Abstract
Premature ovarian failure (POF) is a genetically and phenotypically heterogeneous disorder that includes individuals with manifestations ranging from primary amenorrhea to loss of menstrual function prior to age 40. POF presents as hypergonadotropic hypogonadism and can be part of a syndrome or occur in isolation. Here, we studied 3 sisters with primary amenorrhea, hypothyroidism, and hypergonadotropic hypogonadism. The sisters were born to parents who are first cousins. SNP analysis and whole-exome sequencing revealed the presence of a pathogenic variant of the minichromosome maintenance 8 gene (MCM8, c.446C>G; p.P149R) located within a region of homozygosity that was present in the affected daughters but not in their unaffected sisters. Because MCM8 participates in homologous recombination and dsDNA break repair, we tested fibroblasts from the affected sisters for hypersensitivity to chromosomal breaks. Compared with fibroblasts from unaffected daughters, chromosomal break repair was deficient in fibroblasts from the affected individuals, likely due to inhibited recruitment of MCM8 p.P149R to sites of DNA damage. Our study identifies an autosomal recessive disorder caused by an MCM8 mutation that manifests with endocrine dysfunction and genomic instability.
Introduction
Premature ovarian failure (POF) affects 1%–4% of women and is defined as a cessation of menses prior to age 40, with elevated follicle-stimulating hormone (FSH) and low serum estradiol levels (1). Women with POF present with amenorrhea (primary or secondary) and hypoestrogenic symptoms (i.e., hot flashes, vaginal dryness, premature osteoporosis). POF is genetically heterogeneous (2), with few genes identified, and can be idiopathic and nonsyndromic or part of a genetic syndrome.
Minichromosomal maintenance proteins MCM2–7 participate in DNA replication elongation and prereplication complex formation (3), but have not been associated with human disorders. MCM8 and its physical partner MCM9 are newly discovered members of the MCM family and were initially implicated in DNA replication (4). We show that an autosomal recessive pathogenic variant in MCM8 can cause POF and increased genomic instability.
Results and Discussion
Three sisters (IV-1, IV-6, and IV-9) from a consanguineous family presented for clinical evaluation of hypergonadotropic primary amenorrhea (Figure 1A). Detailed clinical findings are provided in Supplemental Table 1 (supplemental material available online with this article; doi:10.1172/JCI78473DS1). All 3 sisters have a normal 46,XX karyotype, elevated FSH levels, infantile uteri, and small ovaries (Supplemental Figure 1). Secondary sexual characteristics were delayed. All 3 probands are currently being treated with estrogen and progesterone replacement therapy and experiencing regular menstrual cycles. All 3 patients were also diagnosed with hypothyroidism and responded to thyroxine. The mother (III-2) entered menarche at age 14, reported normal pubertal development, and regular menstrual periods (26–28 day menstrual cycles) until her mid-40s, and her last recorded menstrual period was at the age of 49. There is no known family history of anemia, blood dyscrasias, photosensitivity, immunodeficiency, or malignancies. We ruled out autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED) syndrome (MIM 240300) by the absence of mucocutaneous candidiasis, hypocalcemia, hypoglycemia, hypotension, vitiligo, alopecia, anemia, or hepatitis in the affected daughters. We did not identify pathogenic variants in the gene that causes APECED: AIRE (MIM 607358). Also, we did not detect antithyroid or antiadrenal gland antibodies in the affected daughters. In summary, the 3 daughters had idiopathic hypergonadotropic primary amenorrhea with hypothyroidism, atrophic ovaries, and normal female karyotype.
