Abstract
Autophagy is a well-conserved catabolic process essential for cellular homeostasis. First described in yeast as an adaptive response to starvation, this pathway is also present in higher eukaryotes, where it is triggered by stress signals such as damaged organelles or pathogen infection. Autophagy is characterized at the cellular level by the engulfment of portions of the cytoplasm in double-membrane structures called autophagosomes. Autophagosomes fuse with lysosomes, resulting in degradation of the inner autophagosomal membrane and luminal content. This process is coordinated by complex molecular systems, including the ATG8 ubiquitin–like conjugation system and the ATG4 cysteine proteases, which are implicated in the formation, elongation, and fusion of these autophagic vesicles. In this Review, we focus on the diverse functional roles of the autophagins, a protease family formed by the four mammalian orthologs of yeast Atg4. We also address the dysfunctional expression of these proteases in several pathologic conditions such as cancer and inflammation and discuss potential therapies based on their modulation.
Introduction
Originally identified as an adaptive response triggered by fasting and other bioenergetic demands in yeast, autophagy (derived from Greek, meaning “self-eating”) was later described as an evolutionarily conserved pathway that is also essential for cellular homeostasis in higher eukaryotes. This catabolic route, in which cytoplasmic components are degraded by lysosomal hydrolases, plays an important role in the response to both extracellular and intracellular stress signals such as damaged organelles, misfolded proteins, or pathogenic infections (1, 2). Dysfunctions in the autophagic pathway have recently been described in several pathologic conditions including inflammation, cancer, and aging-associated diseases (3–8). Given the increasing relevance of autophagy in these processes, modulation of its activity has emerged as a potential therapeutic target, either through activation or inhibition of key components of the pathway.
There are at least three major autophagic pathways, macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA), which primarily differ in the method by which the cargo is transferred to the lysosome (9). In microautophagy the lysosomal membrane directly engulfs cytoplasmic components (10), whereas in CMA, cytoplasmic proteins are selectively delivered to the lysosome in a process dependent on the recognition of a sequence motif in the target protein by specific lysosomal receptors (11). Macroautophagy (herein referred to as autophagy) involves the formation and elongation of a membrane sac called the phagophore that develops into a double-membrane vesicle, the autophagosome, in which the cytoplasmic cargo is sequestered. Autophagosomes eventually fuse with lysosomes, allowing for the degradation of the cargo and the autophagosome’s inner membrane. Once degraded, the contents of the autophagosome is returned to the cytoplasm as basic building blocks (mainly amino acids, lipids, sugars, and nucleotides) that can be reused by the cell to synthesize new biomolecules (12). Autophagy execution requires the activation of complex molecular machinery that includes regulatory signaling cascades, the formation and completion of autophagosomes, the transport of autophagosomes, and final fusion of autophagosomes with lysosomes (13). Although the intricate molecular machinery of this multistep pathway is not yet fully understood, some of its components have been identified as essential effectors required for a correct autophagic response. This is the case of proteins such as Atg5, Atg7, or the ubiquitin-like protein Atg8 and its conjugation system, which is indispensable for the correct expansion and closure of the preautophagosomal double membrane.
Proteases have recently been described as major signaling pathway initiators due to their ability to perform highly regulated proteolytic processing reactions for a variety of protein substrates (14, 15). In this regard, some proteolytic enzymes are known to indirectly modulate the autophagic response. Cleavage of the autophagy regulator beclin 1 (the mammalian ortholog of yeast Atg6) by different caspases abrogates its proautophagic activity, while the fragment resulting from this cleavage acquires proapoptotic activity (16, 17). Likewise, calpain-mediated processing of ATG5 switches autophagy to apoptosis in different cell types (18). Several caspases and calpains also have the ability to cleave in vitro a wide range of autophagic proteins that could be implicated in the molecular crosstalk between apoptosis and autophagy (19). Among these cell death proteases, caspase-3 targets the Crohn’s disease T300A variant of ATG16L1 and causes defective autophagy, which in turn contributes to disease progression by sustaining cellular stress and facilitating pathogen expansion (20). Reversible ubiquitylation, a process involving different proteases such as proteasome components and deubiquitinating enzymes (DUBs) (21), also contributes to selective autophagy of proteins and organelles (22–25). In fact, DUBs have been shown to control selective autophagy levels in basal conditions (26–28), and their activity could be crucial in different pathologies such as Parkinson’s disease and pathogen infections associated with autophagy dysfunction (29, 30). Additionally, a large number of lysosomal proteases are implicated in the final degradative stages of the autophagic process (31). However, there is only one protease among the numerous Atg proteins originally identified in yeast and directly involved in autophagy: the cysteine protease Atg4, whose function is essential for the Atg8 conjugation system (32).
In this Review we focus on this Atg4 protease and its four mammalian orthologs, termed “autophagins,” which have been identified in multiple species for which genome sequence data are available (33). We also address the functional relevance of this protein family in physiologic and pathologic processes and discuss the emerging importance of ATG4 proteins as potential targets of new therapeutic strategies in human diseases.
The yeast cysteine protease Atg4
The Atg8 ubiquitin-like conjugation system holds a pivotal role in the proper expansion of the phagophore (Figure 1). In yeast, Atg8 is synthesized as an inactive soluble protein and needs to be posttranslationally processed in order to be covalently attached to a molecule of phosphatidylethanolamine (PE) from the preautophagosomal membrane (13). This activation process starts with the cleavage of its C-terminal region by the cysteine protease Atg4 (34), exposing a glycine residue that is essential for subsequent interactions, including the final conjugation with the amino group of PE (Figure 1). Once Atg8 is activated, it undergoes several reactions similar to those involved in protein ubiquitylation, requiring the participation of additional autophagic proteins such as Atg7 (E1-ubiquitin ligase–like enzyme), Atg3 (E2-ubiquitin ligase–like enzyme), and the Atg12-Atg5-Atg16 complex (E3-like enzyme), which finally promotes the formation of the Atg8-PE amide bond. Atg8-PE recruits the Atg12-Atg5-Atg16 complex to the membrane, assembling a scaffold that is critical for phagophore biogenesis (35). This conjugation reaction is essential to retain Atg8 on the expanding structure, making membrane tethering and hemifusion possible (36).
