphosphorylated paratarg-7 for MGUS/MM/WM in all 3 ethnic groups. Since the control group (median age 40 years) was 26 years younger than the patients with paraproteins (66 years), we cannot exclude that the healthy carriers will develop an anti–HSP90-SUMO1–specific paraprotein with longer follow-up. However, if this were the case, it would only further increase the OR of healthy carriers. Testing for an HSP90-SUMO1 carrier state can identify and exclude members of the respective families who have an increased risk of developing MGUS or MM, which might be helpful in the clinical situation.
SUMO proteins are a family of small proteins that covalently bind to amino acid residues of target proteins to modify their function (11–13). Sumoylated proteins are involved in various cellular processes, including protein stability, response or stress, DNA damage response, nuclear-cytoplasmic transport, transcriptional regulation, cell growth, survival, and apoptosis (11, 14, 15). Sumoylated autoantigens have been described as a novel and independent class of autoantigens in primary biliary cirrhosis (16). Whether the permanent sumoylation of HSP90 has functional consequences in the respective carriers remains to be determined. However, since WT HSP90 is still present at a relevant level in the respective cells in HSP90-SUMO1 carriers, we would assume that the presence of HSP90-SUMO1 should not result in major functional changes of cells coexpressing WT HSP90 and HSP90-SUMO1. However, one should keep in mind that the WT HSP90 molecule is a dimer and each protomer can be divided into structurally and functionally distinct domains. Growing evidence suggests that HSP90 interacts asymmetrically with clients and some cochaperones and that each HSP90 protomer can hydrolyze ATP independently (17). HSP90 is subject to numerous posttranslational modifications (18), including sumoylation (19), yet the impact of sumoylation on HSP90 function remains to be determined. Mollapour et al. (20) recently reported that asymmetric sumoylation of HSP90 at K191 facilitates both recruitment of ATPase-activating cochaperone Aha1 and the binding of HSP90 inhibitors. From these findings, we cannot exclude that sumoylation of HSP90 at K559 modifies its function in a way similar to that described for sumoylation at K191 (20).
Sumoylation is a reversible dynamic process balanced by activating (E1), conjugating (E2), and ligating (E3) enzymes and is readily reversed by SENP in humans (21, 22). We demonstrated a defect of desumoylating HSP90-SUMO1 in patients and healthy controls carrying HSP90-SUMO1 and identified SENP2 as the enzyme responsible for desumoylating HSP90-SUMO1. The analysis of SENP2 revealed no differences in DNA sequence, RNA, and protein expression between carriers of HSP90-SUMO1 and WT HSP90, and there was no indication for up- or downregulation of sumoylation enzymes; therefore, other reasons for the (specific) failure of SENP2 to desumoylate HSP90-SUMO1 in the respective carriers must be investigated. As shown in Supplemental Figure 7, there was no indication for up- or downregulation of sumoylation enzymes, including UBC9, SAE1, SAE2, and others (Supplemental Figure 7) in WT HSP90 compared with HSP90-SUMO1 carriers. This together with the observation that HSP90-SUMO1 is desumoylated when coincubated with cellular extracts from healthy carriers of WT HSP90 strongly suggests that HSP90-SUMO1 expression in the respective carriers is not due to enhanced sumoylation, but rather to the specific inability of SENP2 to desumoylate HSP90-SUMO1 (Figure 5A). Among other possible reasons, this substrate-specific inability could be due to a dislocation of HSP90-SUMO1 to other cellular compartments or by steric inhibition of SENP2 by HSP90-SUMO1.
After paratarg-7, HSP90-SUMO1 is the second example of a frequent autoantigenic paratarg that is posttranslationally modified and in which the posttranslational modification is an autosomal-dominantly inherited risk factor explaining cases of familial MGUS/M/WM. There are, however, differences between the family of hyperphosphorylated paratargs and HSP90-SUMO1: the hyperphosphorylated paratargs completely substitute their WT pendants in all tissues (including erythrocytes) of the respective carriers, while HSP90-SUMO1 is expressed in addition to its WT variant, with a restriction to hematopoietic cells and peripheral B cells being the only cells with a stronger expression of the sumoylated compared with the WT variant in HSP90-SUMO1 carriers.
