Abstract
Activation of CD4+ T cells results in rapid proliferation and differentiation into effector and regulatory subsets. CD4+ effector T cell (Teff) (Th1 and Th17) and Treg subsets are metabolically distinct, yet the specific metabolic differences that modify T cell populations are uncertain. Here, we evaluated CD4+ T cell populations in murine models and determined that inflammatory Teffs maintain high expression of glycolytic genes and rely on high glycolytic rates, while Tregs are oxidative and require mitochondrial electron transport to proliferate, differentiate, and survive. Metabolic profiling revealed that pyruvate dehydrogenase (PDH) is a key bifurcation point between T cell glycolytic and oxidative metabolism. PDH function is inhibited by PDH kinases (PDHKs). PDHK1 was expressed in Th17 cells, but not Th1 cells, and at low levels in Tregs, and inhibition or knockdown of PDHK1 selectively suppressed Th17 cells and increased Tregs. This alteration in the CD4+ T cell populations was mediated in part through ROS, as N-acetyl cysteine (NAC) treatment restored Th17 cell generation. Moreover, inhibition of PDHK1 modulated immunity and protected animals against experimental autoimmune encephalomyelitis, decreasing Th17 cells and increasing Tregs. Together, these data show that CD4+ subsets utilize and require distinct metabolic programs that can be targeted to control specific T cell populations in autoimmune and inflammatory diseases.
Introduction
CD4+ T lymphocytes are critical to mediating or suppressing normal immunity as well as inflammatory or autoimmune diseases. Antigen exposure initially leads to T cell activation, inducing rapid growth and proliferation. Depending on the cytokine environment during activation, CD4+ T cells then differentiate into effector (Teff) (Th1 and Th17) or Treg subsets. Each of these subsets plays a unique role in the adaptive immune system, with Teffs driving immunity and inflammation while Tregs play an opposing role, suppressing Teffs to limit excessive inflammatory responses (1). The balance between Teffs and Tregs is crucial to providing sufficient immune protection without promoting autoimmunity. Indeed, many autoimmune diseases, including multiple sclerosis and inflammatory bowel disease (IBD), involve an imbalance of Teffs to Tregs or decreased Treg function (2). Th17, in particular, plays a key proinflammatory role in many autoimmune diseases, including experimental autoimmune encephalomyelitis (EAE), IBD, and graft-versus-host disease (3–5). Identifying characteristics of each T cell population to allow the balance of Teffs and Tregs to be modulated may therefore provide a way to prevent or suppress autoimmunity.
Emerging evidence has suggested that CD4+ T cell metabolism is highly dynamic and may allow targeting of select T cell populations. T cell activation leads to a demand for biosynthetic precursors and adequate energy for effector function (6, 7). This reprogramming involves decreased lipid oxidation and an increase in glucose uptake and glycolysis as well as amino acid transport and glutaminolysis (8–12). Importantly, metabolic reprogramming of Teffs and Tregs leads to distinct metabolic programs. While Teffs utilize large amounts of glucose and a high rate of glycolysis to support their energetic needs, Tregs can oxidize lipids and are able to expand and function even in the absence of glucose (13, 14). The specific metabolic substrate usage and metabolic programs that are required for activation and differentiation of Teffs and Tregs, however, are uncertain.
Pyruvate metabolism is one regulatory point in glucose metabolism that may play a central role in the glycolytic or oxidative roles of Teffs and Tregs. Pyruvate dehydrogenase (PDH) catalyzes the conversion of cytosolic pyruvate into mitochondrial acetyl-CoA for oxidative metabolism. PDH is inhibited by PDH kinase (PDHK) to suppress pyruvate oxidation and instead promote its conversion to lactate via lactate dehydrogenase (LDH) (15). PDHK has 4 isoforms that are regulated by oncogenic signaling and hypoxia (16, 17). Indeed, cancer cells are also highly glycolytic, and targeting PDHK with dichloroacetate (DCA) can suppress glycolysis and lead to increased oxidative metabolism and production of ROS (15, 18). DCA has also been shown to reduce proinflammatory cytokine production and promote FoxP3 expression in vitro and in several in vivo inflammatory models (19–21), although the mechanisms involved have not been established.
Here, we examine the metabolic programs of each T cell subset in vivo and in vitro to identify distinct metabolic pathways that will allow selective targeting of different T cell populations. We show that Tregs oxidize not only lipids at a high rate, but also glycolysis-derived pyruvate. Teffs require glycolytic metabolism for proliferation, differentiation, and survival, while Tregs require oxidative metabolism for these processes. Gene expression and metabolomics data suggest that PDHK isoform 1 (PDHK1) is an important modulator of Th17 and Treg metabolism. Indeed, suppression of PDHK1 expression or inhibition with DCA caused ROS accumulation and selectively inhibited the survival and proliferation of Th17 cells. DCA was also capable of suppressing Th17 cells and CD4+ T cell–mediated inflammation in both IBD and EAE models. Together, these metabolic data have identified PDH as a key regulatory point in CD4+ T cell subset fate and show that inhibition of PDHK1 leads to selective ROS generation and targeting of Th17 cells in vivo in inflammatory and autoimmune disorders.
Results
T cells undergo metabolic reprogramming toward a glycolytic phenotype in vivo.
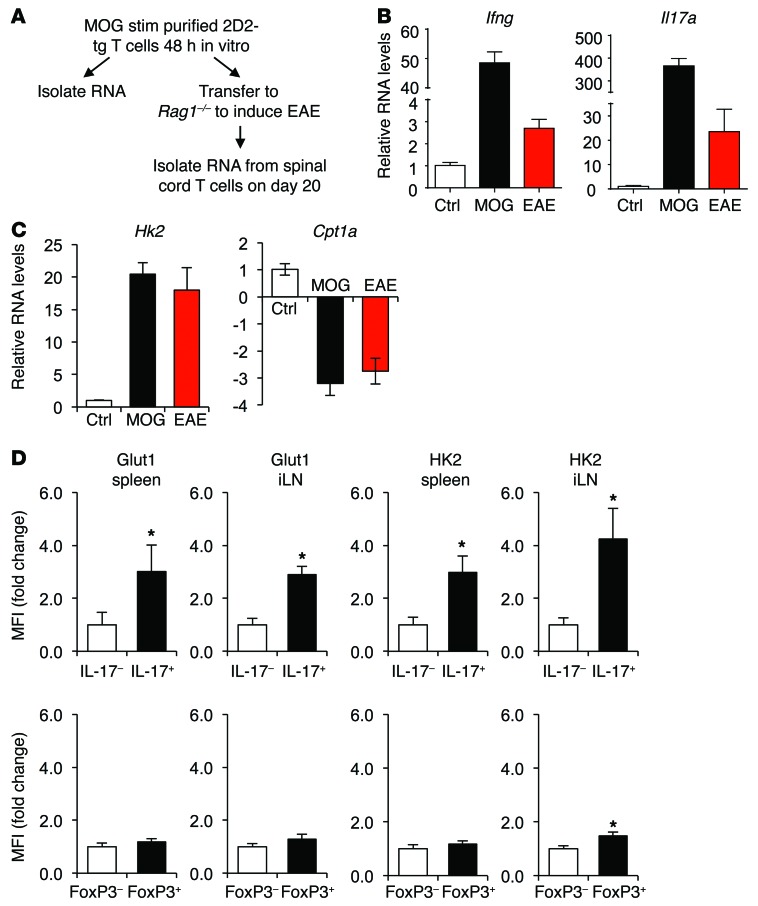
While T cells undergo a metabolic reprogramming toward glycolysis in vitro, little is known regarding the metabolism of inflammatory T cell subsets in vivo. The metabolic phenotype of 2D2 TCR transgenic T cells specific for myelin oligodendrocyte glycoprotein (MOG) was, therefore, observed upon activation and adoptive transfer to induce EAE (Figure 1A). Immunologic and metabolic gene expression was compared between in vitro antigen-activated 2D2 T cells and 2D2 T cells directly isolated from spinal cords of mice with EAE. Ifng and Il17 were sharply induced upon in vitro stimulation and also were significantly elevated in T cells at the disease site in EAE (Figure 1B). Similar to in vitro metabolic reprogramming to glycolytic from oxidative metabolism, T cells from the spinal cord of mice with EAE also had elevated expression of the glycolytic gene Hexokinase 2 (Hk2) and decreased expression of the lipid oxidation gene Carnitine palmitoyltransferase 1A (Cpt1a, Figure 1C). The metabolic profile of in vivo–differentiated T cells was next examined directly by flow cytometry in IL-17–producing or FoxP3-expressing cells in EAE. IL-17–producing cells showed elevated levels of both Glucose transporter 1 (GLUT1) and HK2 proteins in the spleen and inguinal lymph nodes during active EAE disease (Figure 1D). In contrast, neither GLUT1 nor HK2 was elevated in FoxP3-expressing cells. Teffs in inflammatory disease sites in vivo, therefore, appear to be glycolytic, while Tregs utilize a distinct metabolic program.
