Abstract
Autophagy is a survival-promoting pathway that captures, degrades, and recycles intracellular proteins and organelles in lysosomes. Autophagy preserves organelle function, prevents the toxic buildup of cellular waste products, and provides substrates to sustain metabolism in starvation. Although in some contexts autophagy suppresses tumorigenesis, in most contexts autophagy facilitates tumorigenesis. Cancers can upregulate autophagy to survive microenvironmental stress and to increase growth and aggressiveness. Mechanisms by which autophagy promotes cancer include suppressing induction of the p53 tumor suppressor protein and maintaining metabolic function of mitochondria. Efforts to inhibit autophagy to improve cancer therapy have thereby attracted great interest.
Autophagy is a tissue-specific regulator of homeostasis and survival
Autophagy captures and degrades intracellular components such as proteins and organelles to sustain metabolism and homeostasis. Low levels of basal autophagy prevent the gradual accumulation of damaged proteins and organelles in tissues that is toxic over time; thus, autophagy plays an important role in protein and organelle quality control (1). Identification of the autophagy substrates that are deregulated in autophagy-deficient cells and tissues is important to understand the biological role and tissue specificity of autophagy. Determination of the global impact of autophagy on the cellular proteome would be a significant advance, as we are only beginning to understand the broad scope of autophagy substrates and the functional consequence of deregulating their degradation and recycling.
Some tissues such as liver, brain, and muscle are particularly dependent on autophagy to prevent the buildup of damaged mitochondria and protein aggregates containing the autophagy substrate p62/SQSTM1 (p62) and ubiquitin (1). Accumulation of defective mitochondria that results from impaired autophagy can perturb metabolism and generate oxidative stress (2). Autophagy also mitigates ER stress, and autophagy defects can produce accumulation of chaperone proteins that increase the unfolded protein burden (3, 4). Select protein elimination by autophagy is also important for homeostasis. The toxicity of autophagy defects in liver is partly ameliorated by deficiency in the autophagy substrate p62, but this is not the case for the brain (5, 6). The recent revelation that autophagy is important for recycling iron complexed with ferritin through ferritinophagy may be critical for iron homeostasis in many tissues, potentially including brain (7).
The accumulation of, or imbalance in, levels of some cellular components caused by autophagy defects may be indirect. For example, lipid accumulation in autophagy-deficient liver and some lung tumors can arise from defects in the autophagy of lipid droplets through lipophagy (8) or indirectly from defects in mitochondrial fatty acid oxidation (FAO) that repress lipid catabolism (9). These findings collectively demonstrate the broad effects of autophagy on cellular homeostasis at the level of substrate removal, maintenance of organelle function, and detoxification. Thus, autophagy dependence is tissue specific, not only for the general requirement for the level of autophagic activity, but also for the nature of the substrates that require autophagy-mediated elimination or recycling.
Acute autophagy induction is critical for yeast and normal mammalian cells and mammals to survive starvation, which is attributed to the recycling of intracellular components into metabolic pathways (2). Autophagy thereby functions to promote metabolic homeostasis and survival that is essential during nutrient deprivation. While it is generally appreciated that autophagy-mediated degradation of intracellular proteins and organelles provides metabolic substrates during starvation, the exact substrates that are important and the metabolic pathways they support remain to be identified (2, 10). The metabolic role for autophagy partly overlaps with the protein and organelle quality control function and further broadens the impact autophagy has on mammalian physiology and disease.
The role of autophagy in normal cells and tissues is clearly complex and tissue dependent (1). Autophagy deficiency is thought to contribute to the pathogenicity in many diseases including neurodegenerative diseases, liver disease, and aging (11). Autophagy has been reported to either inhibit or promote cancer cell proliferation or tumorigenesis in model systems, suggesting that the role of autophagy in cancer is context dependent (12). It is worth exploring these mechanisms, as they may reveal insights into novel means for regulating cancer growth.