The fact that HSP90-SUMO1 carriership is only associated with MGUS/MM/WM, but not other malignant or autoimmune diseases, supports a specific role of HSP90-SUMO1 in the pathogenesis of these BCR-expressing malignancies rather than being just an autoimmunogenic epiphenomenon in MGUS/MM/WM. The fact that HSP90-SUMO1–responding B cells at different stages of their development can undergo premalignant or malignant transformation (into IgA/IgG MGUS eventually resulting in MM or IgM MGUS resulting in WM) underlines the strength of this autoantigenic stimulus. With hyperphosphorylated paratarg-7 and HSP90-SUMO1, strong molecularly defined autosomal risk factors are now known in nearly one-third of European and nearly one-half of African-American MGUS/MM/WM patients. This high prevalence of autosomal-dominant inheritance is not in conflict with the observation that first-degree relatives with (manifest) MGUS/MM were found in only 6% of the cases, with a relative risk of a relative of a patient with MGUS/MM ranging between 2.36 and 3.7 (23). Despite the fact that carriership of HSP90-SUMO1 represents a very strong risk factor for developing MGUS/MM, the risk of an HSP90-SUMO1 carrier compared with a noncarrier is 14.8 higher for European, 6.2 for African-American, and 7.4 for Japanese healthy carriers for MGUS/MM/WM. This means that only few carriers develop manifest MGUS/MM/WM, and there might be several generations in a pedigree where one does not find overt MGUS/MM/WM, despite the fact that there is at least one HSP90-SUMO1 carrier in each generation. Therefore, this dominantly inherited risk factor might only be recognized if more than first-degree relatives of MGUS/MM patients with HSP90-SUMO1–specific paraproteins and more than one or two generations are included in such studies.
Our findings not only support the notion that inherited risk factors play an important role in the pathogenesis of MGUS/MM/WM by entertaining chronic antigenic stimulation; they should also facilitate more detailed studies of the role of chronic antigenic stimulation in these diseases and enable genome-wide association studies to identify the genetic variant responsible for the HSP90-SUMO1–specific desumoylation defect in HSP90-SUMO1 carriers.
Methods
Patients and controls.
Materials from patients and healthy persons were obtained during routine diagnostic or therapeutic procedures and stored at –80°C. For healthy donors, the repetition of serum electrophoresis with immunofixation as well as the light-chain ratio excluded the presence of monoclonal Igs in the serum of these individuals (data not shown).
Sumoylation of proteins on macroarray membranes.
High-density protein arrays Unipex 1 and Unipex 2 were obtained from Source Bioscience Life Sciences. In brief, each membrane (Unipex 1 or Unipex 2) consists of 15,300 spotted UniPEx expression clones derived from human fetal brain, T cells, lung, and colon, which, after induction of expression, represent 7,390 distinct recombinant human proteins (for details, see http://www.lifesciences.sourcebioscience.com/products/antibodies/proteomics/protein-arrays/). Expression clones pETE12 and pRHSUMO, functional for the sumoylation machinery in E. coli, were provided by M. Mencía and V. de Lorenzo (24). In detail, the 2 E. coli cultures of the sumoylation machinery were induced for 4 hours with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) before the cells were harvested by centrifugation. Cells were lysed by sonication in 1 ml of 2× PBS plus 1% Triton X-100, 1 mM β-mercaptoethanol, and 1 mM PMSF. Each lysate was centrifuged, and the pellet was discarded. The 2 extracts were mixed 1:2 to get a functional sumoylation-competent mixture (SUMOextract), which was used for sumoylation of membrane-bound proteins. Unipex membranes and controls (RanGAP model clones provided by T. Sixma and K. Schwamborn; ref. 25) spotted on PVDF were incubated for 30 minutes at 37°C with the SUMOextract diluted 1:5 in reaction buffer (50 mM NaCl, 5 mM MgCl2, 0.2 M DTT, 1% BSA, 3% Tween 20, and 5 mM ATP). Nonspecifically bound proteins were then washed off using TBST followed by a second wash with 1% SDS, 0.1 % β-mercaptoethanol, and 100 mM Na2PO4. Nonspecific binding was blocked by incubation with 10% (w/v) nonfat dry milk powder in TBST (TBS, 0.1% [v/v] Tween 20) at 4°C overnight.