Figure 1. Teffs, but not Tregs, upregulate glycolytic metabolism during inflammatory processes in vivo.
(A–C) EAE was induced in 2D2 TCR transgenic mice, and RNA was extracted from spinal cords of mice with active disease or in vitro MOG-stimulated (MOG stim) 2D2 T cells as indicated (A) for real-time PCR of (B) inflammatory and (C) metabolic gene expression. (D) EAE was induced in wild-type mice and CD4+ T cells in spleens, and inguinal lymph nodes of mice with active disease were examined using flow cytometry. Data are representative of 2 experiments (A–C, n = 10; D, n = 5) and shown as mean ± SD. *P < 0.05.
Teffs and Tregs utilize different metabolic pathways and have distinct fuel capacities.
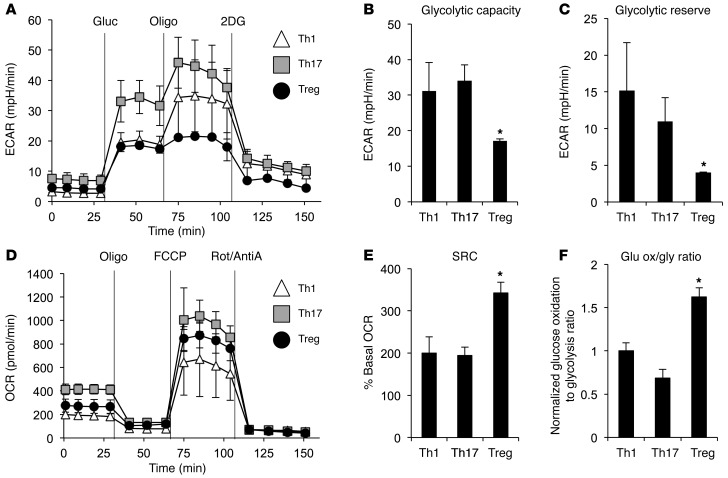
Although previous studies (13, 14) and in vivo analyses here (Figure 1) have identified basic metabolic differences between Teffs (Th1 and Th17) and Tregs, the underlying metabolic features of CD4+ T cell subsets are uncertain. To examine the detailed metabolic phenotype of the CD4+ T cell subsets, Teffs and Tregs were differentiated in vitro and oxygen consumption and lactate production were measured using an extracellular metabolic flux analyzer. Cells were cultured in the absence of glucose, glutamine, or lipids, and the extracellular acidification rate (ECAR), a measurement of lactate production, was determined upon readdition of glucose (Figure 2A). All CD4+ T cell subsets had increased ECAR following glucose addition, although Th1 and Treg had less of an increase than Th17 cells. Oligomycin was then added to block mitochondrial ATP production and promote maximal rates of glycolysis. Importantly, Th1 and Th17 cells each showed a robust increase in ECAR following oligomycin treatment, but Tregs were largely unchanged. These data indicate that Tregs were performing glycolysis at maximal rates following glucose addition and have limited capacity to increase this pathway. Teffs, in contrast, generate lactate at a high rate and can further elevate glycolytic rate when required to generate ATP. The glycolytic capacity and glycolytic reserve were both severely impaired in Tregs compared with Th1 and Th17 cells (Figure 2, B and C). Therefore, when glucose is the only fuel available, Teffs efficiently perform glycolysis while Tregs are unable to increase their glycolytic capacity.
Figure 2. Teffs and Tregs utilize different metabolic pathways and have distinct fuel capacities.
CD4+CD25– T cells were polarized in vitro for 3 days, split 1:2, and cultured with IL-2 alone for an additional 2 days to generate induced Th1 or Th17 cells or Tregs. (A–C) T cells were cultured in base DMEM media with no glucose or glutamine. ECAR was assessed after the addition of 25 mM glucose (gluc) and in response to the metabolic inhibitors oligomycin (oligo) and 2DG. Shown are the (A) time course and calculations of (B) glycolytic capacity and (C) glycolytic reserve. (D and E) T cells were cultured in base DMEM media with 25 mM glucose. OCR was assessed basally and in response to the mitochondrial inhibitors oligomycin, FCCP, and rotenone and antimycin A (Rot/AntiA). Shown are the (D) time course and (E) calculation of SRC. (F) Glucose oxidation was measured in the T cell subsets, and the ratio of glucose oxidation to glycolysis was graphed. Data are shown as mean ± SD of triplicate samples (B, C, E, and F), and all data are representative of at least 3 independent experiments. *P < 0.05.
Mitochondrial and oxidative metabolism can play a key role in supporting T cell activation and proliferation (22, 23). While Tregs have high rates of lipid oxidation (13), mitochondrial oxidation of pyruvate has not been previously examined. Mitochondrial oxygen consumption rate (OCR) was therefore measured in each CD4+ subset in media containing glucose. Prior to addition of metabolic inhibitors, Tregs had an intermediate level of oxygen consumption relative to Teffs, with Th17 cells maintaining the highest basal rate of oxygen consumption (Figure 2D). Oligomycin treatment to inhibit mitochondrial ATP production suppressed oxygen consumption in each subset to an equivalently low level, indicating that oxygen consumption was tightly coupled to ATP generation for all T cell subsets. Upon the addition of the protonophore carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) to uncouple oxidative phosphorylation from electron transport and allow maximal respiration, Tregs and Th17 greatly upregulated oxygen consumption. When compared with the basal rates of oxygen consumption, these data show that Tregs have the greatest spare respiratory capacity (SRC) of the subsets (Figure 2E). Thus, Tregs have low glycolytic production of lactate and cannot further elevate this pathway, yet have the greatest capacity for mitochondrial oxidation when glucose is the only available fuel.
These data suggested that Teffs and Tregs may differentially utilize glucose, with Teffs preferentially converting glucose-derived pyruvate to lactate, while Tregs instead oxidize this fuel in the mitochondria. We have previously shown that Tregs preferentially oxidize lipids, but relative glucose oxidation rates in T cell subsets have not been directly tested (13). To measure the fate of glycolysis-derived pyruvate, CD4+ T cell subsets were provided radiolabeled glucose and glycolytic flux was measured through enolase production of phosphoenolpyruvate and glucose oxidation to CO2. The ratio of glucose oxidation to glycolytic flux was higher in Tregs, supporting the conclusion that this subset preferentially oxidizes glucose rather than converting pyruvate to lactate (Figure 2F). Together, these data indicate that Tregs utilize mitochondrial oxidative pathways using both lipids (13) and glucose as fuels while Teffs have low levels of mitochondrial oxidative metabolism for either fuel and primarily perform aerobic glycolysis, converting pyruvate to lactate.
Inhibition of glycolysis or mitochondrial oxidation selectively affects Teff or Treg proliferation, differentiation, and survival.
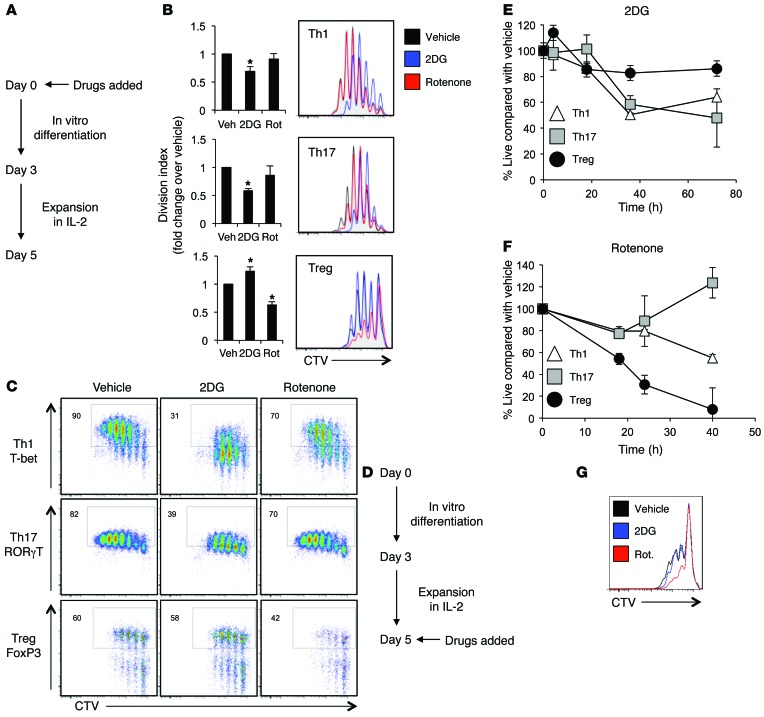
Although extracellular flux analysis revealed that Teffs and Tregs utilize pyruvate through distinct glycolytic or mitochondrial oxidative metabolic pathways, the reliance of these subsets on each mode of metabolism was not clear. To determine whether each CD4+ subset required these programs, CD4+ T cell subsets were labeled with the proliferation indicator dye CellTrace Violet (CTV), differentiated in vitro, and treated with either low-dose 2-deoxyglucose (2DG) or rotenone to inhibit glycolysis or electron transport, respectively (Figure 3A and Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI76012DS1). 2DG treatment inhibited the proliferation of each Teff subset and instead increased Treg proliferation (Figure 3B). Conversely, rotenone did not affect Teffs, but sharply reduced Treg proliferation.