Autophagy as a tumor suppressor
Autophagy was initially thought to be a tumor-suppression mechanism. This concept derived from early reports that the essential autophagy gene ATG6/BECN1 was monoallelically lost in 40% to 75% of human prostate, breast, and ovarian cancers (13–15). Indeed, in certain cell-based assays, autophagy suppression promotes cancer cell growth, and Becn1 heterozygous mutant mice are prone to development of liver and lung tumors and lymphomas with long latency (16, 17). In contrast, mosaic or liver-specific autophagy deficiency through deletion of the essential autophagy genes Atg5 or Atg7 in mice produces only benign liver tumors (18). These findings called into question the role of autophagy in tumor suppression in tissues other than liver and whether the potential role of BECN1 in other cancer is autophagy related.
The possibility that BECN1 is a tumor suppressor gene based on its allelic loss is confounded by its location adjacent to breast and ovarian tumor suppressor breast cancer 1, early onset (BRCA1) on human chromosome 17q21. Mutations in BRCA1 are known drivers in human breast and ovarian cancers. Moreover, hereditary breast and ovarian cancer arises from missense mutations in BRCA1 with subsequent loss of the wild-type allele that may or may not include deletion of BECN1 (19). In genetically engineered mouse models (GEMMs) for hereditary breast cancer, allelic loss of Becn1 promotes p53 activation and reduces tumorigenesis, which is the opposite result expected if Becn1 is acting as a tumor suppressor (20). Note that large-scale genomic analysis of human cancers to date has failed to identify recurrent mutations in BECN1 or other essential autophagy genes (21, 22).
The mutational status of BECN1 was assessed in the human tumor sequencing data from 10,000 tumors with matched normal tissue in The Cancer Genome Atlas (TCGA; https://tcga-data.nci.nih.gov/tcga/) and other databases. Large deletions of both BRCA1 and BECN1, and deletions of only BRCA1 but not BECN1, were found in breast and ovarian cancers, consistent with BRCA1 loss being the driver mutation in these cancers (19). Furthermore, there was no evidence for statistically significant recurrent missense mutations in BECN1 in breast or ovarian cancers or mutation or loss in any other cancer, including prostate cancer (19). Thus, loss of BECN1 in human cancers cannot be disassociated from the loss of BRCA1, indicating that BECN1 is not a tumor suppressor in most human cancers (19). There may, however, be tumor types or subtypes that have not yet been sufficiently characterized at the genomic level, such as hepatomas, where autophagy genes may be mutated and where their loss of function may promote cancer.
The observation that autophagy deficiency in mice results in benign hepatomas (18) suggests that autophagy may be important in liver to suppress tumor initiation, but also that autophagy may be required for progression from benign to malignant disease. Possible mechanisms by which autophagy may promote tumor initiation have been identified (12), although demonstration that any of these occur in human cancers is so far lacking.
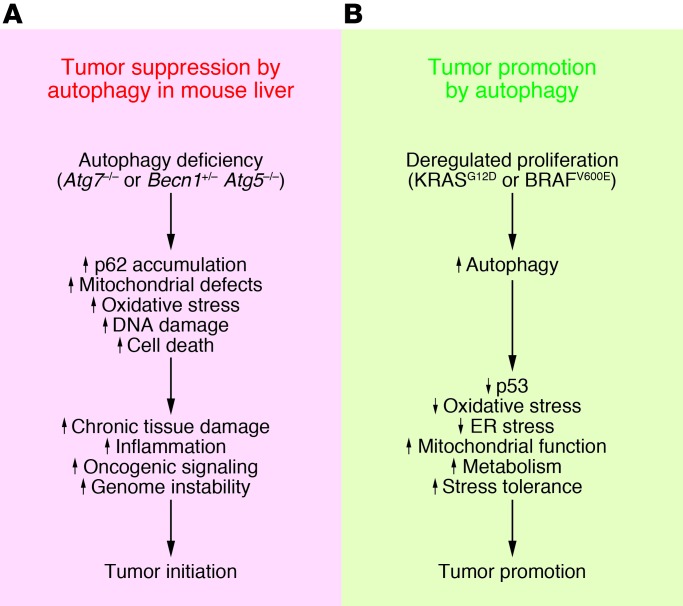