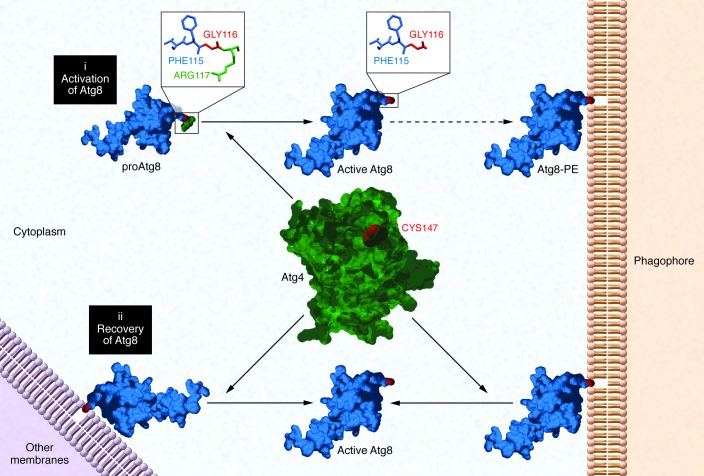
Figure 1. Atg4 functions.
(i) Yeast Atg4 is a cysteine protease required for the activation of immature Atg8 protein, cleaving its C-terminal region and exposing a glycine residue (GLY116) that is essential for its attachment to the phagophore. (ii) Atg4 is also responsible for the recovery of Atg8 via delipidation of this protein from mature autophagosomes or organelle membranes, where it can be erroneously attached.
The lipidation of Atg8 is a reversible process because the protein can be deconjugated and released back to the cytosol (34). Interestingly, the recovery of Atg8 is also Atg4 dependent, as this protease is able to cleave the amide bond with PE, disassembling the scaffold (ref. 35 and Figure 1). This additional role of Atg4 has been described as an important process for the dynamism of autophagy, as it provides a new source of active Atg8 to maintain a longer autophagic response (37). Moreover, it allows the recycling of Atg8 molecules that have been attached to inappropriate membranes (38). Finally, the release of Atg8 from mature autophagosomes facilitates the fusion of these structures with lysosomes, allowing the completion of the autophagic process (39).
The mammalian Atg4 family
Autophagy is evolutionarily well conserved, and the Atg8 ubiquitin–like conjugation system has been described in higher eukaryotes (40–42). Accordingly, four mammalian orthologs of yeast protease Atg4 were identified and cloned in our laboratory, which led us to define the autophagin protease family (ref. 42 and Figure 2). These four proteases were called autophagin-1/ATG4B, autophagin-2/ATG4A, autophagin-3/ATG4C, and autophagin-4/ATG4D and contained all the residues required for the catalytic activity of cysteine proteases, including the conserved cysteine residue within the catalytic site. Intriguingly, the mammalian ATG8 protein family of putative ATG4 substrates is more complex; it is divided into two main subfamilies that differ in both structural features and functional role in the formation of the autophagosome (43). Thus, the microtubule-associated protein 1–light chain 3 (MAP1-LC3) subfamily (including MAP1-LC3A, MAP1-LC3B, and MAP1-LC3C) is involved in the elongation of the initial phagophore, while the GATE-16/GABARAP subfamily (including GABARAP, GABARAPL1/ATG8L, and GABARAPL2/GATE-16) is required during the final maturation of the double-membrane vesicle. Notably, the latter subfamily is preferred by the Unc-51–like autophagy-activating kinase (ULK) complex as the scaffold necessary for its recruitment in the phagophore, an essential step for the efficient formation and maturation of the autophagosome (44).
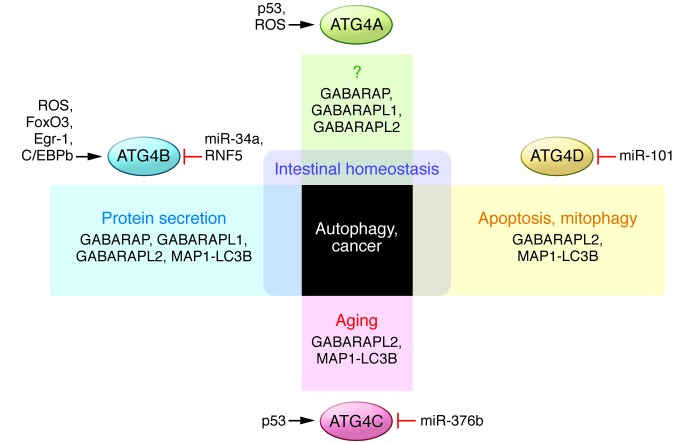
Figure 2. Roles and regulation of the ATG4 protease family.
The complexity of the mammalian ATG8 protein system is reflected in the substrate specificity of ATG4 proteases and their particular roles. Besides the autophagic response, all ATG4 proteases have been involved in cancer, and defects in ATG4A, ATG4B, and ATG4D are also linked to intestinal disorders. Variants of ATG4C have been identified in aging free of major diseases, and ATG4D might be essential in the crosstalk between autophagy and apoptosis and is required for correct mitophagy during erythropoiesis. ATG4B has been widely described as necessary for the proper function of secretory cells. These proteases must be finely controlled by different regulators to coordinate this wide range of functions. Thus transcription factors such as p53, FOXO3, EGR11, or C/EBPb and several microRNAs such as miR-101, miR-376b, and miR-34a are responsible for the modulation of Atg4 genes. Additionally, ATG4 proteases can also be posttranslationally regulated by ROS-dependent oxidation or by E3 ubiquitin ligases such as RNF5.