Figure 3. Inhibition of glycolysis or mitochondrial electron transport selectively affects Teff or Treg survival, proliferation, and function.
(A) Schematic of drug treatments for B and C. (B and C) CD4+CD25– T cells were labeled with CTV and polarized in vitro for 3 days to generate Th1 or Th17 cells or Tregs. Cells were treated with 250 μM 2DG or 5 nM rotenone, and (B) proliferation or (C) transcription factor staining was assessed by flow cytometry after 72 hours. (D) Schematic of drug treatments for E and F. (E and F) CD4+CD25– T cells were polarized in vitro for 3 days, split 1:2, and cultured with IL-2 alone for an additional 2 days and then incubated with (E) 250 μM 2DG or (F) 5 nM rotenone; survival was determined by propidium iodide exclusion relative to vehicle-treated control. (G) CD4+CD25+ natural Tregs were labeled with CTV and activated in the presence of 250 μM 2DG or 5 nM rotenone, and proliferation was assessed by CTV dilution. Data are shown as mean ± SD of triplicate samples (B, E, and F), and all data are representative of at least 3 independent experiments. *P < 0.05.
Differentiation of each CD4+ T cell subset is characterized by induction of specific transcription factors as cells undergo DNA replication and division (1). Given the reliance of Teffs on glycolysis and Tregs on mitochondrial metabolism for proliferation, it was possible that the ability of subsets to express these transcription factors was also metabolically sensitive. Consistent with this notion, 2DG treatment suppressed induction of the Th1-specific transcription factor T-bet in cells cultured under Th1 conditions as well as retinoid-related orphan receptor γ T cell (RORγT), which is required for Th17, in T cells cultured under Th17 conditions (Figure 3C). Importantly, this effect was not dependent on cell division, as T-bet and RORγT were inhibited at each equivalent cell-cycle division. Likewise, expression of the T-bet and RORγT transcriptional targets, IFN-γ and IL-17, respectively, were also suppressed by 2DG at each equivalent cell division (Supplemental Figure 1B). Aerobic glycolysis was previously shown to be required for maximal IFN-γ production in activated T cells (22), and 2DG also suppressed IFN-γ production and proliferation of T cells activated in the absence of skewing conditions (Supplemental Figure 2A). Induction of the Treg transcription factor FoxP3, however, was not affected by 2DG. In contrast, the electron transport inhibitor rotenone selectively reduced FoxP3 expression in Tregs at each equivalent cell division and only minimally suppressed T-bet and RORγT expression by Th1 and Th17 cells (Figure 3C) or IFN-γ production or proliferation of activated T cells (Supplemental Figure 2A). FoxP3+ natural Tregs (nTregs) isolated from splenocytes had reduced proliferation, but not cell survival, following rotenone treatment (Supplemental Figure 2, B and C). Importantly, Teffs were not affected by 2DG and rotenone following differentiation, demonstrating that 2DG affects Teff differentiation and cytokine production primarily during early activation and subset specification (Supplemental Figure 3, A and B). Likewise, there was no effect of rotenone on FoxP3 expression in differentiated Tregs.
CD4+ subsets may rely on specific metabolic programs, not only for proliferation and differentiation, but also for survival. Teff and Treg subsets were therefore established in vitro and treated with 2DG or rotenone following differentiation (Figure 3D). Tregs survived well in the presence of 2DG, as measured by propidium iodide exclusion, and were only moderately affected by glycolytic inhibition (Figure 3E). In contrast, Th1 and Th17 were sensitive to 2DG treatment and showed increased cell death (Figure 3E and Supplemental Figure 3C). Teffs were relatively insensitive to rotenone, and rotenone instead enhanced Th17 relative to vehicle (Figure 3F). Tregs, however, were sensitive to rotenone treatment, with reduced viability (Figure 3F and Supplemental Figure 3C). This effect was not specific to induced Tregs, as reduced proliferation was also observed in rotenone-treated natural Tregs (Figure 3G). Overall, these data suggest that specific metabolic programs of Teffs and Tregs are fundamentally linked to CD4+ T cell proliferation, differentiation, and survival.
Th17 cells and Tregs have distinct metabolic phenotypes.
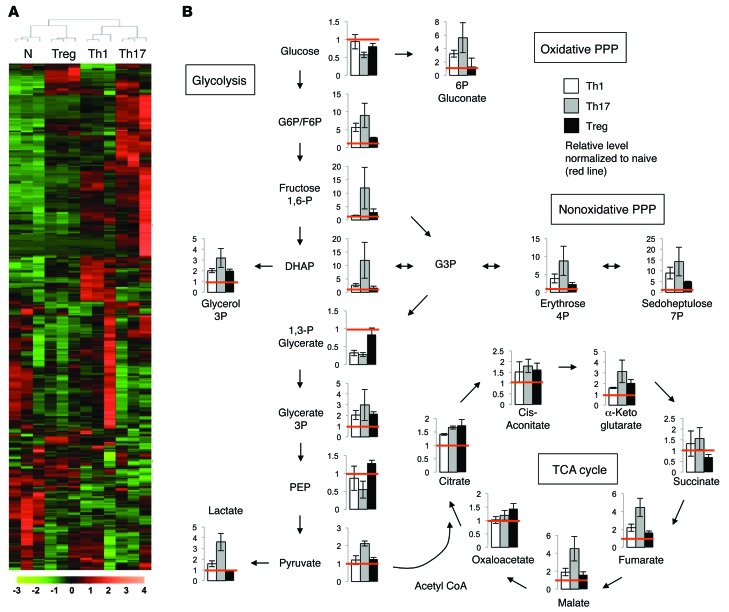
The distinct metabolic dependencies of Teffs and Tregs suggested detailed metabolic profiling could reveal key regulatory points of CD4+ T cell subsets. We therefore examined metabolite levels and metabolic gene expression of Th1 and Th17 cells and Tregs in detail, as the balance of these subtypes is crucial in a variety of inflammatory settings. Unsupervised clustering of steady-state levels of approximately 400 metabolites measured by high-resolution nontargeted Q exactive–mass spectrometry (QE-MS) metabolomics (24) showed that Teffs and Tregs had broad metabolic differences (Figure 4A and Supplemental Table 1). Importantly, Tregs were dissimilar from Th1 and Th17 cells, which clustered together. A separate liquid chromatography/gas chromatography–MS (LC/GC-MS) analysis of approximately 200 metabolites found similar results (Supplemental Figure 4 and Supplemental Table 2). Th17 cells and Tregs were the most distinct, and Th17 cells had high levels of early glycolytic and pentose phosphate pathway intermediates (Figure 4B). Late glycolytic intermediates were more similar, although Th17 had high pyruvate and higher still levels of lactate, suggesting preferential pyruvate conversion to lactate. Despite lower levels of pyruvate, Tregs had TCA-cycle intermediates equivalent to those of Th17 cells up to α-ketoglutarate, which may indicate preferential pyruvate oxidation in Tregs and glutamine oxidation by Th17.
Figure 4. Metabolomic profiling of CD4+ subsets shows distinct metabolic profiles.
Naive CD4+CD25– T cells were collected or polarized in vitro for 3 days, split 1:2, and cultured with IL-2 alone for an additional 2 days to generate Th1, Th17, or Tregs. T cell subset lysates from 3 biological replicate samples were extracted and analyzed using high-resolution LC-QE-MS for determination of cellular metabolites. (A) Heat map showing relative levels of each metabolite and unsupervised hierarchical clustering from independent biological replicates. (B) Relative levels of each metabolite in the glycolysis and TCA-cycle pathways are shown. Samples are normalized to naive T cells, as indicated by red lines. Data are shown as mean ± SD of triplicate samples.