Screening of sumoylated high-density protein membranes for immunoreactivity.
Sumoylated membranes were blocked in 10% (w/v) nonfat dry milk powder in TBST at 4°C overnight, washed twice in TBST, and incubated for 1 hour with the paraprotein-containing sera diluted to 1 × 106. After subsequent incubation with biotinylated goat anti-human IgA or anti-human IgG, anti-human Fc, anti-human κ-light chain, anti-human λ-light chain (1:2500; Dako), and Strep-POX (1:15,000) in 2% (w/v) milk/TBST, the filters were washed in TBST, followed by detection using Pharmacia’s ECL System (GE Health Care). Clones with positive signals were obtained from Source Bioscience Life Sciences.
Isoelectric focusing, Western blotting, and coimmunoprecipitation.
The analyses were performed as described (26). The following antibodies were used: anti-HSP90 (1:2000, antibodies-online), anti-SUMO (1:1000, Biozol), and anti-SENP2 (1:1000, antibodies-online).
ELISA.
ELISA was performed as described (26), with the modification that anti-HSP90 (antibodies-online) was coated (4°C overnight) onto Nunc MaxiSorp plates (Nalge Nunc International) before the plates were blocked with 1.5% gelatin/PBS.
BCR cloning and expression.
Bone marrow smears from MM patients were used for the isolation of genomic DNA using a QIAGEN Blood DNA purification kit (QIAGEN). Variable regions of Ig heavy and light chains were amplified as described previously (27). PCR products were sequenced and adapted to pCES vector for expression of His6-tagged proteins in E. coli TG1, as previously described (2). After lysis with PBS, the Fab products were purified by immobilized metal ion affinity (IMAC) chromatography (QIAGEN), concentrated, and stored at –20°C until use.
Competition ELISA.
Nunc MaxiSorp plates were coated with recombinant HSP90-SUMO1, followed by incubation with recombinant His6-tagged anti–HSP90-SUMO1 Fab. Increasing amounts of paraprotein purified by protein A/G chromatography from patients’ sera were added to replace the bound Fab.
Site-directed mutagenesis.
Using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies) and a HSP90-SUMO1 (NM_007355.2) DNA fragment coding for C-terminal FLAG-tagged amino acids 155 to 724, mutants were constructed in which the lysines were changed to arginines. These mutants were stably transfected into HEK293 cells. Fragments of HSP90 were amplified by PCR and HSP90 as template followed by cloning into pSfi-FLAG as expression vector. All primers are listed in Supplemental Methods.
shRNA knockdown of SENP proteins.
For the screen and subsequent knockdown experiments, LCLs were transfected with 1 μg of 1 out of 4 individual SENP shRNA constructs in pSUPER specific for the knockdown of the specific SENP. The primers used are listed in Supplemental Methods.
Expression of SENP in LCLs.
ORFs were amplified by PCR using primers that introduced an EcoRV restriction site before the start codon and replaced the stop codon with another EcoRV site. Blunt-end cloning into the EcoRV site of a subcloning construct (pSfiExpress) then provided a C-terminal FLAG-tag and 2 flanking SfiI sites for further cloning into pRTS constructs. The SfiI fragment was then cloned into the SfiI sites of pRTS (10, 11).
Cell culture and transient transfections.
LCLs were established by infection of PBMCs with EBV and cultured as described previously (28). Transfections and analyses were performed as described (26).
Complementation assay.
PBMCs were transfected with a construct expressing HSP90-FLAG. After 3 days of culture, cells were lysed and inactivated (80°C, 15 minutes). A second healthy blood sample was used for the preparation of native enzyme extracts (lysed in 10 mM Tris pH 8 and centrifuged). Both extracts were mixed and incubated (37°C overnight), followed by analysis by SDS-PAGE and immunodetection using anti-FLAG mAb.
SENP2 expression and activity test.