Additionally, these Atg8 orthologs are differentially cleaved by the different Atg4 family members, with ATG4B representing the most potent and promiscuous of all of them in terms of substrate specificity (ref. 45 and Figure 2). In fact, ATG4B is able to process a wide range of Atg8 orthologs (46), and the analysis of their structures has been very useful to understand the interaction between both enzyme and substrate (47–50). ATG4A has also been described as an effective protease with the ability to target GABARAPL2 (51), although it is unable to cleave MAP1-LC3B, which is one of the most important orthologs of yeast Atg8. Finally, ATG4C and ATG4D efficiently bind these substrates but show reduced catalytic activity against them, as they can only process MAP1-LC3B and GABARAPL2. In fact, human ATG4D must be cleaved by caspase-3 to increase its activity against GABARAPL1 (52). ATG4C also presents the canonical cleavage site DEVD, which allows its caspase-mediated activation in vitro (19). However, ATG4A and ATG4B lack this motif and do not seem to require any caspase-mediated cleavage for their activation, which could explain their high proteolytic activity compared with that of other ATG4 family members. Notably, the cleavage of ATG4D exposes a BH3-like domain that contributes to the recruitment of the protein to the mitochondria, where it induces apoptosis. Thus, ATG4D could be a crucial component in the crosstalk between autophagy and apoptosis, while it has also been proposed to be involved in the mitochondrial clearance by mitophagy during erythropoiesis (53, 54).
The complex ATG8 conjugation system observed in mammals, formed by several orthologs of both Atg4 and Atg8 proteins, points to the existence of a complex network of protease-substrate interactions with specific spatiotemporal modulation at different levels, although the specific regulatory mechanisms are not yet fully understood (55). Interestingly, the binding between an ATG4 autophagic protease and its corresponding substrate seems to be the main regulator of its activity. It has been shown that ATG4B is autoinhibited in its free form by two domains that hide the catalytic cysteine: a regulatory loop that prevents the entry of the substrate, and the N-terminal tail that blocks its exit (49). However, the interaction of the ATG4 enzyme with MAP1-LC3 induces conformational changes that displace the loop and open the tail, uncovering the active site and permitting the cleavage of the substrate (48). Moreover, this open conformation of ATG4B can be stabilized by the interaction of the N-terminal tail with adjacent non-substrate MAP1-LC3, facilitating the membrane targeting of this protease required for MAP1-LC3 delipidation. This additional binding is mediated by the LC3-interacting region present in the N-terminal tail of ATG4B (56).
Transcriptionally, ATG4 proteases are part of intricate signal transduction pathways, complicating the identification of transcription factors with the ability to induce their expression (Figure 2). In this regard, it has been shown that p53 can bind and regulate Atg4a and Atg4c (among other several Atg genes such as Atg7, Atg10, Ulk1, and Ulk2) in response to DNA damage, inducing an autophagic response that could contribute to tumor suppression by enhancing p53-dependent apoptosis (57). Moreover, FoxO3 has been described to upregulate Atg4b and other autophagic genes in mouse skeletal muscle (58, 59). FOXO3a can also regulate ATG4 genes in ovarian cancer cells, although it must be dephosphorylated and retained in the nucleus after inhibition of PI3K and Ras/MAPK signaling pathways by the tumor suppressor AHRI (DIRAS3) (60, 61). Expression of human ATG4B also depends on tissue-specific transcription factors, including EGR1 in lung tissue (62) and C/EBPb in differentiating murine 3T3-L1 adipocytes (63). Notably, the activity of the different ATG4 family members can be fine-tuned by microRNAs after transcription. The tumor suppressor miR-101 inhibits autophagy by targeting ATG4D (64), miR-376b modulates human autophagy by regulating intracellular levels of ATG4C (65), and miR-34a targets ATG4B (66).
It has been proposed that ROS may also regulate ATG4 proteins (Figure 2). In fact, hydrogen peroxide can directly inhibit the delipidating activity of ATG4A and ATG4B by oxidation of a non-catalytic residue near the active site (67). Accumulation of H2O2 after amino acid starvation has been described to promote ATG8 lipidation while preventing its release from the autophagosome. ROS can also be generated in autophagosomes and lysosomes, reinforcing the hypothesis of a ROS-mediated protection of ATG8-PE in the autophagosome (68), while recycling of additional ATG8 molecules from other membranes is still possible in the reducing environment of the cytosol. Thus, the oxidative regulation of ATG4 proteases might limit their activities to specific areas, allowing for the proper initiation and completion of autophagy. In vascular smooth muscle, for example, 7-ketocholesterol induces the expression of NADPH oxidase 4 and increases H2O2 levels that inhibit ATG4B delipidating activity, maintaining the autophagic response triggered by ER stress or atherosclerosis (69). The modification of catalytic or non-catalytic residues (including those responsible for the interaction between protease and substrate that, once oxidized, can alter the structural conformation of the enzyme) controls the ability of these proteins to cleave substrates at specific sites under different conditions.
Additional effectors might also contribute to the spatiotemporal regulation of ATG4 proteins (Figure 2). The E3 ligase RNF5 spatially controls ATG4B stability by inducing its ubiquitination and proteasome-mediated degradation at membrane domains like the phagophore. This mechanism thus represents a new regulatory layer of basal autophagy, blocking LC3 priming at the beginning of the process (70). In yeast, an Atg18-Atg21 complex is a key regulator during the phagophore formation (71), recruiting and protecting Atg8-PE. This new scaffold could also be present in higher organisms, acting as a barrier to ATG4 that prevents the access of the enzyme to the substrate until the autophagosome is mature, when the complex disassociates and allows the recycling of ATG8.
Given the wide diversity of orthologs and activities described in the mammalian Atg8 system, it has been hypothesized that Atg4 autophagic proteases may be involved in different physiologic processes. Historically, knockout mice have been research tools with important limitations for global in vivo studies on autophagy, as proteins like ATG5 or ATG7 are essential for the viability of these animals (72, 73). However, the apparent functional redundancy of the ATG4 family has made possible the generation of Atg4-deficient mice, which exhibit impaired autophagic flux but are perfectly viable. Accordingly, these animals represent excellent models to assess the in vivo roles of autophagy. By using Atg4c-deficient mice, we have found that under prolonged periods of starvation, ATG4C is necessary for maximal autophagy activation in tissues dependent on continuous energy consumption, such as the diaphragm (74). We have also found that mutant mice lacking ATG4B show altered secretion and assembly of otoconial components in the inner ear vestibular regions, ultimately leading to balance disorders (75, 76). Moreover, Atg4b-deficient mice show impaired release of lysozyme granules in Paneth cells during dextran sodium sulfate–induced (DSS-induced) experimental colitis, resulting in exacerbated inflammation that leads to the death of these animals (77). This surprising role of ATG4B in protein secretion has also been described in bone resorption by osteoclasts (78), pointing to a novel function of autophagy in secretory cells (79) and further supporting the idea that ATG proteins are involved in non-canonical autophagic processes (80–82). Similarly to the described intestinal phenotype, Atg4b-deficient mice subjected to an endotoxemia model show increased mortality and lung inflammation caused by sequestration of the antiinflammatory transcription factor ATF3 (83). Paradoxically, autophagy impairment in these mutant mice ameliorates the inflammatory response to mechanical ventilation and decreases lung injury by blockade of the NF-κB pathway (84), indicating that autophagic functions of ATG4 proteases in pathologic conditions may be dual and highly dependent on the cell and tissue context.