Autophagy deficiency causes oxidative stress, activation of the DNA damage response, and genome instability, a known cause of cancer initiation and progression (refs. 4, 23, 24, and Figure 1A). This increased oxidative stress activates the master regulator of antioxidant defense, nuclear factor, erythroid-2–like 2 (NRF2), which may also stimulate tumor growth (see below) (25). Loss of autophagy in liver is also toxic, producing chronic cell death of hepatocytes and inflammation (4, 6), which are known drivers of liver cancer (ref. 26 and Figure 1A). p62 deficiency reduces both the toxicity and tumorigenesis caused by defective autophagy, indicating that aberrant p62 accumulation that results from autophagy deficiency is partly the cause (Figure 1A). In other settings, p62 expression promotes oxidative stress and tumor growth (4), and induction of p62 expression due to amplification of chromosome 5q has been implicated as a contributing factor in the growth and pathogenicity of clear cell renal cell carcinoma (27). Conversely, p62 deficiency suppresses tumorigenesis in GEMMs for KRASG12D-driven lung cancer (28). How deregulation of p62 contributes to tumorigenesis is not known, but it is associated with increased oxidative stress. p62 is also a signaling adaptor that regulates many oncogenic pathways, including those involving NRF2, mTOR, and NF-κB (29). It will be of great interest to define how induction of p62 in autophagy-deficient cells alters cell function and possibly promotes tumorigenesis.
Figure 1. Proposed tumor-suppressing and tumor-promoting roles for autophagy in cancer.
(A) Proposed mechanisms by which autophagy may suppress tumorigenesis. Autophagy defects impair tissue health, leading to chronic tissue damage and regeneration that may create an environment that promotes cancer. (B) Proposed mechanisms by which autophagy promotes cancer by limiting stress responses and supporting metabolism and survival.
Autophagy as a tumor promoter
Cancer cells also rely on autophagy; in many cases, they are more autophagy dependent than normal cells and tissues. This is likely due to inherent deficiencies in the microenvironment and to the increased metabolic and biosynthetic demands imposed by deregulated proliferation (Figure 1B and ref. 2). For example, basal autophagy is upregulated in hypoxic tumor regions, where it is essential for tumor cell survival (30). Autophagy is also upregulated in RAS-transformed cancer cells and promotes their growth, survival, tumorigenesis (31–33), invasion, and metastasis (34). Mitochondrial metabolic defects and the resulting susceptibility to stress that result from autophagy deficiency in RAS-driven cancers are implicated as the underlying mechanism. These findings led to the concept that RAS-driven cancers may be “autophagy addicted” (31, 33, 35). ATG17/FIP200 deficiency also inhibits the growth of mammary cancers in mice driven by polyoma middle T antigen (36), suggesting that the role of autophagy in promoting tumorigenesis and the concept of autophagy addiction in cancer may be more broadly applicable. The genetic context that creates autophagy dependency in cancer is still poorly understood and remains to be investigated further.
While informative, these in vitro, transplantable, and virally induced cancer models do not recapitulate many facets of tumorigenesis, particularly those that relate to the role of autophagy in tumor-host interactions. In contrast, autochthonous GEMMs for spontaneous cancers driven by the activation of cellular oncogenes and the loss of tumor suppressor genes in an immune-competent host provide a more physiologic setting with which to address the role of autophagy in cancer. Given the tissue-specific, homeostatic function of autophagy, addressing the role of autophagy in cancer in physiologic settings, such as those where the immune system is intact and where tumors evolve within the relevant microenvironment, is absolutely critical.
Role of autophagy in GEMMs for RAS-driven cancers.