SENP2 derived from PBMCs was purified using immobilized anti-SENP2 and used for incubation with GST-RanGAP-SUMO prepared as described above. GST-RanGAP-SUMO1 (100 μM) was incubated with SENP2 (50 μM) in 50 mM Tris-HCl pH 7.8/100 μg/ml BSA at 37°C for 3 hours. Reaction was stopped by addition of gel-loading buffer and analyzed by PAGE followed by immunodetection.
Statistics.
Each ELISA experiment was repeated a minimum of twice. Data shown in Figure 3 represent mean value ± SD of 3 independent measurements. In Table 1, the OR, its SEM, and the 95% CI are calculated according to Altman (ref. 29; MedCalc, http://www.medcalc.org/calc/odds_ratio.php). The significance level was P < 0.05.
Study approval.
This study was approved by the local ethical review board (Ethikkommission der τrztekammer des Saarlandes) and conducted according to the Declaration of Helsinki. Recombinant DNA work was performed with the permission and according to the regulations of local authorities (Government of Saarland). Blood and tissue samples from patients and healthy persons were obtained after written, informed consent.
Supplementary Material
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG), Sander Stiftung (a charity organization), and HOMFOR (faculty grant). We thank the patients and their families for participating in this study; Leon Bernal-Mizrachi, John D. Roback, and Susan Sunay (all Atlanta, Georgia, USA) for providing samples from African-American patients and healthy controls; and Shinsuke Iida, Yoshiaki Kuroda, Akira Sakai, and Ryuzo Ueda for providing samples from Japanese patients and healthy controls.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(1):316–323. doi:10.1172/JCI76802.
References
- 1.Preuss KD, et al. A frequent target of paraproteins in the sera of patients with multiple myeloma and MGUS. Int J Cancer. 2009;125(3):656–661. doi: 10.1002/ijc.24427. [DOI] [PubMed] [Google Scholar]
- 2.Grass S, et al. Hyperphosphorylated paratarg-7: a new molecularly defined risk factor for monoclonal gammopathy of undetermined significance of the IgM type and Waldenstrom macroglobulinemia. Blood. 2011;117(10):2918–2923. doi: 10.1182/blood-2010-09-306076. [DOI] [PubMed] [Google Scholar]
- 3.Zwick C, et al. Over one-third of African-American MGUS and multiple myeloma patients are carriers of hyperphosphorylated paratarg-7, an autosomal dominantly inherited risk factor for MGUS/MM. Int J Cancer. 2014;135(4):934–938. doi: 10.1002/ijc.28731. [DOI] [PubMed] [Google Scholar]
- 4.Grass S, et al. Association of a dominantly inherited hyperphosphorylated paraprotein target with sporadic and familial multiple myeloma and monoclonal gammopathy of undetermined significance: a case-control study. Lancet Oncol. 2009;10(10):950–956. doi: 10.1016/S1470-2045(09)70234-7. [DOI] [PubMed] [Google Scholar]
- 5.Preuss KD, et al. Hyperphosphorylation of autoantigenic targets of paraproteins is due to inactivation of PP2A. Blood. 2011;118(12):3340–3346. doi: 10.1182/blood-2011-04-351668. [DOI] [PubMed] [Google Scholar]
- 6.Grass S, et al. Paraproteins of familial MGUS/multiple myeloma target family-typical antigens: hyperphosphorylation of autoantigens is a consistent finding in familial and sporadic MGUS/MM. Blood. 2011;118(3):635–637. doi: 10.1182/blood-2011-01-331454. [DOI] [PubMed] [Google Scholar]
- 7.Pruijn GJ, Wiik A, van Venrooij WJ. The use of citrullinated peptides and proteins for the diagnosis of rheumatoid arthritis. Arthritis Res Ther. 2010;12(1):203. doi: 10.1186/ar2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quarles RH. Myelin-associated glycoprotein (MAG): past, present and beyond. J Neurochem. 2007;100(6):1431–1448. doi: 10.1111/j.1471-4159.2006.04319.x. [DOI] [PubMed] [Google Scholar]
- 9.Barton DE, Arquint M, Roder J, Dunn R, Francke U. The myelin-associated glycoprotein gene: mapping to human chromosome 19 and mouse chromosome 7 and expression in quivering mice. Genomics. 1987;1(2):107–112. doi: 10.1016/0888-7543(87)90002-4. [DOI] [PubMed] [Google Scholar]