Dysregulation of Atg4 proteases in disease
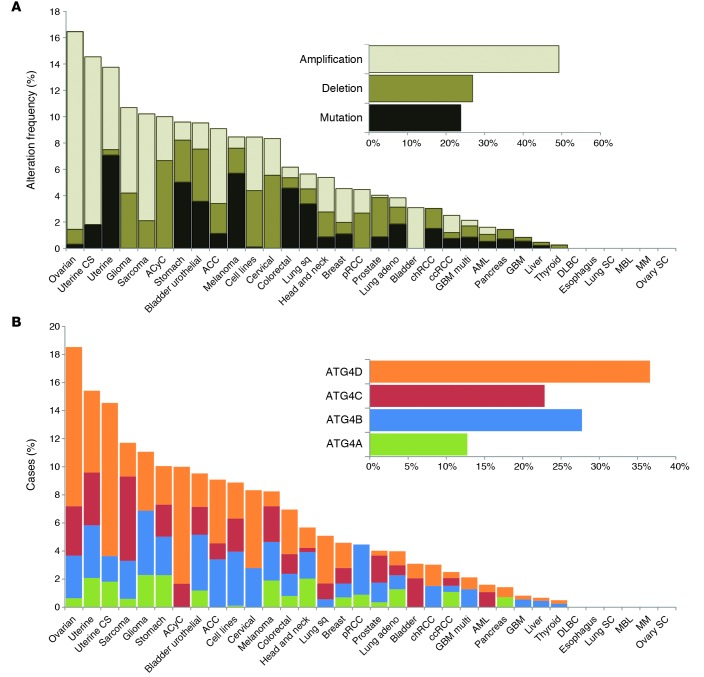
Dysfunctional autophagic responses have been reported in several pathologies, including cancer and pathogen infection (1–5). In some cases, this aberrant autophagy is associated with a dysregulation of members of the ATG4 family during the development or progression of these diseases. As for the relevance of ATG4 proteases in cancer, different structural alterations in the genes encoding these enzymes have been found in several malignancies, with copy number amplification representing the most common of these modifications (Figure 3A). Furthermore, a large proportion of these alterations have been detected in female reproductive tissue tumors, including ovarian serous cystadenocarcinoma and uterine cancers. Intriguingly, hypomethylation of ATG4A in ovarian tumor–initiating cells increases their stem properties and is linked to poor prognosis in ovarian cancer patients (85). ATG4A expression has also been reported to be essential for breast cancer stem cells, regulating their tumorigenicity in vivo (86). Nevertheless, ATG4D seems to be the most frequently altered ATG4 gene in human cancer, followed by ATG4B and ATG4C (Figure 3B). ATG4B overexpression has been described in linearly patterned programmed cell necrosis, a special type of cell death observed during early tumor growth of aggressive melanomas (87). Even though there is substantial genetic evidence, little is known about the specific roles of these autophagic proteases in cancer progression. Loss of Atg4c in a chemically induced murine model of fibrosarcoma suggests a tumor suppressor role for this protease (74). Nevertheless, further in vivo studies with other Atg4-deficient models are needed, especially in light of the dual role of autophagy in cancer, either promoting cancer cell survival or suppressing tumorigenesis by maintaining cellular homeostasis in a context-dependent manner (88).
Figure 3. ATG4 proteases in human cancer.
(A) Structural alterations of ATG4 genes in several malignancies. (B) Percentage of genetic alterations in the different ATG4 genes found in human tumors. ACC, adrenocortical carcinoma; ACyC, adenoid cystic carcinoma; adeno, adenocarcinoma; AML, acute myeloid leukemia; ccRCC, kidney renal clear cell carcinoma; chRCC, kidney chromophobe; CS, carcinosarcoma; DLBC, lymphoid neoplasm diffuse large B cell lymphoma; GBM, glioblastoma; GBM multi, GBM multiforme; MBL, medulloblastoma; MM, multiple myeloma; pRCC, kidney renal papillary cell carcinoma; SC; small cell carcinoma; Sq, squamous cell carcinoma. Data were obtained from cBioPortal database (133, 134), and those human tumors with structural alterations in ATG4 genes were compiled and used to construct both graphs.
ATG4 mutations are also linked to some inflammatory bowel diseases (IBDs), as genetic variants in ATG4A and ATG4D have been proposed to contribute to granuloma formation in Crohn’s disease (89).
Additionally, patients with IBD show low expression of ATG4B in colon (77). These observations of altered ATG4 function in IBD are consistent with the fact that disrupted autophagy results in impaired processing of bacterial components, triggering the exacerbated inflammatory responses that characterize these disorders (90–94).
Autophagic proteins, including several ATG4 proteases, have also been implicated in the pathogenesis of infectious diseases (3). This is the case for HIV infection, which induces the expression of ATG4D and other ATG genes during viral morphogenesis and propagation (95). Hepatitis C virus is another example of a pathogen that evades and exploits autophagy, using autophagic components such as ATG4B to initiate its replication (96). Infections by parasites including Trypanosoma cruzi, which causes Chagas disease, or Leishmania major, responsible for leishmaniasis, are different from those caused by other pathogens, as these eukaryotic organisms also utilize autophagy (97). In fact, the activity of their Atg4 orthologs is essential for their survival, differentiation, and virulence, and new therapeutic strategies targeting these proteases could block or slow the infection (98–101).
Consistent with the growing relevance of autophagy in the modulation of aging (6, 8, 102, 103), it is remarkable that ATG4C variants have been linked to both aging in the absence of major diseases and increased longevity in a GWAS (104).