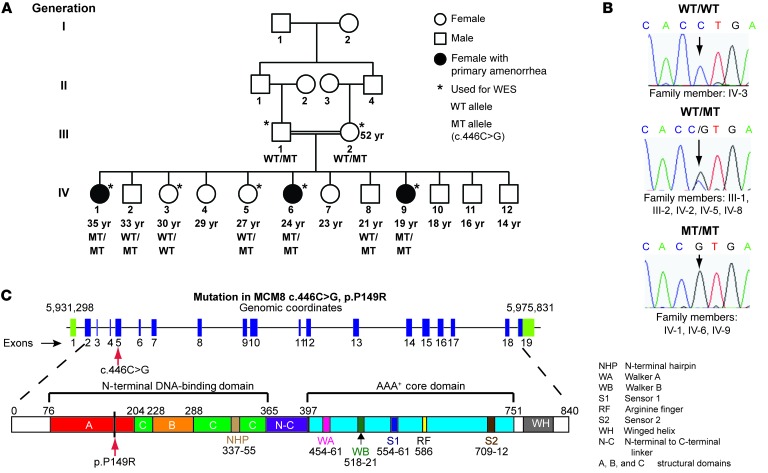
Figure 1. Pedigree of a family with 3 daughters afflicted by premature ovarian failure and homozygous for the MCM8 c.446C>G variant.
(A) Family members are designated by Arabic numerals. Horizontal lines between individuals represent marriage. Double horizontal lines indicate consanguinity in a marriage. Vertical lines represent lineage. Below each individual, the individual’s current age (if known) and MCM8 genotype are provided. (B) Sanger sequencing was used to validate genotypes, and representative chromatograms are shown. Individuals who are heterozygous for the c.446C>G MCM8 variant show overlapping C and G peaks (middle graph). Individuals homozygous for the c.446C>G MCM8 variant have a single G peak (bottom graph). (C) MCM8 is encoded on chromosome 20: 5,931,298-5,975,831 (NCBI37/hg19), and the c.446C>G variant in exon 5 is shown (red arrow). Full boxes represent exons (blue denotes coding sequences; green denotes noncoding sequences), and introns are indicated by lines. MCM8 consists of an N-terminal DNA-binding domain and a AAA+ core domain. The c.446C>G substitution caused a change in the amino acid sequence p.P149R within the predicted DNA-binding domain (red arrow). All domains are color coded with the homology model (Supplemental Figure 3).
SNP analyses identified a 3.3-Mb region of homozygosity on chromosome 20p13-p12.3 flanked by rs1547618 and rs1012891, present only in affected subjects (Supplemental Table 2). No known POF genes lay in this interval, and we performed whole-exome sequencing (WES) to identify pathogenic variants. WES revealed 2 nonsynonymous variants, FERMT1 (MIM 607900) and MCM8 (MIM 608187), which met autosomal recessive inheritance filter criteria and mapped to the region of homozygosity with the highest LOD score on chromosome 20. The FERMT1 variant (NM_017671:c.293G>A, p.R98H) was previously reported (rs137862671) in public databases and is likely a benign variant. Pathogenic variants in FERMT1 cause Kindler syndrome (MIM 173650), and none of the manifestations of Kindler syndrome (i.e., congenital blistering, skin atrophy, photosensitivity, skin fragility, scaling) were reported in the family we studied here.
The MCM8 (NM_032485) c.446C>G missense variant remained as the only candidate for the observed phenotype. MCM8 c.446C>G has not been reported in either the Exome Variant Server or 1000 Genomes databases. The MCM8 c.446C>G variant was verified in the family by Sanger sequencing (Figure 1B) and was absent in 200 fertile women. The resulting protein mutation, p.P149R, occurs at a highly conserved residue located in the N-terminal DNA-binding domain of MCM8 (Figure 1C and Supplemental Figure 2).