To determine the role of autophagy in spontaneously arising lung cancer in an immune-competent setting, the essential autophagy gene Atg7 was deleted concurrently with KRASG12D activation in mouse models for non–small-cell lung cancer (NSCLC). It is important to note that autophagy deficiency in this and other GEMMs occurs only in the tumor cells themselves, so these models address the tumor cell–autonomous role of autophagy and not its role in the microenvironment or elsewhere throughout the animal. Tumor-specific Atg7 deficiency causes tumors to accumulate dysfunctional mitochondria and to prematurely induce p53, proliferative arrest, and cell death, which reduces tumor burden (ref. 9 and Figure 1B). Most importantly, Atg7 loss alters tumor fate from that of adenomas and carcinomas to oncocytomas, which are rare, benign neoplasms characterized by the accumulation of defective mitochondria (37). Despite the reduction in tumor burden and the generation of benign instead of malignant neoplasms, there is no life span extension in animals bearing Atg7-deficient tumors because the mice die of pneumonia instead of cancer (9). Autophagy defects, including those in tumors, are known to stimulate inflammation (30, 38) that may trigger pneumonia. Alternatively, pneumonia may merely be the consequence of lungs bearing Atg7-deficient oncocytomas containing many dying tumor cells (9). It will be interesting to test whether autophagy deficiency in tumors directly activates pro-inflammatory pathways and an antitumor immune response.
The accelerated induction of p53, growth arrest, and cell death in NSCLCs lacking Atg7 suggested that augmentation of p53 function may limit tumor growth (9). To test this hypothesis, Trp53 was deleted concurrently with KRASG12D activation in the lung without and with Atg7 deletion, and the consequence on NSCLC was assessed. Although removing p53 clearly eliminates a barrier that limits the growth of Atg7-deficient tumors, tumor cells nonetheless accumulate damaged mitochondria, undergo growth arrest and cell death, and become oncocytomas rather than carcinomas (9). In contrast to the setting with p53 intact, in the setting of p53 deficiency, pneumonia does not readily occur in response to tumor-specific autophagy deficiency, and the reduction in tumor burden extends mouse life span (9). Thus, autophagy deficiency activates p53, which suppresses tumor growth, but clearly autophagy deficiency also limits tumor growth by other mechanisms independent of p53 (ref. 35 and Figure 1B).
A feature of Atg7-deficient lung tumors specific to p53 deletion is the accumulation of lipids (9). In comparison to those with ATG7, ATG7- and p53-deficient tumor-derived cell lines (TDCLs) from the aforementioned GEMMs are defective for survival in starvation in vitro and form lipidic cysts instead of tumors when transplanted back into mice (9). This suggests that autophagy loss causes defective utilization of lipid stores. Atg7 deficiency impairs FAO and increases sensitivity to FAO inhibition, indicating that without p53, RAS-driven tumors require autophagy to retain mitochondrial function for lipid catabolism. It is likely that this inability to catabolize lipids contributes to defective survival in starvation and may also impair tumor cell fitness in the context of tumorigenesis. These autophagy-defective TDCLs are also highly dependent on exogenously supplied glutamine to sustain mitochondrial respiration and to survive starvation (9). This suggests that, in addition to the role of autophagy in the preservation of mitochondrial lipid catabolism, autophagy-supplied substrates from protein degradation (particularly glutamine) may sustain tumor metabolism and growth.
These findings collectively indicate that autophagy is required for carcinoma fate in RAS-driven NSCLC, and that autophagy defects may underlie the genesis of human oncocytomas. This also suggests that autophagy inactivation can divert progression of lung carcinomas to benign disease, which would be a therapeutic advantage if it could be achieved in the clinic with an autophagy inhibitor. Moreover, this suggests that different oncogenic events can alter the autophagy requirement for distinct roles in metabolism (35). Deletion of Atg5 in RAS-driven NSCLC reduces tumorigenesis similarly to Atg7 deficiency, indicating that tumor promotion is autophagy mediated and does not result from autophagy-independent activity of Atg7 (39). Although Atg5 deficiency did not extend life span in the absence of p53, this may be due to Atg5 loss producing a less effective autophagy block than Atg7 loss. Alternatively, the comparison between Atg5–/– and Atg5–/+ and not Atg5+/+ tumor-bearing mice may have masked the phenotype (39).