Therapeutic options targeting Atg4 proteases
The increasing evidence that dysregulation of ATG4 proteins occurs in a number of different diseases has opened new possibilities for the development of therapies targeting these proteases. Moreover, the essential role of ATG4 proteases in autophagy suggests that their chemical regulation may help to control the autophagic response in some contexts. In fact, ATG4 levels are correlated with autophagic flux, and synthetic substrates have been developed to measure ATG4 activity and monitor autophagy both in cultured cells and in vitro (105, 106). Similar approaches have recently been used to screen for specific inhibitors of these enzymes, which could be useful for the treatment of pathologic conditions associated with excess autophagic protease activity (107, 108). Another strategy to inhibit autophagy, either for research or therapeutic purposes, is the utilization of inactive mutant forms of ATG4 proteases with the ability to sequester their corresponding substrates (109–111).
In recent years, attenuation of ATG4 protease activity in different contexts has produced promising results. Overexpression of an inactive mutant of ATG4B in hepatocellular carcinoma cells reduced their viability (112), and autophagy inhibition by miR-101, which targets ATG4D, enhanced cisplatin-induced apoptosis in these tumor cells (113). Disruption of the autophagic response has also been proposed as a therapeutic option in cisplatin-resistant patients with squamous cell carcinoma, in which ATG4A is upregulated (114), and chronic myeloid leukemia, in which ATG4B expression is involved in resistance of CD34+ cells to imatinib mesylate (66). Apart from the interest in ATG4 inhibition in cancer, a treatment based on the blockade of these proteases has also been suggested for type 1 diabetes mellitus, as some autophagic proteins (including ATG4A) could be related to neural injury of young patients with early neuronal deficits and diabetic ketoacidosis (115).
Strategies based on the induction of ATG4 autophagic proteases have also provided relevant results in several cellular models. In fact, treatment of human breast carcinoma MCF-7 cells with BBP (N-benzoyl-O-[N′-(1-benzyloxycarbonyl-4-piperidiylcarbonyl)-d-phenylalanyl]- d-phenylalaninol), a novel asperphenamate derivative, requires the upregulation of ATG4B activity (116). This modulation is JNK dependent and involves the regulation of ATG4 proteins by ROS. Overexpression of these proteases is also a potential therapeutic strategy in lung infections such as chronic obstructive pulmonary disease (62), ischemia and reperfusion injury in liver and heart (117, 118), and neurodegenerative diseases such as Huntington’s disease (119), in which in vitro stimulation of the autophagic response ameliorates the effects of this pathologic condition. However, the role of ATG4 proteases in most diseases is context dependent, and the consequences of their modulation could differ greatly. For example, it has been suggested that changes in the expression of ATG4B in prostate cancer could either amplify the action of both chemotherapy and radiotherapy or contribute to the development of treatment resistance (120). Furthermore, inhibition of ATG4 proteases in radiation therapies sensitizes resistant carcinoma cells in most cases, although it also promotes resistance in some conditions (121), demonstrating the importance of a more comprehensive understanding of ATG4 function.
Nevertheless, the development of new therapies based on the modulation of ATG4 proteases is still at a very preliminary stage. To date, no modulators of these enzymes have been successfully tested in clinical trials, and specific inhibitors for them are still being characterized (122). This could be partially due to the fact that autophagins are cysteine proteases, which have been historically difficult to target when compared with other proteases, due to the metabolic instability and lack of specificity of small-molecule drugs (123). Moreover, our current knowledge of the activity and regulation of ATG4 proteases is still limited, and specific modulators should be able to discriminate between distinct ATG4 enzymes in order to avoid unexpected effects. Ideally, priming and delipidation steps should also be specifically targeted, although we still do not fully understand these processes. Consequently, new genetic and proteomic approaches will be required for the development of ATG4-based treatments.
Conclusions and perspectives
Autophagy is a well-conserved pathway that has gained functional complexity throughout evolution. The identification of four mammalian orthologs of yeast protease Atg4 has helped to dissect the complex ATG8 conjugation system present in mammals, which consists of a large number of substrate orthologs of these proteolytic enzymes. This complexity contrasts with other key autophagic components such as Atg3, Atg5, or Atg7, for which unique genes have been described in mammals. The reasons behind this genetic redundancy in mammalian orthologs of yeast Atg4 and Atg8 remain elusive, but it may reflect the existence of specific functions for these proteins that are not limited to the canonical autophagic response observed in yeast. In fact, members of the GATE-16/GABARAP subfamily were first described in membrane trafficking processes (124–126), and MAP1-LC3B was initially characterized because of its interaction with microtubule-associated proteins MAP1A and MAP1B (127, 128). Accordingly, mammalian ATG4 autophagic proteases are also involved in several physiologic processes distinct from macromolecular recycling, including protein secretion and apoptosis.
Dysregulation of the activity of these cysteine proteases has been associated with several diseases such as cancer, inflammatory disorders, and vertigo, supporting their importance as emerging therapeutic targets. However, many questions remain concerning the specific functions of ATG4 proteases, including their potential dual roles in cancer, which can complicate future strategies aimed at targeting these enzymes in human malignancies. Moreover, expression of the different ATG4 family members depends on intricate signaling pathways, and their specific spatiotemporal regulation remains largely unknown. Further understanding of this protein family is still necessary to develop efficient treatments while avoiding undesired side effects. In this regard, the generation and characterization of additional gain- or loss-of-function animal models would be a valuable tool for the study of these proteases. Mutant mice lacking some of these enzymes such as ATG4B and ATG4C are viable due to the functional redundancy among members of this protease family, thereby resulting in very useful models to analyze the role of autophagy in vivo (129, 130). Atg4b-deficient mice, for example, show minor basal autophagy defects but impaired induced autophagic response after diverse stress signals. This alteration is comparable to that described in several pathologies in which the pathway is not completely disrupted but attenuated (131), which reinforces the value of these mutant mice as models of human diseases with autophagy deficiency. Furthermore, the generation of specific ATG4 inhibitors is currently ongoing and may contribute to the development of valuable tools to clarify the roles of these proteolytic enzymes in health and disease (132). Thus, ATG4 proteases are emerging as potential pharmaceutical targets for the treatment of dysfunctional autophagy or specific alterations involving these enzymes, but additional efforts will be needed to elucidate their role in physiologic and pathologic processes and to develop new therapies for human diseases associated with dysregulation of ATG4 proteolytic enzymes.