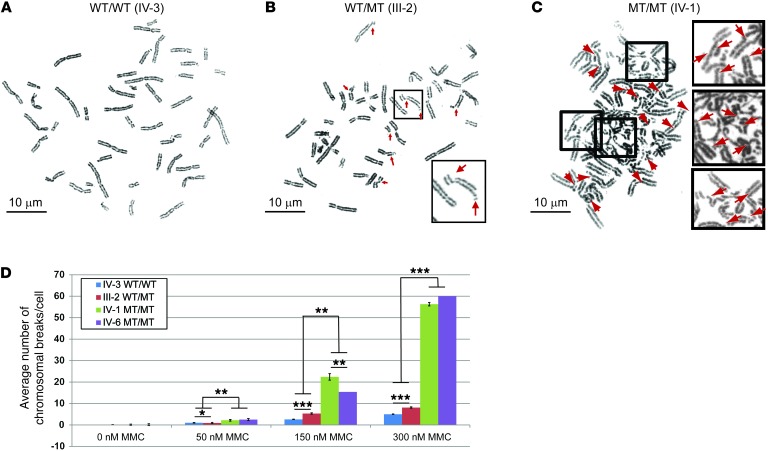
Fibroblasts from Mcm8-deficient mice showed hypersensitivity to agents that induce dsDNA breaks and DNA crosslinks, resulting in a higher number of broken chromosomes, a hallmark of genomic instability (5). We assayed the DNA repair capabilities of cultured fibroblasts derived from affected and unaffected family members (Figure 2, A–C, and ref. 6). Fibroblasts from unaffected sister IV-3 (WT genotype) showed few chromosomal breaks at 150-nM and 300-nM concentrations of mitomycin C (MMC) (Figure 2, A and D). Cells from III-2 (unaffected mother, heterozygous WT/MT) showed a significantly increased number of chromosomal breaks per cell when compared with cells from IV-3 (unaffected sister, WT genotype) at all concentrations of MMC (Figure 2, B and D): 50 nM (0.9 ± 0.3 vs. 0.1 ± 0.1, P = 0.02), 150 nM (5.3 ± 0.4 vs. 2.6 ± 0.3, P < 0.001), and 300 nM (8.1 ± 0.5 vs. 5.0 ± 0.4, P < 0.001). In both affected sisters (IV-1 and IV-6; homozygous MT/MT), the number of chromosomal breaks per cell was at least 8–10 fold higher than that in heterozygous carriers (Figure 2, C and D, P < 0.01). At 150 nM MMC, an average of 22.4 ± 1.5 and 15.4 ± 0.8 chromosome breaks per cell were observed for affected sisters IV-1 and IV-6, respectively (P < 0.001). We observed more than 50 chromosomal breaks per cell at 300 nM MMC exposure in cells from affected sisters IV-1 and IV-6 (P < 0.001). In the majority of cells homozygous for the mutation, we observed multiple complex chromosomal aberrations (Figure 2, C and D). Therefore, MCM8 c.446C>G homozygous mutation impedes the repair of MMC-induced chromosomal breaks. Exposure of peripheral lymphocytes to diepoxybutane (0.1 μg/ml) did not cause chromosomal breakage in cells from the same family members (IV-1 and IV-6) homozygous for the MCM8 c.446C>G mutation. We evaluated 10 metaphase cells per patient, but there were fewer than than 0.2 breaks per cell (Supplemental Table 3). Diepoxybutane results suggest that MCM8 is not part of the Fanconi anemia DNA repair pathway. WES data analysis from affected sisters did not reveal additional variants that would affect protein function in genes known to be associated with chromosomal instability and ovarian failure (Supplemental Table 4).
Figure 2. MCM8 mutation impairs DNA break repair.
Cells from homozygous MCM8 c.446C>G individuals have an impaired ability to repair dsDNA breaks induced by MMC. A total of 4 experimental groups were treated with MMC for each sample. Representative metaphase cells treated with 300 nM MMC are shown from (A) a healthy and fertile individual with the MCM8 WT/WT genotype (family member IV-3), (B) an unaffected WT/MT genotype individual (family member III-2), and (C) an affected female homozygous for the MCM8 c.446C>G pathogenic variant (MT/MT genotype; family member IV-1). Arrows point to chromosomal breaks. Original magnification, ×63/1.4 for chromosomal spreads and ×4 for inserts from the spreads. Scale bars: 10 μm. (D) At least 10 metaphase cells were used to calculate the total number of chromosomal breaks per cell for each MMC concentration. The number of chromosomal breaks counted was limited to 60 per cell, and those with more were indicated as 60+ breaks per cell. Error bars represent SEM. *P < 0.01, **P < 0.001, and ***P < 0.0001 by 2-tailed t test.