Th17 cell and Treg subsets were next examined for levels of amino acids and acyl-carnitines, which are intermediates that reflect lipid oxidation products and available mitochondrial fuels. No clear T cell–subtype patterns were observed with regard to amino acid levels, except that Tregs had elevated levels of aspartate compared with Th17 (Supplemental Figure 5A). Acyl-carnitines were variable between Th17 cells and Tregs, and Tregs had increased C2 and C4-OH carnitine levels relative to Th17 (Supplemental Figure 5, B and C). These metabolites equilibrate with and reflect acetyl and β-hydroxybutryl CoA species, respectively. We also observed a drop in oleyl-carnitine (C18:1). This fall in a long-chain acyl-carnitine coupled with the rise in C4-OH (hydroxybutyrylcarnitine) is consistent with increased fatty acid oxidation (25) and supports the model that Tregs utilize fatty acids to a greater extent than Th17. Together, these metabolic data suggest that each CD4+ T cell subset has a unique metabolic phenotype and point to specific metabolic distinctions in fuel consumption patterns and dependencies between the Treg and Teff subsets.
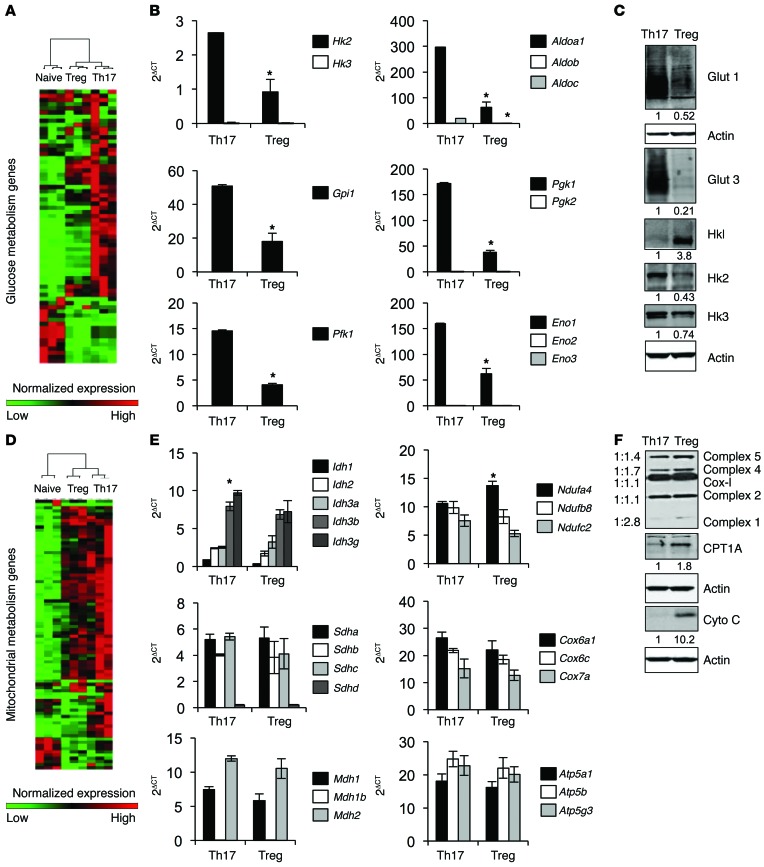
Expression of metabolic genes and select proteins was next determined in Th17 cells and Tregs. Gene-clustering analysis showed that Th17 had broadly higher expression of glucose metabolism genes, followed by Tregs and naive T cells (Figure 5A, Supplemental Figure 6, and Supplemental Tables 3 and 4). Specifically, Th17 had higher levels of expression of many glycolytic genes compared with Tregs (Figure 5B). Interestingly, CD4+ T cells preferentially expressed a single isoform of each glycolytic gene (Figure 5B). In addition, CD4+ T cells expressed several glucose transporters, including GLUT1 and GLUT3. Expression of GLUT1 protein was elevated in Th17 compared with Tregs, and Tregs did not express detectable levels of GLUT3 (Figure 5C and Supplemental Figures 6 and 7). After uptake, glucose is phosphorylated by hexokinase (HK), of which there are 4 isoforms. Surprisingly, Tregs expressed primarily HK1 protein, while Th17 mainly expressed HK2 and HK3 (Figure 5C and Supplemental Figure 7). Interestingly, although HK1 protein levels were elevated in Tregs, the RNA levels of HK1–3 were decreased relative to Th17 (Figure 5C and Supplemental Figure 7). Unsupervised clustering of mitochondrial genes showed that both Th17 cells and Tregs expressed higher levels of mitochondrial metabolism genes compared with naive T cells (Figure 5D, Supplemental Figure 6, and Supplemental Tables 4 and 5). The expression of genes specifically involved in the TCA cycle and electron transport were similar between Th17 cells and Tregs (Figure 5E). Additionally, Tregs had elevated levels of many of the electron transport chain proteins as well as higher levels of the fatty acid transporter CPT1A and electron transport chain component cytochrome c compared with Th17 (Figure 5F and Supplemental Figure 7). Together, these data show distinct genetic and metabolite differences that likely mediate the specific metabolic programs and dependencies of Teffs and Tregs.
Figure 5. Th17 cells and Tregs have distinct metabolic gene and protein expression.
CD4+CD25– T cells were polarized in vitro for 3 days, split 1:2, and cultured with IL-2 alone for an additional 2 days to generate Th17 or Tregs for (A, B, D, and E) real-time PCR or (C and F) immunoblot. (A, B, D, and E) Data shown are mean ± SD of 3 biological replicates and are shown as 2ΔCT normalized to the geometric mean of the reference genes Tbp and Bgu. Data are representative of 2 (A, B, D, and E) or 2 (Hk3), 3 (Glut1, Glut3, Hk1, Hk2, OxPhos), 4 (Cyto c), or 5 (Cpt1a) (C and F) independent experiments. *P < 0.05.
PDHK is required for Th17, but not Treg, function in vitro.
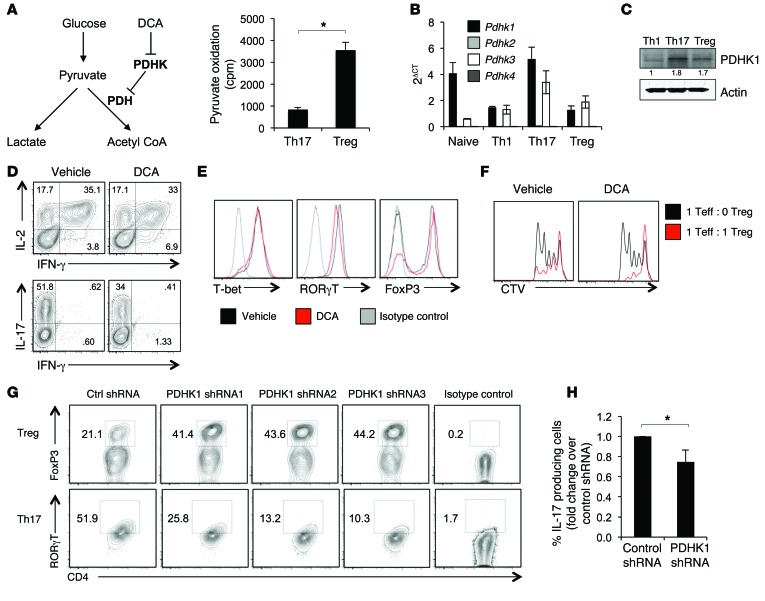
The finding of distinct substrate utilization patterns suggested that the bifurcation point for pyruvate to lactate or conversion to acetyl-CoA via the PDH complex for mitochondrial oxidation may be an important regulatory node for Th17 and Treg CD4+ subsets. To measure the flux of pyruvate through PDH, Th17 cells and Tregs were provided radiolabeled pyruvate and its oxidation to CO2 was measured. Pyruvate oxidation was higher in Tregs, demonstrating that Tregs preferentially direct pyruvate to mitochondrial metabolism (Figure 6A). PDH is a highly regulated multisubunit complex that is controlled, in part, by PDHK, which phosphorylates and inhibits PDH to direct pyruvate to lactate rather than to acetyl-CoA. Expression of the 4 Pdhk isoforms was therefore examined in the CD4+ T cell subsets. T cells expressed Pdhk1 and Pdhk3, with Pdhk1 being the predominant isoform (Figure 6B). At both the RNA and protein levels, Th17 expressed the highest levels of PDHK1, followed by Tregs, while Th1 had little PDHK1 expression (Figure 6C and Supplemental Figure 8A). Therefore, PDHK1 is differentially expressed in the CD4+ T cell subsets and may play a role in controlling T cell metabolism.
Figure 6. PDHK is required for Th17, but not Treg, function in vitro.
CD4+CD25– T cells were polarized in vitro for 3 days, split 1:2, and cultured with IL-2 alone for an additional 2 days to generate Th1, Th17, or Tregs. (A) Schematic showing the different fates of pyruvate and mechanism of DCA inhibition and measurement of 14C-pyruvate oxidation in Th17 cells and Tregs. (B) Real-time PCR and (C) immunoblots are shown, representative of 4 independent experiments. (D–F) Cells were treated with 10 mM DCA, and then cytokine production (D) and transcription factor staining (E) were determined by flow cytometry. (F) Treg function was assessed by an in vitro Treg-suppression assay. (G and H) T cells were polarized and infected with lentivirus expressing PDHK1 shRNA. (G) transcription factor staining for FoxP3 and RORγT or (H) intracellular cytokine staining for IL-17 was performed. Data are shown as mean ± SD of triplicate samples (A, B, and H), and all data are representative of at least 3 independent experiments. *P < 0.05.