Acknowledgments
We are grateful to José M.P. Freije and Guillermo Mariño for their helpful comments. The work in our laboratory is supported by grants from Ministerio de Economía y Competitividad and Red Temática de Investigación Cooperativa en Cáncer – Instituto de Salud Carlos III. The Instituto Universitario de Oncología is supported by Obra Social Cajastur-Asturias. C. López-Otín is an Investigator of the Botin Foundation.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(1):33–41. doi:10.1172/JCI73940.
References
- 1.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(7):651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 2.Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157(1):65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baixauli F, López-Otín C, Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol. 2014;5:403. doi: 10.3389/fimmu.2014.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider JL, Cuervo AM. Autophagy and human disease: emerging themes. Curr Opin Genet Dev. 2014;26C:16–23. doi: 10.1016/j.gde.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Marino G, Lopez-Otin C. Autophagy: molecular mechanisms, physiological functions and relevance in human pathology. Cell Mol Life Sci. 2004;61(12):1439–1454. doi: 10.1007/s00018-004-4012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahu R, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20(1):131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22(8):407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13(5):495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2):124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turk B, Turk D, Turk V. Protease signalling: the cutting edge. EMBO J. 2012;31(7):1630–1643. doi: 10.1038/emboj.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Otin C, Hunter T. The regulatory crosstalk between kinases and proteases in cancer. Nat Rev Cancer. 2010;10(4):278–292. doi: 10.1038/nrc2823. [DOI] [PubMed] [Google Scholar]
- 16.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene. 2010;29(12):1717–1719. doi: 10.1038/onc.2009.519. [DOI] [PubMed] [Google Scholar]
- 18.Yousefi S, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8(10):1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 19.Norman JM, Cohen GM, Bampton ET. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy. 2010;6(8):1042–1056. doi: 10.4161/auto.6.8.13337. [DOI] [PubMed] [Google Scholar]
- 20.Murthy A, et al. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506(7489):456–462. doi: 10.1038/nature13044. [DOI] [PubMed] [Google Scholar]
- 21.Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31(19):2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 22.Schreiber A, Peter M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim Biophys Acta. 2014;1843(1):163–181. doi: 10.1016/j.bbamcr.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 23.Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20(1):21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuang E, Qi J, Ronai Z. Emerging roles of E3 ubiquitin ligases in autophagy. Trends Biochem Sci. 2013;38(9):453–460. doi: 10.1016/j.tibs.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 26.Rott R, et al. α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc Natl Acad Sci U S A. 2011;108(46):18666–18671. doi: 10.1073/pnas.1105725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornelissen T, et al. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet. 2014;23(19):5227–5242. doi: 10.1093/hmg/ddu244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bingol B, et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510(7505):370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 29.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 30.Mesquita FS, Thomas M, Sachse M, Santos AJ, Figueira R, Holden DW. The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog. 2012;8(6):e1002743. doi: 10.1371/journal.ppat.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Repnik U, Stoka V, Turk V, Turk B. Lysosomes and lysosomal cathepsins in cell death. Biochim Biophys Acta. 2012;1824(1):22–33. doi: 10.1016/j.bbapap.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Kaminskyy V, Zhivotovsky B. Proteases in autophagy. Biochim Biophys Acta. 2012;1824(1):44–50. doi: 10.1016/j.bbapap.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Quesada V, Ordonez GR, Sanchez LM, Puente XS, Lopez-Otin C. The Degradome database: mammalian proteases and diseases of proteolysis. Nucleic Acids Res. 2009;37(Database issue):D239–D243. doi: 10.1093/nar/gkn570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirisako T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151(2):263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaufmann A, Beier V, Franquelim HG, Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156(3):469–481. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 36.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130(1):165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 37.Nair U, et al. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy. 2012;8(5):780–793. doi: 10.4161/auto.19385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakatogawa H, Ishii J, Asai E, Ohsumi Y. Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy. 2012;8(2):177–186. doi: 10.4161/auto.8.2.18373. [DOI] [PubMed] [Google Scholar]
- 39.Yu ZQ, et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy. 2012;8(6):883–892. doi: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu F, Li Y, Wang F, Noda NN, Zhang H. Differential function of the two Atg4 homologues in the aggrephagy pathway in Caenorhabditis elegans. J Biol Chem. 2012;287(35):29457–29467. doi: 10.1074/jbc.M112.365676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshimoto K, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16(11):2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278(6):3671–3678. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 43.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29(11):1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alemu EA, et al. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem. 2012;287(47):39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286(9):7327–7338. doi: 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278(51):51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 47.Kumanomidou T, et al. The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome-forming modifiers. J Mol Biol. 2006;355(4):612–618. doi: 10.1016/j.jmb.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 48.Satoo K, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28(9):1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J Biol Chem. 2005;280(48):40058–40065. doi: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- 50.Hurley JH, Schulman BA. Atomistic autophagy: the structures of cellular self-digestion. Cell. 2014;157(2):300–311. doi: 10.1016/j.cell.2014.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scherz-Shouval R, Sagiv Y, Shorer H, Elazar Z. The COOH terminus of GATE-16, an intra-Golgi transport modulator, is cleaved by the human cysteine protease HsApg4A. J Biol Chem. 2003;278(16):14053–14058. doi: 10.1074/jbc.M212108200. [DOI] [PubMed] [Google Scholar]