Autophagy also promotes tumor growth in a mouse model of RAS-driven pancreatic cancer by suppressing p53 activation (40). In contrast to the lung cancer model, Trp53 loss in RAS-driven pancreatic cancer ameliorates tumor growth defects produced by loss of autophagy. It has been proposed that p53 loss specifically increases glycolysis and reduces oxidative metabolism in the pancreatic cancer model, thereby relieving the requirement for autophagy (40), indicating that there may be cell type–specific differences in autophagy dependency in cancer. It is also important to note that the lung and pancreatic cancer models are very different, which may influence the p53 dependence of autophagy. For example, in the lung model, cancer is induced sporadically in lung cells in adult mice through the inhalation of adenovirus expressing Cre recombinase that activates RAS and deletes Trp53. In contrast, the pancreas cancer model uses a developmental Cre that induces cancer during embryogenesis. Thus, the entire environment of cancer development (adult versus embryo, individual cells versus large fields of cells) is vastly different in the lung and pancreas cancer models. There is also a difference in the time course of disease, which takes more than four months in the lung cancer model, whereas all mice with pancreatic cancer succumb to the disease within four weeks of age (9, 40).
Given the differences in the lung and pancreatic cancer models, it will be interesting to test how autophagy deficiency alters RAS-driven pancreatic cancer when Trp53 loss is initially heterozygous and when cancer develops more gradually in adult animals with Trp53 loss of heterozygosity (LOH) in a manner that more closely resembles pancreatic cancer in humans. Indeed, this was recently done, and Atg5 deficiency blunted tumor growth and extended life span of mice with KRASG12D-driven pancreatic cancers in the context of Trp53 LOH (41). Moreover, autophagy inactivation genetically or pharmacologically with hydroxychloroquine (HCQ), which blocks lysosome function and the degradation of autophagy cargo, inhibits survival and tumorigenesis of human pancreatic cancer cell lines (33) and transplanted human pancreatic cancers (41). As human pancreatic cancers have mutations that inactivate p53 or the p53 pathway with high frequency, this suggests that autophagy promotes pancreatic cancer independent of the functional status of p53 (33, 41), contrary to findings in ref. 40.
Role of autophagy in GEMMs for BRAF-driven cancers.
To address how activation of an oncogene other than RAS in the lung would alter the requirement for autophagy, Atg7 was deleted in a mouse model of BRAFV600E-induced lung cancer in the presence or absence of Trp53 (25). Atg7 deletion transiently induces oxidative stress and accelerates tumor cell proliferation in a manner indistinguishable from ablation of Nrf2, the master regulator of antioxidant defense (25). Combined deletion of Atg7 and Nrf2 has no additive effect, suggesting that both genes promote initial tumorigenesis by regulating oxidative stress. Whether or not this happens in human cancers remains to be investigated, but this identifies another possible antitumor mechanism of autophagy: suppression of oxidative stress and thereby oncogenic signaling pathways and mutagenesis (Figure 1A).
At later stages of tumorigenesis, Atg7 deficiency causes p53 activation, accumulation of defective mitochondria, proliferative defects, reduced tumor burden, conversion of adenomas and adenocarcinomas to oncocytomas, and increased mouse survival (25). Thus, defects in autophagy may be the basis for the genesis of oncocytomas independent of the oncogenes and tumor suppressor genes involved in tumor development. As with the RAS-driven lung cancers, Atg7 deficiency accelerates p53 induction, and deletion of Trp53 only partly relieves the anticancer effects of autophagy deficiency (25). It is important to note that although autophagy deficiency initially stimulates early tumor growth, the dominant overall effect of autophagy deficiency is inhibition of tumor growth (25). Autophagy-deficient TDCLs are also defective for mitochondrial respiration and survival in starvation and are dependent on exogenously supplied glutamine. This suggests that as in RAS-driven lung cancers, autophagy-supplied substrates from protein degradation may sustain BRAFV600E tumor metabolism and growth (25, 42). Autophagy inhibition also sensitizes melanomas to leucine deprivation, which suggests that autophagy impairment creates multiple metabolic vulnerabilities (43).