PDHK is a target of the inhibitor compound DCA (Figure 6A and ref. 26), which has been previously shown to affect cytokine production and Tregs (19–21). To determine the effect of PDHK inhibition on CD4+ T cell fate and function, CD4+ T cells were differentiated in vitro in the presence of DCA or treated with DCA following 3 days of differentiation. DCA treatment did not affect Th1 differentiation or function, as IFN-γ production and T-bet expression were similar regardless of treatment (Figure 6, D and E, and Supplemental Figure 8, B–D). In contrast, DCA inhibited the production of IL-17 in cells cultured in Th17-skewing conditions and suppressed expression of the Th17 transcription factor RORγT (Figure 6, D and E, and Supplemental Figure 8, B–D). Conversely, treatment of Tregs with DCA increased the expression of FoxP3 compared with vehicle and maintained or potentially increased the in vitro suppressive capacity of Tregs (Figure 6, D–F, and Supplemental Figure 8E). We next genetically targeted Pdhk1 using 3 different lentiviral shRNA constructs (Supplemental Figure 9A). Pdhk1 deficiency inhibited RORγT and IL-17 expression in Th17 cells and increased FoxP3 expression, mimicking the in vitro effects seen with DCA (Figure 6, G and H, and Supplemental Figure 9, B and C). Lentiviral transduction itself had no effect on metabolism (Supplemental Figure 9, D and E). These data demonstrate that pyruvate metabolism and PDHK1 play a key role in modulating Th17 cells and Tregs.
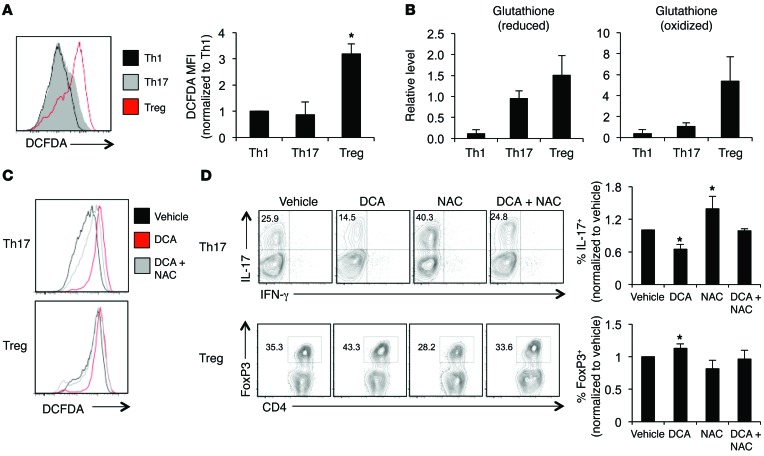
One outcome of DCA treatment to promote pyruvate oxidation is a potential increase in the generation of ROS. Indeed, DCA can suppress aerobic glycolysis of cancer cells and stimulate ROS production that can lead to cancer cell senescence (15, 18). Previous literature also suggests that Teffs may be more sensitive to ROS stress than Tregs (27–29). ROS levels were therefore examined in the T cell subsets. The ROS indicator dye DCFDA showed that Tregs had higher levels of ROS than the Teff subsets (Figure 7A). Consistent with ROS stress, Tregs also had a large reserve pool of reduced glutathione (GSH) as well as high levels of oxidized glutathione (GSSG) compared with Th1 and Th17 (Figure 7B). This suggests that Tregs have a greater capacity to handle ROS and utilize this GSH pool to a greater extent than Teffs. Tregs have also been shown to have higher levels of the antioxidant thioredoxin-1, also contributing to Treg redox regulation (30). To determine whether DCA acted in part through ROS generation, established Th17 and Treg cultures were treated acutely with DCA. In both cases, DCA increased DCFDA staining to indicate generation of ROS (Figure 7C). We next tested the effects of DCA-induced ROS by cotreatment of Th17 cells and Tregs with DCA and the antioxidant NAC to prevent ROS accumulation. Differentiated T cells treated with DCA showed levels of ROS accumulation similar to those with acute DCA treatment (Supplemental Figure 10A), and 1 mM NAC was sufficient to partially rescue ROS accumulation following DCA treatment (Figure 7C and Supplemental Figure 10B). DCA treatment alone inhibited IL-17 production; however, cotreatment with NAC prevented the decrease in IL-17 (Figure 7D). In contrast, while DCA promoted Tregs, cotreatment with NAC prevented this increase in Tregs. These data suggest that DCA drives glucose oxidation and ROS production that Tregs are programmed to manage.
Figure 7. DCA treatment generates ROS that negatively affects Th17.
CD4+CD25– T cells were polarized in vitro for 3 days, split 1:2, and cultured with IL-2 alone for an additional 2 days to generate Th1, Th17, or Tregs. (A) ROS production was measured by flow cytometry using the indicator dye DCFDA. (B) Relative glutathione levels were measured in the T cell subsets using LC/MS. (C) ROS production in Th17 cells and Tregs treated with vehicle, DCA, or DCA/NAC was measured using DCFDA. (D and E) Tregs and Th17 cells were treated with 10 mM DCA, 1 mM NAC, or both in combination, and (D) IL-17 production and FoxP3 expression were examined after 3 days. Data are shown as mean ± SD of triplicate samples (A, B, and D), and all data are representative of at least 2 independent experiments. *P < 0.05.
Inhibition of PDHK in vivo differentially affects Th17 cells and Tregs to inhibit the progression of colitis and EAE.
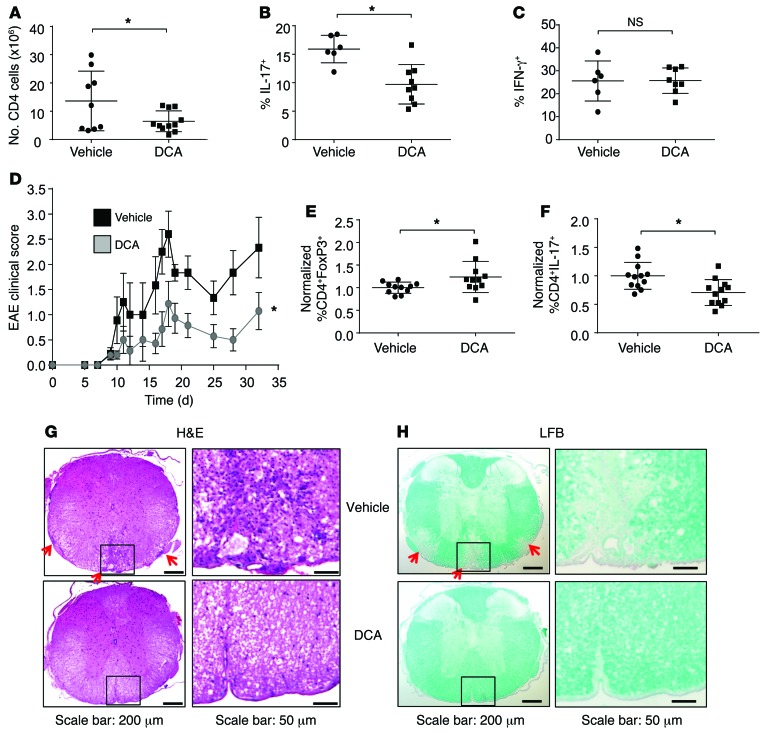
In vitro, PDHK inhibition by DCA treatment does not affect Th1, but has a selective deleterious effect on Th17 function while favoring Treg differentiation. While DCA has been previously shown to suppress inflammation in arthritis, asthma, and alloreactivity (19–21), the specific effect on the differentiated CD4+ T cell subsets has not been examined in vivo. We therefore tested the ability of DCA to suppress the Th17-mediated inflammatory diseases. IBD is driven by both Th1 and Th17 and alleviated by Tregs, which allow functional examination of each subset in vivo (31). Naive T cells were adoptively transferred into immunodeficient recipients in the absence of Tregs, and mice were then given normal or DCA-containing drinking water. IBD was initiated upon generation of Teffs by encounter with gut microbiota following exposure to the clinically relevant NSAID piroxicam 2 weeks after the T cell transfer. Significantly fewer CD4+ cells were found in the mesenteric lymph nodes and infiltrating into the gut tissue of mice treated with DCA relative to those receiving vehicle water (Figure 8A and Supplemental Figure 11A). While Th17 cells were reduced, DCA treatment did not lead to a reduction of Th1 cells, as measured by the percentage of IFN-γ–producing CD4+ T cells (Figure 8, B and C, and Supplemental Figure 11B). These data suggest that PDHK inhibition selectively affects Th17 but not Th1 proliferation and function in vivo. Despite inhibition of Th17 cells, DCA treatment did not prevent intestinal inflammation or disease progression, likely due to the functional Th1 response (Supplemental Figure 11C).