- 52.Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122(pt 14):2554–2566. doi: 10.1242/jcs.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Betin VM, MacVicar TD, Parsons SF, Anstee DJ, Lane JD. A cryptic mitochondrial targeting motif in Atg4D links caspase cleavage with mitochondrial import and oxidative stress. Autophagy. 2012;8(4):664–676. doi: 10.4161/auto.19227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Betin VM, Singleton BK, Parsons SF, Anstee DJ, Lane JD. Autophagy facilitates organelle clearance during differentiation of human erythroblasts: evidence for a role for ATG4 paralogs during autophagosome maturation. Autophagy. 2013;9(6):881–893. doi: 10.4161/auto.24172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Klionsky DJ.The regulation of autophagy — unanswered questions. J Cell Sci. 2011. 124pt 2161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wild P, McEwan DG, Dikic I. The LC3 interactome at a glance. J Cell Sci. 2014;127(pt 1):3–9. doi: 10.1242/jcs.140426. [DOI] [PubMed] [Google Scholar]
- 57.Kenzelmann Broz D, et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27(9):1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 59.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Lu Z, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118(12):3917–3929. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu Z, et al. ARHI (DIRAS3) induces autophagy in ovarian cancer cells by downregulating the epidermal growth factor receptor, inhibiting PI3K and Ras/MAP signaling and activating the FOXo3a-mediated induction of Rab7. Cell Death Differ. 2014;21(8):1275–1289. doi: 10.1038/cdd.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen ZH, et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS One. 2008;3(10):e3316. doi: 10.1371/journal.pone.0003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo L, et al. Transactivation of Atg4b by C/EBPβ promotes autophagy to facilitate adipogenesis. Mol Cell Biol. 2013;33(16):3180–3190. doi: 10.1128/MCB.00193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frankel LB, et al. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011;30(22):4628–4641. doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8(2):165–176. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 66.Rothe K, et al. The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood. 2014;123(23):3622–3634. doi: 10.1182/blood-2013-07-516807. [DOI] [PubMed] [Google Scholar]
- 67.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26(7):1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kubota C, et al. Constitutive reactive oxygen species generation from autophagosome/lysosome in neuronal oxidative toxicity. J Biol Chem. 2010;285(1):667–674. doi: 10.1074/jbc.M109.053058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He C, et al. 7-Ketocholesterol induces autophagy in vascular smooth muscle cells through Nox4 and Atg4B. Am J Pathol. 2013;183(2):626–637. doi: 10.1016/j.ajpath.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuang E, et al. Regulation of ATG4B stability by RNF5 limits basal levels of autophagy and influences susceptibility to bacterial infection. PLoS Genet. 2012;8(10):e1003007. doi: 10.1371/journal.pgen.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nair U, Cao Y, Xie Z, Klionsky DJ. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J Biol Chem. 2010;285(15):11476–11488. doi: 10.1074/jbc.M109.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 73.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282(25):18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 75.Marino G, et al. Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120(7):2331–2344. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cabrera S, Marino G, Fernandez AF, Lopez-Otin C. Autophagy, proteases and the sense of balance. Autophagy. 2010;6(7):961–963. doi: 10.4161/auto.6.7.13065. [DOI] [PubMed] [Google Scholar]
- 77.Cabrera S, et al. ATG4B/autophagin-1 regulates intestinal homeostasis and protects mice from experimental colitis. Autophagy. 2013;9(8):1188–1200. doi: 10.4161/auto.24797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeSelm CJ, et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011;21(5):966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Till A, Subramani S. A balancing act for autophagin. J Clin Invest. 2010;120(7):2273–2276. doi: 10.1172/JCI43238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013;14(2):143–151. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hwang S, et al. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon γ. Cell Host Microbe. 2012;11(4):397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15(7):713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aguirre A, et al. Defective autophagy impairs ATF3 activity and worsens lung injury during endotoxemia. J Mol Med (Berl). 2014;92(6):665–676. doi: 10.1007/s00109-014-1132-7. [DOI] [PubMed] [Google Scholar]
- 84.Lopez-Alonso I, et al. Impairment of autophagy decreases ventilator-induced lung injury by blockade of the NF-κB pathway. Am J Physiol Lung Cell Mol Physiol. 2013;304(12):L844–L852. doi: 10.1152/ajplung.00422.2012. [DOI] [PubMed] [Google Scholar]
- 85.Liao YP, et al. Hypomethylation signature of tumor-initiating cells predicts poor prognosis of ovarian cancer patients. Hum Mol Genet. 2014;23(7):1894–1906. doi: 10.1093/hmg/ddt583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolf J, et al. A mammosphere formation RNAi screen reveals that ATG4A promotes a breast cancer stem-like phenotype. Breast Cancer Res. 2013;15(6):R109. doi: 10.1186/bcr3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han C, et al. A pilot study on morphology and the mechanism involved in linearly patterned programmed cell necrosis in melanoma. Oncol Lett. 2010;1(5):821–826. doi: 10.3892/ol_00000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brinar M, et al. Genetic variants in autophagy-related genes and granuloma formation in a cohort of surgically treated Crohn’s disease patients. J Crohns Colitis. 2012;6(1):43–50. doi: 10.1016/j.crohns.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 90.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8(6):458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 91.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39(2):207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 92.Henckaerts L, et al. Genetic variation in the autophagy gene ULK1 and risk of Crohn’s disease. Inflamm Bowel Dis. 2011;17(6):1392–1397. doi: 10.1002/ibd.21486. [DOI] [PubMed] [Google Scholar]