The requirement for autophagy to promote the growth of BRAF-driven lung cancers is consistent with increased dependence of BRAF-driven melanomas on oxidative metabolism and autophagy (42). Human melanomas with activating mutations in BRAF have high levels of basal autophagy that correlate with aggressive disease and sensitivity to autophagy inactivation (44). Indeed, autophagy is a resistance mechanism to BRAF inhibition in BRAFV600E-mutant human melanomas (45). In this setting autophagy mitigates ER stress caused by BRAFV600E inhibition, highlighting the additional importance of therapy-induced autophagy as a resistance mechanism (46). Indeed, there are other examples of therapy-induced autophagy functioning as a resistance mechanism, suggesting that augmentation of the anticancer activity of autophagy inhibitor therapeutics may be generally applicable (47–50).
Challenges, unknowns, and therapeutic opportunities
The studies described above raise the possibility that autophagy inhibition may be therapeutically advantageous for cancer therapy, but they also raise additional questions and concerns. We have learned that autophagy inhibits oxidative stress, inflammation, p62 accumulation, and genome instability that may contribute to tumor suppression in some model systems, but it remains to be determined whether these events contribute to the suppression of human cancer. If so, essential autophagy genes should be represented among those genes with recurrent inactivating mutations in human cancers. Based on the currently available data, this does not appear to be the case. The possibility remains that loss of tumor suppression by autophagy in cancer occurs indirectly, for example, through activation of mTOR or Her2/Neu signaling (51, 52).
We have also learned that tumor-specific autophagy inactivation inhibits tumor growth and survival in part by promoting p53 activation, but also does so by p53-independent mechanisms (9, 25, 41). How autophagy deficiency activates p53 is not yet known, but p53 is activated by metabolic deficiency and AMPK activation or by oxidative stress and induction of DNA damage (53). Metabolic defects caused by accumulation of defective mitochondria would be expected to compromise tumor growth and fitness independent of p53, as is observed in lung and pancreas cancer models (9, 25, 41, 54). Identification of the autophagy substrates and the metabolic pathways they support for cellular survival in the face of oncogenic or microenvironmental stress will be important. Data indicate that generation of glutamine from protein degradation or of fatty acids from lipid catabolism may be important (9, 25). Perhaps impairment of ferritinophagy and iron homeostasis critical for the function of many cellular enzymes could also influence tumor growth (7). The role of autophagy in mitigation of ER stress and maintenance of the functional pool of mitochondria needs to be addressed further. It is important to note that inactivation of the essential mitochondrial transcription factor, mitochondrial transcription factor A (TFAM), prevents the growth of RAS-driven lung cancers demonstrating the general importance of mitochondrial function for tumor growth (55).
One major limitation of the work to date is that all the cancer models have addressed the role of autophagy only in tumors and not with direct comparison to autophagy deficiency in normal tissues. Since we know autophagy is important for some normal tissues, a critical question is whether systemic autophagy inactivation will be sufficiently selective to impair cancer growth while sparing normal tissues from the deleterious consequences. Indeed, this has recently been done genetically — systemic Atg7 ablation was selectively detrimental to established lung tumors compared with normal tissue (54). Thus, we will need to determine the best means to therapeutically inhibit autophagy for cancer therapy, and to determine which patient subpopulations would most benefit from this approach. HCQ is currently being used in the clinic to interfere with lysosome function and the degradation of autophagy cargo, but it is not yet clear whether this will be selective and sufficiently effective or specific (46). Targeting essential components of the autophagy machinery may be required to address whether or not this approach is therapeutically advantageous.
Acknowledgments
I wish to thank all of the members of the White laboratory for their thoughtful comments and discussions. The White laboratory acknowledges support from the NIH grants R01CA163591 and R01CA130893, the Val Skinner Foundation, the New Jersey Commission for Cancer Research, and the Rutgers Cancer Institute of New Jersey.