- 10.Lanemo Myhrinder A, et al. A new perspective: molecular motifs on oxidized LDL, apoptotic cells, and bacteria are targets for chronic lymphocytic leukemia antibodies. Blood. 2008;111(7):3838–3848. doi: 10.1182/blood-2007-11-125450. [DOI] [PubMed] [Google Scholar]
- 11.Pinder JB, Attwood KM, Dellaire G. Reading, writing, and repair: the role of ubiquitin and the ubiquitin-like proteins in DNA damage signaling and repair. Front Genet. 2013;4:45. doi: 10.3389/fgene.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bologna S, Ferrari S. It takes two to tango: Ubiquitin and SUMO in the DNA damage response. Front Genet. 2013;4:106. doi: 10.3389/fgene.2013.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Hakim A, et al. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst). 2010;9(12):1229–1240. doi: 10.1016/j.dnarep.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135(6 pt 1):1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88(1):97–107. doi: 10.1016/S0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 16.Janka C, Selmi C, Gershwin ME, Will H, Sternsdorf T. Small ubiquitin-related modifiers: a novel and independent class of autoantigens in primary biliary cirrhosis. Hepatology. 2005;41(3):609–616. doi: 10.1002/hep.20619. [DOI] [PubMed] [Google Scholar]
- 17.Cunningham CN, Krukenberg KA, Agard DA. Intra- and intermonomer interactions are required to synergistically facilitate ATP hydrolysis in Hsp90. J Biol Chem. 2008;283(30):21170–21178. doi: 10.1074/jbc.M800046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mollapour M, Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim Biophys Acta. 2012;1823(3):648–655. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pountney DL, Raftery MJ, Chegini F, Blumbergs PC, Gai WP. NSF, Unc-18-1, dynamin-1 and HSP90 are inclusion body components in neuronal intranuclear inclusion disease identified by anti-SUMO-1-immunocapture. Acta Neuropathol. 2008;116(6):603–614. doi: 10.1007/s00401-008-0437-4. [DOI] [PubMed] [Google Scholar]
- 20.Mollapour M, et al. Asymmetric Hsp90 N domain SUMOylation recruits Aha1 ATP-competitive inhibitors. Mol Cell. 2014;53(2):317–329. doi: 10.1016/j.molcel.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11(12):861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8(12):947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg AJ, Rajkumar SV, Vachon CM. Familial monoclonal gammopathy of undetermined significance and multiple myeloma: epidemiology, risk factors, and biological characteristics. Blood. 2012;119(23):5359–5366. doi: 10.1182/blood-2011-11-387324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mencía M, de Lorenzo V. Functional transplantation of the sumoylation machinery into Escherichia coli. Protein Expr Purif. 2004;37(2):409–418. doi: 10.1016/j.pep.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Schwamborn K, et al. SUMO assay with peptide arrays on solid support: insights into SUMO target sites. J Biochem. 2008;144(1):39–49. doi: 10.1093/jb/mvn039. [DOI] [PubMed] [Google Scholar]
- 26.Preuss KD, Fadle N, Regitz E, Held G, Pfreundschuh M. Inactivation of protein-phosphatase 2A causing hyperphosphorylation of autoantigenic paraprotein targets in MGUS/MM is due to an exchange of its regulatory subunits. Int J Cancer. 2014;135(9):2046–2053. doi: 10.1002/ijc.28864. [DOI] [PubMed] [Google Scholar]
- 27.van Dongen JJ, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 28.Neumann F, et al. Identification of an epitope derived from the cancer testis antigen HOM-TES-14/SCP1 presented by dendritic cells to circulating CD4+ T cells. Blood. 2005;106(9):3105–3113. doi: 10.1182/blood-2005-04-1487. [DOI] [PubMed] [Google Scholar]
- 29. Altman DG. Practical Statistics For Medical Research. Chapman & Hall/CRC Texts in Statistical Science (Book 12). London, United Kingdom: Chapman and Hall; 1990. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.