- 93.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39(7):830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39(5):596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang X, et al. HIV-1 and HIV-2 infections induce autophagy in Jurkat and CD4+ T cells. Cell Signal. 2012;24(7):1414–1419. doi: 10.1016/j.cellsig.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 96.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106(33):14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Duszenko M, et al. Autophagy in protists. Autophagy. 2011;7(2):127–158. doi: 10.4161/auto.7.2.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alvarez VE, Niemirowicz GT, Cazzulo JJ. The peptidases of Trypanosoma cruzi: digestive enzymes, virulence factors, and mediators of autophagy and programmed cell death. Biochim Biophys Acta. 2012;1824(1):195–206. doi: 10.1016/j.bbapap.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 99.Williams RA, Mottram JC, Coombs GH. Distinct roles in autophagy and importance in infectivity of the two ATG4 cysteine peptidases of Leishmania major. J Biol Chem. 2013;288(5):3678–3690. doi: 10.1074/jbc.M112.415372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alvarez VE, Kosec G, Sant’Anna C, Turk V, Cazzulo JJ, Turk B. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J Biol Chem. 2008;283(6):3454–3464. doi: 10.1074/jbc.M708474200. [DOI] [PubMed] [Google Scholar]
- 101.Alvarez VE, Niemirowicz GT, Cazzulo JJ. Metacaspases, autophagins and metallocarboxypeptidases: potential new targets for chemotherapy of the trypanosomiases. Curr Med Chem. 2013;20(25):3069–3077. doi: 10.2174/0929867311320250004. [DOI] [PubMed] [Google Scholar]
- 102.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marino G, et al. Premature aging in mice activates a systemic metabolic response involving autophagy induction. Hum Mol Genet. 2008;17(14):2196–2211. doi: 10.1093/hmg/ddn120. [DOI] [PubMed] [Google Scholar]
- 104.Walter S, et al. A genome-wide association study of aging. Neurobiol Aging. 2011;32(11):2109.e15–2109.e28. doi: 10.1016/j.neurobiolaging.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choi KM, et al. A monitoring method for Atg4 activation in living cells using peptide-conjugated polymeric nanoparticles. Autophagy. 2011;7(9):1052–1062. doi: 10.4161/auto.7.9.16451. [DOI] [PubMed] [Google Scholar]
- 106.Shu CW, Drag M, Bekes M, Zhai D, Salvesen GS, Reed JC. Synthetic substrates for measuring activity of autophagy proteases: autophagins (Atg4). Autophagy. 2010;6(7):936–947. doi: 10.4161/auto.6.7.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shu CW, et al. High-throughput fluorescence assay for small-molecule inhibitors of autophagins/Atg4. J Biomol Screen. 2011;16(2):174–182. doi: 10.1177/1087057110392996. [DOI] [PubMed] [Google Scholar]
- 108.Nguyen TG, et al. Development of fluorescent substrates and assays for the key autophagy-related cysteine protease enzyme, ATG4B. Assay Drug Dev Technol. 2014;12(3):176–189. doi: 10.1089/adt.2013.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dupont N, et al. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol. 2014;24(6):609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fujita N, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19(11):4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ramirez-Peinado S, et al. Glucose-starved cells do not engage in prosurvival autophagy. J Biol Chem. 2013;288(42):30387–30398. doi: 10.1074/jbc.M113.490581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Toshima T, et al. Autophagy enhances hepatocellular carcinoma progression by activation of mitochondrial beta-oxidation. J Gastroenterol. 2014;49(5):907–916. doi: 10.1007/s00535-013-0835-9. [DOI] [PubMed] [Google Scholar]
- 113.Xu Y, et al. miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2013;29(5):2019–2024. doi: 10.3892/or.2013.2338. [DOI] [PubMed] [Google Scholar]
- 114.Huang Y, Guerrero-Preston R, Ratovitski EA. Phospho-ΔNp63α-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle. 2012;11(6):1247–1259. doi: 10.4161/cc.11.6.19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hoffman WH, Shacka JJ, Andjelkovic AV. Autophagy in the brains of young patients with poorly controlled T1DM and fatal diabetic ketoacidosis. Exp Mol Pathol. 2012;93(2):273–280. doi: 10.1016/j.yexmp.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Y, et al. JNK-dependent Atg4 upregulation mediates asperphenamate derivative BBP-induced autophagy in MCF-7 cells. Toxicol Appl Pharmacol. 2012;263(1):21–31. doi: 10.1016/j.taap.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 117.Singh KK, et al. Autophagy gene fingerprint in human ischemia and reperfusion. J Thorac Cardiovasc Surg. 2014;147(3):1065–1072.e1061. doi: 10.1016/j.jtcvs.2013.04.042. [DOI] [PubMed] [Google Scholar]
- 118.Wang JH, et al. Autophagy suppresses age-dependent ischemia and reperfusion injury in livers of mice. Gastroenterology. 2011;141(6):2188–2199.e2186. doi: 10.1053/j.gastro.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Proenca CC, et al. Atg4b-dependent autophagic flux alleviates Huntington’s disease progression. PLoS One. 2013;8(7):e68357. doi: 10.1371/journal.pone.0068357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tran E, et al. Context-dependent role of ATG4B as target for autophagy inhibition in prostate cancer therapy. Biochem Biophys Res Commun. 2013;441(4):726–731. doi: 10.1016/j.bbrc.2013.10.117. [DOI] [PubMed] [Google Scholar]
- 121.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68(5):1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 122.Gorski SM, Ries J, Lum JJ. Targeting autophagy: the Achilles’ heel of cancer. Autophagy. 2012;8(8):1279–1280. doi: 10.4161/auto.20828. [DOI] [PubMed] [Google Scholar]
- 123.Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 2006;5(9):785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 124.Sagiv Y, Legesse-Miller A, Porat A, Elazar Z. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 2000;19(7):1494–1504. doi: 10.1093/emboj/19.7.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kittler JT, et al. The subcellular distribution of GABARAP and its ability to interact with NSF suggest a role for this protein in the intracellular transport of GABA(A) receptors. Mol Cell Neurosci. 2001;18(1):13–25. doi: 10.1006/mcne.2001.1005. [DOI] [PubMed] [Google Scholar]
- 126.Chen C, Li JG, Chen Y, Huang P, Wang Y, Liu-Chen LY. GEC1 interacts with the kappa opioid receptor and enhances expression of the receptor. J Biol Chem. 2006;281(12):7983–7993. doi: 10.1074/jbc.M509805200. [DOI] [PubMed] [Google Scholar]
- 127.Kuznetsov SA, Gelfand VI. 18 kDa microtubule-associated protein: identification as a new light chain (LC-3) of microtubule-associated protein 1 (MAP-1). FEBS Lett. 1987;212(1):145–148. doi: 10.1016/0014-5793(87)81574-0. [DOI] [PubMed] [Google Scholar]
- 128.Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem. 1994;269(15):11492–11497. [PubMed] [Google Scholar]
- 129.Starr T, et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11(1):33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rodriguez-Muela N, Germain F, Marino G, Fitze PS, Boya P. Autophagy promotes survival of retinal ganglion cells after optic nerve axotomy in mice. Cell Death Differ. 2012;19(1):162–169. doi: 10.1038/cdd.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ravikumar B, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90(4):1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 132.Byrne BG, Dubuisson JF, Joshi AD, Persson JJ, Swanson MS.Inflammasome components coordinate autophagy and pyroptosis as macrophage responses to infection. MBio. 201341e00620-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]