Figure 8. PDHK is selectively required for Th17, but not Treg, expansion and function in vivo.
(A–C) Rag1–/– mice were injected with naive Teffs (CD4+CD25–CD45RBhi), and colitis was induced. Mice were given 2 g/l of DCA (n = 10) or vehicle (n = 10) in the drinking water for the duration of the experiment. (A) The number of CD4+ T cells or (B and C) the percentages of IFN-γ– and IL-17–producing cells in the spleen and mesenteric lymph nodes were determined using flow cytometry. (D–H) EAE was induced in wild-type mice with or without DCA treatment (10 mice per group). (D) A time course of clinical scores is shown. (E and F) The percentages of (E) CD4+FoxP3+ and (F) CD4+IL-17+ T cells in the draining lymph nodes on day 9 were determined using flow cytometry. (G) H&E and (H) LFB staining were performed on spinal cord sections from mice with active disease. Data are shown as mean ± SD (A–C, E, and F), and data are representative of 3 independent experiments. *P < 0.05.
To independently test the role of PDHK on Th17 and Treg populations, we next examined the effects of DCA in EAE. While Th1 cells can contribute, this model is largely Th17 dependent, as RORγT-knockout mice are disease resistant (32, 33). EAE was induced in wild-type mice by MOG immunization, and DCA was given in the drinking water throughout the course of the experiment; clinical signs of EAE were assessed at regular intervals. PDHK inhibition by DCA treatment significantly alleviated EAE clinical symptoms throughout the course of disease progression (Figure 8D). T cells isolated from the inguinal draining lymph nodes were reduced in number and showed an increase in FoxP3+ Tregs and a decrease in IL-17–producing and CD44hi T cells, consistent with our in vitro findings (Figure 8, E and F, and Supplemental Figure 11D). Importantly, DCA treatment also impaired infiltration of T cells into the spinal cord and prevented demyelination, as measured by H&E and Luxol Fast Blue (LFB) staining, respectively (Figure 8, G and H). Therefore, PDHK inhibition in vivo selectively modulates the balance between Th17 cells and Tregs and suppresses autoimmunity. These data suggest that the distinct metabolic programs of CD4+ subsets are essential for each subset and can be exploited to target specific T cell populations in inflammatory diseases.
Discussion
It is increasingly evident that metabolic reprogramming plays a crucial role in T cell activation, differentiation, and function. Activated CD4+ T cells require high rates of glucose uptake and glycolysis, glutaminolysis, and lipid synthesis to support proliferation and function (10–12, 34). We show, for what we believe is the first time, a detailed analysis of the metabolic programs underlying Teffs (Th1 and Th17) and Tregs. Both in vivo and in vitro, our data show that inflammatory Teffs and CD4+ Tregs have distinct metabolic programs and dependencies. Specifically, Teffs are reliant on glycolysis, while Tregs have greater fuel flexibility and oxidize glucose in addition to lipids. While Th1 and Th17 have a similar need for glucose, metabolic differences in downstream pathways and regulation of each T cell subset allowed for selective targeting of Th17 cells. Here, we identify PDHK as a selective regulator of CD4+ T cell differentiation and inflammation and show that PDHK inhibition specifically impairs Th17 while sparing Th1 and promoting Tregs.
Through detailed metabolic analyses, distinct programs were found for Th1 and Th17 cells and Tregs. Each subset expressed different levels of metabolic enzymes and metabolites. While both Th1 and Th17 were highly glycolytic, Th17 cells had higher expression levels of glycolytic genes and levels of glycolytic intermediates. Th1 cells were also dependent on glycolysis, but appeared to regulate this pathway posttranscriptionally without accumulation of glycolytic intermediates. Glutamine metabolism may also be preferentially increased in Th17 cells. These metabolic distinctions are consistent with differential regulatory mechanisms of Th1 and Th17, such as selective requirement of Th17 cells for HIF1α (14, 35). In contrast, Tregs were dependent on mitochondrial metabolism and showed flexibility to oxidize lipids or glucose. While steady-state levels of metabolites do not necessarily reflect pathway flux rates, our combined data support a model in which Th1 and Th17 cells rely on glycolysis and potentially glutaminolysis, while Tregs oxidize pyruvate and lipids.
While Teffs efficiently convert glucose to lactate, Tregs have a higher respiratory capacity and preferentially oxidize glucose-derived pyruvate. The reduced expression of glucose transporters and glycolytic components in Tregs may partially restrict Treg glucose metabolism. High expression of CPT1A and lipid oxidation, however, provides Tregs with multiple fuel sources. This fuel flexibility may be important to support T cell suppression in vivo. Increased respiratory capacity has been associated with increased cell viability (36), while glycolysis may restrict cell longevity (37). There is also increasing evidence that Tregs may utilize metabolic strategies to suppress Teffs (33). These strategies include degrading ATP through upregulating the expression of CD39, which converts extracellular ATP/ADP to AMP, and directly competing with Teffs for cysteine uptake (34). Tregs can also induce dendritic cells to consume essential amino acids, which inhibits T cell proliferation (35). It may be advantageous, therefore, for Tregs to have a metabolic profile distinct from that of Teffs, and the metabolic differences between Teffs and Tregs may be necessary for the ability of Tregs to function and suppress inflammation.
In vitro treatment with 2DG can impair glycolysis and prevent Teff proinflammatory cytokine production, survival, proliferation, and even differentiation (14). In contrast, Tregs were not affected by 2DG and instead were sensitive to mitochondrial inhibitors. Previous literature has demonstrated that the glycolytic enzyme GAPDH can bind to IFNG mRNA and inhibit its translation to link glycolysis with inflammatory cytokine production (22). Here, we show that 2DG treatment also inhibited Teff proliferation and expression of the effector transcription factors T-bet and RORγT, demonstrating a different pathway by which glycolytic inhibition can affect T cell differentiation. Mitochondrial inhibition, in contrast, suppressed Tregs. This may have been partly due to reduced ATP generation or altered ROS regulation, given the modulation of Tregs by the antioxidant NAC.
Given that Tregs utilize mitochondrial oxidation while Teffs convert glucose-derived pyruvate to lactate, the branch point between glycolysis and glucose oxidation was identified as a potential target to modulate the balance between Teffs and Tregs. PDHK is a primary regulator of this branch point and inhibits PDH to suppress glucose oxidation and instead promote lactate production and glycolysis. Interestingly, PDHK1 was differentially expressed in T cell subsets, with robust expression in Th17, but little expression in Th1 and intermediate expression in Tregs. Naive CD4+ T cells also expressed PDHK1, although these cells are predominantly oxidative rather than glycolytic. DCA inhibits PDHK and is currently under investigation in several different types of cancer (38–42), where it may be effective by inhibition of aerobic glycolysis by suppressing generation of lactate and instead directing glucose to be oxidized in the mitochondria. DCA also can promote ROS in several cancer models that may drive cancer cell senescence (15, 38, 42). In the immune system, treatment of human peripheral blood mononuclear cells (PBMCs) with DCA inhibited proinflammatory cytokine production and promoted FoxP3 expression in the setting of asthma and alloreactivity (19–21). However, these studies did not directly examine the role of PDHK or DCA in T cell subsets or establish a mechanism of action.
Our data show that DCA inhibits Teff differentiation and cytokine production and does so by inhibition of PDHK1 and subsequent alteration of glucose utilization and ROS. Promoting glucose oxidation through PDHK1 inhibition selectively suppresses Th17 generation; this is mediated in part through the generation of ROS. In contrast, Th1 cells are not regulated by PDHK1 and instead maintain a glycolytic program through an independent pathway. Several recent reports suggest that Teffs are more sensitive to ROS stress than Tregs (27, 28, 30). Deletion of the antioxidant molecule peroxiredoxin II in T cells increased Tregs and prevented dextran sulfate sodium–induced colitis (28). In our study, Tregs were found to have high levels of the antioxidant glutathione. While Tregs also have high levels of ROS compared with Teffs, Tregs likely produce ROS during mitochondrial oxidation and are equipped with antioxidant molecules to handle this ROS generation. Our data suggest that Th17 cells are less able to handle ROS stress generated by the activation of PDH and subsequent glucose oxidation, as DCA inhibited IL-17 production. Consistent with this model, neutralization of ROS with the antioxidant NAC normalized IL-17 production. Therefore, PDHK selectively modulates the balance between Th17 cells and Tregs, in part through ROS generation.