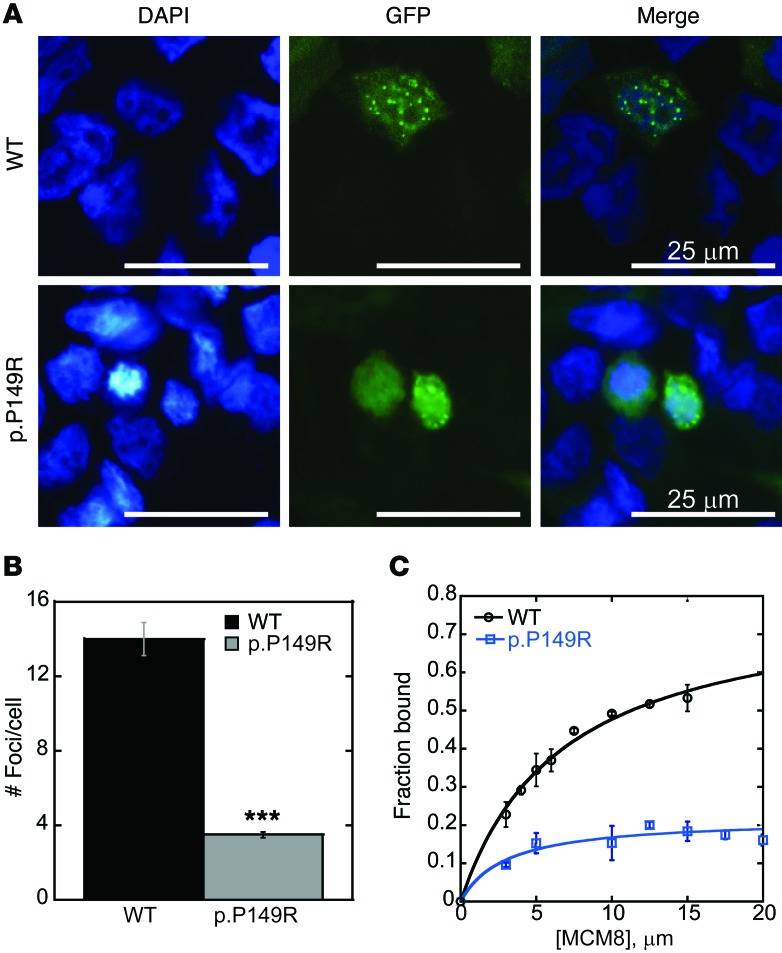
MCM8-containing complexes form foci at sites of DNA damage (7). We generated an MCM8-GFP construct containing the c.446C>G/p.P149R mutation to determine whether foci formation was affected. Cells were transfected with the MCM8-GFP constructs and treated with 300 nM MMC to induce DNA damage. We found that GFP expression was primarily nuclear, with an average of 14 ± 0.9 nuclear GFP foci per cell formed at sites of DNA damage in cells expressing wild-type MCM8-GFP (Figure 3, A and B). We found that significantly fewer nuclear GFP foci formed, averaging 3.5 ± 0.2 foci per cell (P < 0.001; Figure 3, A and B), in cells expressing mutant MCM8-GFP. GFP fluorescence was more diffuse throughout the nucleus and the cytoplasm (Figure 3A). These experiments show that the c.446C>G/p.P149R mutation inhibits MCM8 recruitment to sites of DNA damage.
Figure 3. MCM8 mutation disrupts MCM8 foci formation and DNA binding.