Footnotes
Conflict of interest: Eileen White has a financial interest in Forma Therapeutics.
Reference information:J Clin Invest. 2015;125(1):42–46. doi:10.1172/JCI73941.
References
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330(6009):1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14(9):1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 4.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137(6):1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 6.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 7.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509(7498):105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo JY, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27(13):1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White E. Exploiting the bad eating habits of Ras-driven cancers. Genes Dev. 2013;27(19):2065–2071. doi: 10.1101/gad.228122.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aita VM, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59(1):59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 14.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(7):651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 15.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 16.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25(8):795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laddha SV, Ganesan S, Chan CS, White E. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Mol Cancer Res. 2014;12(4):485–490. doi: 10.1158/1541-7786.MCR-13-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huo Y, et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer Discov. 2013;3(8):894–907. doi: 10.1158/2159-8290.CD-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karantza-Wadsworth V, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21(13):1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathew R, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21(11):1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strohecker AM, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3(11):1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun B, Karin M. Inflammation and liver tumorigenesis. Front Med. 2013;7(2):242–254. doi: 10.1007/s11684-013-0256-4. [DOI] [PubMed] [Google Scholar]
- 27.Li L, et al. SQSTM1 is a pathogenic target of 5q copy number gains in kidney cancer. Cancer Cell. 2013;24(6):738–750. doi: 10.1016/j.ccr.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duran A, et al. The signaling adaptor p62 is an important NF-κB mediator in tumorigenesis. Cancer Cell. 2008;13(4):343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Moscat J, Diaz-Meco MT. p62: a versatile multitasker takes on cancer. Trends Biochem Sci. 2012;37(6):230–236. doi: 10.1016/j.tibs.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10(1):51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25(5):460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lock R, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22(2):165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25(7):717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lock R, Kenific CM, Leidal AM, Salas E, Debnath J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 2014;4(4):466–479. doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155(6):1216–1219. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25(14):1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasparre G, Romeo G, Rugolo M, Porcelli AM. Learning from oncocytic tumors: why choose inefficient mitochondria? Biochim Biophys Acta. 2011;1807(6):633–642. doi: 10.1016/j.bbabio.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao S, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun. 2014;5:3056. doi: 10.1038/ncomms4056. [DOI] [PubMed] [Google Scholar]
- 40.Rosenfeldt MT, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504(7479):296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 41.Yang A, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4(8):905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strohecker AM, White E. Targeting mitochondrial metabolism by inhibiting autophagy in BRAF-driven cancers. Cancer Discov. 2014;4(7):766–772. doi: 10.1158/2159-8290.CD-14-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheen JH, Zoncu R, Kim D, Sabatini DM. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell. 2011;19(5):613–628. doi: 10.1016/j.ccr.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma XH, et al. Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res. 2011;17(10):3478–3489. doi: 10.1158/1078-0432.CCR-10-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma XH, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124(3):1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amaravadi RK, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17(4):654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amaravadi R. Autophagy can contribute to cell death when combining targeted therapy. Cancer Biol Ther. 2009;8(21):130–133. doi: 10.4161/cbt.8.21.10416. [DOI] [PubMed] [Google Scholar]
- 48.Bray K, et al. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PLoS One. 2012;7(7):e41831. doi: 10.1371/journal.pone.0041831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorr JR, et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature. 2013;501(7467):421–425. doi: 10.1038/nature12437. [DOI] [PubMed] [Google Scholar]
- 50.Xie X, White EP, Mehnert JM. Coordinate autophagy and mTOR pathway inhibition enhances cell death in melanoma. PLoS One. 2013;8(1):e55096. doi: 10.1371/journal.pone.0055096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lozy F, et al. ERBB2 overexpression suppresses stress-induced autophagy and renders ERBB2-induced mammary tumorigenesis independent of monoallelic Becn1 loss. Autophagy. 2014;10(4):662–676. doi: 10.4161/auto.27867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126(pt 8):1713–1719. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20(7):427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karsli-Uzunbas G, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4(8):914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107(19):8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]