Previous studies have described potential therapeutic strategies to modulate the balance between Teffs and Tregs. Each subset plays a different role in immunity, and it may be therapeutically useful to inhibit one of the subtypes without affecting the others. The finding that Th1 cells express little PDHK1 provides the potential to fine tune CD4+ T cell differentiation and the immune response. Inhibition of PDHK1 did not affect T-bet expression or IFN-γ production in Th1 cells, but suppressed RORγT and IL-17 production in vitro. In addition, treatment of mice with DCA in the context of IBD suppressed IL-17, but preserved IFN-γ, production. Our data therefore suggest that targeting PDHK1 inhibits Th17 differentiation and cytokine production while leaving Th1 unaffected. This may be important in the context of Th17-driven autoimmune disorders, where it would be favorable to suppress the Th17 lineage without affecting Th1 immunity. Overall, this work identifies key metabolic differences between the CD4+ T cell subsets that establish PDHK1 as a selective regulator of the Th17/Treg balance and inflammatory disease.
Methods
Mice.
Six- to eight-week-old sex-matched C57BL/6J mice (Jackson Laboratory) were used for all experiments unless otherwise indicated.
T cell isolation and differentiation.
Naive CD4+CD25– T cells were isolated ex vivo, and T helper cell subsets were generated as described previously (13). Briefly, CD4+CD25– T cells were cultured on irradiated (30 Gy) splenocytes with 2.5 μg/ml of anti-CD3 antibody at a ratio of 5:1 in RPMI 1640 media supplemented with 10% FBS, sodium pyruvate, penicillin/streptomycin, HEPES, and β-mercaptoethanol. The following cytokines were added to generate each subset: Th1, 10 ng/ml IL-12 (R&D Systems), 10 μg/ml anti–IL-4 (eBioscience, clone 11B11), 1 μg/ml anti–IFN-γ (eBioscience); Th17, 20 ng/ml IL-6 (R&D Systems), 2.5 ng/ml TGF-β (R&D Systems), 10 μg/ml anti–IFN-γ; Tregs, 3 ng/ml TGF-β. On day 3 after stimulation, cells were split 1:2 and replated with 20 ng/ml IL-2 alone for an additional 2 days prior to analysis. In some experiments, cells were treated with 5 nM rotenone or 250 μM 2DG or labeled with CTV (Invitrogen) per the manufacturer’s instructions to assess cell proliferation.
qRT-PCR.
RNA was isolated from naive (Th0), Th1, or Th17 cells or Tregs using RNeasy Plus Mini Kit (QIAGEN) following the manufacturer’s instructions. 1 μg total RNA was subjected to single-strand cDNA synthesis using the RT2 First Strand Kit (QIAGEN). In certain experiments, the cDNA was used according to the manufacturer’s instructions (QIAGEN) in the mitochondrial energy metabolism and glucose metabolism SuperArray RT2 Profiler PCR arrays or a custom array designed to include metabolic genes in pathways important in T cell activation (QIAGEN, CAPM1256). The PCR arrays were assayed on the ViiA 7 Real-Time PCR System (Applied Biosystems). Data were analyzed using the RT2 Profiler Program supplied by QIAGEN and normalized to the reference genes TATA box-binding protein (Tbp) and β-glucuronidase (Bgu), as determined by GeNorm stability analysis.
Immunoblots.
Protein expression was measured as previously described (13). Briefly, cells were lysed in a buffer containing Triton X-100 and SDS with protease and phosphatase inhibitors. The following antibodies were used: GLUT1 (ab652, Abcam), GLUT3 (Millipore), HK1 (Millipore), HK2 (Millipore), HK3 (Abcam), OXPHOS antibody cocktail (Abcam), CPT1A, cytochrome c (BD Biosciences), actin (Abcam), and PDHK1 (Abcam).
Metabolomics and acyl-carnitines.
Nontargeted metabolomic analyses were performed as described using LC Q Exactive Mass Spectrometer (LC-QE-MS) (Thermo Scientific) (24). Additional metabolomics analysis was performed using GC/MS and LC/MS/MS platforms (Metabolon Inc.) for determination of cellular metabolites. Data were then grouped by unsupervised clustering using MetaboAnalyst software. Samples were normalized using Bradford protein concentration and rescaled to set the median to 1. Missing values were imputed with the minimum value. Acyl-carnitines were measured by targeted flow injection–MS/MS, as previously described (43).
Metabolic assays.
T cells were differentiated, and OCR and ECAR were analyzed using the XF24 Extracellular Flux Analyzer (Seahorse Bioscience). OCR and ECAR values were normalized to cell number. Glycolytic capacity was defined as the difference between ECAR following the injection of 1 μM oligomycin (Seahorse Bioscience) and the basal ECAR reading (T cells cultured in base DMEM with no added glucose or glutamine). Glycolytic reserve was defined as the difference in ECAR between the glucose and oligomycin injections. SRC was defined as the percentage increase in OCR between the initial basal readings (T cells cultured in base DMEM with 25 mM glucose added) and the injection of 500 nM of the ionophore FCCP (Seahorse Bioscience) to uncouple oxidative phosphorylation and electron transport. In some experiments, OCR and ECAR were measured in the presence of rotenone (5 nM) and 2DG (250 μM). Glycolytic flux and glucose and pyruvate oxidation were measured as previously described (44). Briefly, glycolytic flux was determined by measuring the detritiation of [3H]-glucose. Glucose and pyruvate oxidation were measured by culture of cells in U-14C glucose or U-14C pyruvate to measure production of 14CO2.
Viability and intracellular staining.
The following antibodies were used: rat anti-mouse CD4+ Pacific Blue, IL-2 phycoerythrin (PE), IL-17 PE, IFN-γ APC, T-bet PE, RORγT PE, FoxP3 PE, and goat anti-rabbit PE (all from eBioscience). Cell death was measured by exclusion of 1 μg/ml propidium iodide. To measure intracellular cytokines, cells were left unstimulated or stimulated for 4 to 5 hours with PMA (50 ng/ml, Sigma-Aldrich) and ionomycin (750 ng/ml, Calbiochem) in the presence of GolgiPlug (IL-2, IFN-γ, IL-17) or GolgiStop (IL-17), permeabilized using Cytofix/Cytoperm Plus (BD), then stained with the appropriate antibodies. Transcription factor staining to identify committed cells was performed using the Mouse Regulatory T Cell Staining Kit (eBioscience) and intracellular staining for T-bet, RORγT, or FoxP3. Intracellular staining for GLUT1 and HK2 was performed as previously described (13). Data were acquired on a MacsQuant Analyzer (Miltenyi Biotec) and analyzed using FlowJo (TreeStar software).
Lentiviral PDHK1 shRNA.
PDHK1 shRNA–expressing and control lentiviruses were purchased from Sigma-Aldrich (puromycin resistance) or Origene (GFP-expressing). CD4+ T cells were isolated and differentiated as described above and infected with PDHK1 shRNA or control lentivirus 24 hours after activation. Polybrene (8 μg/ml) was added to facilitate the infection, and the cells were centrifuged for 90 minutes at 750 g. Where indicated, the cells were split 1:2 after 2 days and treated with 2 μg/ml puromycin for 2 days to select for infected cells.
T cell transfer model of colitis.
Wild-type CD4+ T cells were isolated as described above, and naive effector (CD4+CD25–CD45RBhi) T cells were sorted (FACSVantage, BD Bioscience). Naive Teffs were injected i.p. into 6- to 8-week-old C57BL/6 Rag1–/– recipients (Jackson Laboratories) (4 × 105 cells/mouse). Mice were given normal water or treated with 2 g/l DCA in the drinking water throughout the course of the experiment. Two weeks after T cell transfer, mice were given 200 ppm piroxicam (Sigma-Aldrich) in powdered rodent chow for 5 days to disrupt the intestinal barrier and initiate disease (31).
EAE.
EAE was induced as previously described (45). Briefly, wild-type mice were injected with 100 ng MOG35–55 peptide (New England Peptide) mixed with CFA, including heat-killed Mycobacterium tuberculosis (Sigma-Aldrich) followed by 200 ng pertussis toxin administered by i.p injection on days 0 and 2. Vehicle or DCA (2 g/l) was given to the mice in their drinking water. Clinical signs of EAE were assessed according to the following score: 0, no signs of disease; 1, loss of tone in the tail; 2, hind limb paresis; 3, hind limb paralysis; 4, tetraplegia. Lymphocyte infiltration and spinal cord demyelination were assessed in sections of spinal cords from mice stained with H&E and LFB as previously described (46). In select experiments, splenocytes from 2D2 mice with TCR transgenic T cells specific for MOG were stimulated with 20 μg/ml MOG peptide and 0.5 ng/ml IL-12 in vitro for 48 hours and then transferred into Rag1–/– recipients to induce EAE. T cells were then isolated from spinal cord for qRT-PCR.
Statistics.
Data were analyzed using a 2-tailed Student’s t test, and P < 0.05 was considered significant. For the EAE model, vehicle and DCA-treated mice were analyzed by 2-way ANOVA, and P < 0.05 was considered significant.
Study approval.
All animal studies were approved by the Institutional Animal Care and Use Committee at Duke University and the Novartis Institutes for Biomedical Research review board.
Supplementary Material
Acknowledgments
We would like to acknowledge members of the Rathmell lab for support and helpful discussions and Marshall Nichols for technical assistance. This work was supported by grants from the Kenneth Rainin Foundation (to J.C. Rathmell), the Leukemia and Lymphoma Society (to J.C. Rathmell), the Crohn’s and Colitis Foundation of America (Senior Research Award to J.C. Rathmell; postdoctoral fellowship 284879 to A.N. Macintyre), the Alliance for Lupus Research (Target Identification in Lupus to J.C. Rathmell), and the National Multiple Sclerosis Society (RG4536-A-1 to M.L. Shinohara)and by NIH grants R01HL108006 (to J.C. Rathmell), R56AI102074 (to J.C. Rathmell), R00CA168997 (to J.W. Locasale), and R01AI110613 (to J.W. Locasale).
Footnotes
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Conflict of interest: Marta E. Slawinska, Lea Haeberli, Catherine Huck, Paul A. Smith, and Martin A. Schneider are employees of Novartis Institutes of Biomedical Research. Laurence A. Turka owns equity in, and has a family member employed by, Novartis.
Reference information:J Clin Invest. 2015;125(1):194–207. doi:10.1172/JCI76012.
References
- 1.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buckner JH. Mechanisms of impaired regulation by CD4CD25(+)FOXP3(+) regulatory T cells in human autoimmune diseases. Nat Rev Immunol. 2010;10(12):849–859. doi: 10.1038/nri2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cua DJ, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421(6924):744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 4.Yen D, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116(5):1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serody JS, Hill GR. The IL-17 differentiation pathway and its role in transplant outcome. Biol Blood Marrow Transplant. 2012;18(1 suppl):S56–S61. doi: 10.1016/j.bbmt.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5(11):844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 8.Frauwirth KA, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16(6):769–777. doi: 10.1016/S1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 9.Maciver NJ, Jacobs SR, Wieman HL, Wofford JA, Coloff JL, Rathmell JC. Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J Leuk Biol. 2008;84(4):949–957. doi: 10.1189/jlb.0108024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs SR, et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J Immunol. 2008;180(7):4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol. 2013;14(5):500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35(6):871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michalek RD, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186(6):3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208(7):1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplon J, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498(7452):109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- 16.Hitosugi T, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell. 2011;44(6):864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korotchkina LG, Patel MS. Site specificity of four pyruvate dehydrogenase kinase isoenzymes toward the three phosphorylation sites of human pyruvate dehydrogenase. J Biol Chem. 2001;276(40):37223–37229. doi: 10.1074/jbc.M103069200. [DOI] [PubMed] [Google Scholar]
- 18.Michelakis ED, et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med. 2010;2(31):31ra4. doi: 10.1126/scitranslmed.3000677. [DOI] [PubMed] [Google Scholar]
- 19.Bian L, et al. Dichloroacetate alleviates development of collagen II-induced arthritis in female DBA/1 mice. Arthritis Res Ther. 2009;11(5):R132. doi: 10.1186/ar2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eleftheriadis T, et al. Dichloroacetate at therapeutic concentration alters glucose metabolism and induces regulatory T-cell differentiation in alloreactive human lymphocytes. J Basic Clin Physiol Pharmacol. 2013;24(4):271–276. doi: 10.1515/jbcpp-2013-0001. [DOI] [PubMed] [Google Scholar]
- 21.Ostroukhova M, et al. The role of low-level lactate production in airway inflammation in asthma. Am J Physiol Lung Cell Mol Physiol. 2012;302(3):L300–L307. doi: 10.1152/ajplung.00221.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153(6):1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sena LA, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38(2):225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Ser Z, Locasale JW. Development and quantitative evaluation of a high-resolution metabolomics technology. Anal Chem. 2014;86(4):2175–2184. doi: 10.1021/ac403845u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schooneman MG, Vaz FM, Houten SM, Soeters MR. Acylcarnitines: reflecting or inflicting insulin resistance? Diabetes. 2013;62(1):1–8. doi: 10.2337/db12-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato M, Li J, Chuang JL, Chuang DT. Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure. 2007;15(8):992–1004. doi: 10.1016/j.str.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mougiakakos D, Johansson CC, Kiessling R. Naturally occurring regulatory T cells show reduced sensitivity toward oxidative stress-induced cell death. Blood. 2009;113(15):3542–3545. doi: 10.1182/blood-2008-09-181040. [DOI] [PubMed] [Google Scholar]
- 28.Won HY, et al. Ablation of peroxiredoxin II attenuates experimental colitis by increasing FoxO1-induced Foxp3+ regulatory T cells. J Immunol. 2013;191(8):4029–4037. doi: 10.4049/jimmunol.1203247. [DOI] [PubMed] [Google Scholar]
- 29.Kim HR, et al. Attenuation of experimental colitis in glutathione peroxidase 1 and catalase double knockout mice through enhancing regulatory T cell function. PLoS One. 2014;9(4):e95332. doi: 10.1371/journal.pone.0095332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mougiakakos D, Johansson CC, Jitschin R, Bottcher M, Kiessling R. Increased thioredoxin-1 production in human naturally occurring regulatory T cells confers enhanced tolerance to oxidative stress. Blood. 2011;117(3):857–861. doi: 10.1182/blood-2010-09-307041. [DOI] [PubMed] [Google Scholar]
- 31.Hale LP, Gottfried MR, Swidsinski A. Piroxicam treatment of IL-10-deficient mice enhances colonic epithelial apoptosis and mucosal exposure to intestinal bacteria. Inflamm Bowel Dis. 2005;11(12):1060–1069. doi: 10.1097/01.MIB.0000187582.90423.bc. [DOI] [PubMed] [Google Scholar]
- 32.Ivanov, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126(6):1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 33.McGeachy MJ, Stephens LA, Anderton SM. Natural recovery and protection from autoimmune encephalomyelitis: contribution of CD4+CD25+ regulatory cells within the central nervous system. J Immunol. 2005;175(5):3025–3032. doi: 10.4049/jimmunol.175.5.3025. [DOI] [PubMed] [Google Scholar]
- 34.Kidani Y, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14(5):489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146(5):772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Windt GJ, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36(1):68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sukumar M, et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest. 2013;123(10):4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stockwin LH, et al. Sodium dichloroacetate selectively targets cells with defects in the mitochondrial ETC. Int J Cancer. 2010;127(11):2510–2519. doi: 10.1002/ijc.25499. [DOI] [PubMed] [Google Scholar]
- 39.Sun RC, Fadia M, Dahlstrom JE, Parish CR, Board PG, Blackburn AC. Reversal of the glycolytic phenotype by dichloroacetate inhibits metastatic breast cancer cell growth in vitro and in vivo. Breast Cancer Res Treat. 2010;120(1):253–260. doi: 10.1007/s10549-009-0435-9. [DOI] [PubMed] [Google Scholar]
- 40.Sutendra G, Michelakis ED. Pyruvate dehydrogenase kinase as a novel therapeutic target in oncology. Front Oncol. 2013;3:38. doi: 10.3389/fonc.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garon EB, et al. Dichloroacetate should be considered with platinum-based chemotherapy in hypoxic tumors rather than as a single agent in advanced non-small cell lung cancer. J Cancer Res Clin Oncol. 2014;140(3):443–452. doi: 10.1007/s00432-014-1583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong F, et al. Dichloroacetate induces protective autophagy in LoVo cells: involvement of cathepsin D/thioredoxin-like protein 1 and Akt-mTOR-mediated signaling. Cell Death Dis. 2013;4:e913. doi: 10.1038/cddis.2013.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An J, et al. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat Med. 2004;10(3):268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- 44.Caro-Maldonado A, et al. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J Immunol. 2014;192(8):3626–3636. doi: 10.4049/jimmunol.1302062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michalek RD, et al. Estrogen-related receptor-alpha is a metabolic regulator of effector T-cell activation and differentiation. Proc Natl Acad Sci U S A. 2011;108(45):18348–18353. doi: 10.1073/pnas.1108856108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inoue M, Williams KL, Gunn MD, Shinohara ML. NLRP3 inflammasome induces chemotactic immune cell migration to the CNS in experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2012;109(26):10480–10485. doi: 10.1073/pnas.1201836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.