(A) MCM8 c.446C>G (p.P149R) mutation inhibits MCM8 foci formation at sites of DNA damage. HEK293T cells were transfected with wild-type MCM8-GFP (WT) or mutant MCM8-GFP (p.P149R; shown in green) and treated with 300 nM MMC for 6 hours. Nuclei were counterstained with DAPI (blue). Foci formed in 293T cells expressing wild-type MCM8, but not in cells expressing mutant MCM8. Four independent experiments for transfection of MCM8-GFP (WT vs. p.P149R) coupled with DNA damage were performed. Representative confocal images are shown. (B) Twenty representative cells per condition were quantified for the number of damage-induced nuclear foci. A 2-tailed t test (assuming unequal variance) revealed a statistically significant difference (***P < 0.001) between wild-type (14 ± 0.9 foci/cell) and c.446C>G-expressing (3.5 ± 0.2 foci/cell) cells. Error bars represent SEM. (C) MCM8 c.446C>G (p.P149R) inhibited DNA binding by EMSA. Wild-type MCM8 or mutant MCM8 (p.P149R) protein was bound to a random 46 nt ssDNA oligonucleotide. Gels were imaged, and quantification of the fraction of band shift was performed. Data were fit to a single-site binding model defined by ΔF[MCM8]/KD + [MCM8], where F is the fraction bound and KD is the dissociation constant. Mutant MCM8 (p.P149R, blue squares) showed a significant reduction in DNA binding affinity for ssDNA at each concentration when compared with that of wild-type MCM8 (WT, black circles). Each point is the average of 3 replicates. Error bars represent SD.
We also compared DNA binding of wild-type and mutant MCM8 protein. Wild-type and mutant MCM8 cDNAs were generated that correspond to the N-terminus of the protein that is predicted to bind DNA (nucleotides 1-1104; aa 1-368). We chose a random 46 nt single-stranded DNA (ssDNA), since RAD51 is recruited to ssDNA ends at sites of damage, and MCM8 is likely to bind at these sites (8). Wild-type MCM8 showed an increase in binding to ssDNA, with a concentration of KD = 6.7 ± 0.8 μM (Figure 3C). Unlike wild-type, mutant MCM8 does not saturate binding over the course of the titration, making calculation of a KD difficult (KD not determined). These results show that the c.446C>G/p.P149R mutation impairs DNA binding ability at the N-terminus of MCM8.
We used SNP arrays and WES in a consanguineous family to identify a homozygous recessive pathogenic variant in MCM8 (c.446C>G) as a cause of a novel syndrome in humans characterized by ovarian insufficiency, hypothyroidism, and genomic instability in somatic cells. The resulting p.P149R substitution occurs in a highly conserved region of MCM8 predicted to bind DNA. Individuals homozygous for this variant show high numbers of chromosomal breaks when exposed to MMC, and the p.P149R mutation inhibits MCM8 foci formation at sites of DNA damage. MCM8 p.P149R mutant protein shows impaired binding to DNA and likely prevents repair at these sites. The heterozygous sisters and their mother have unremarkable medical histories to date, which is consistent with the unaffected carrier state, and there is no known family history of cancers. Future follow-up and phenotyping of additional individuals with MCM8 variants will be of great importance to understand their predisposition to cancer. Given the consanguineous nature of the family, we cannot rule out the possibility that other genes contribute to the observed clinical phenotype.
Mcm8- and Mcm9-deficient mice are infertile and have small gonads due to germ cell depletion (5). Additionally, somatic cells exhibit growth defects and chromosomal instability (5). MCM8 and MCM9 are novel regulators of germ cell survival, are rapidly induced and recruited to DNA damage sites, coregulate each other’s stability, and promote RAD51 recruitment to ssDNA (5, 8). The MCM8/MCM9 complex is likely required for the resolution of dsDNA breaks that occur during homologous recombination in pachytene of meiosis I. Failure to resolve breaks likely leads to oocyte death and small or absent ovaries in women homozygous for these mutations. The phenotype of the affected daughters is similar to that in knockout mice. Ovarian failure was identified in these daughters, and their soma displayed increased sensitivity to MMC. Hypothyroidism was not reported in Mcm8-deficient mice, however, it is unclear whether thyroid function was investigated in these animals. It is possible that the more extensive phenotype observed in this consanguineous family is due to the dysfunction of other genes. Genomic instability syndromes, such as Fanconi anemia, are associated with multiple endocrine dysfunctions (9), including hypothyroidism and gonadal failure. The cause for endocrinopathies in chromosomal instability syndromes is unclear.
A nonsynonymous SNP in exon 9 of MCM8 (rs16991615) is highly associated with the age of natural menopause, and meta-analyses of menopause GWAS studies strongly implicate DNA repair pathways (10–15). The functional relevance of rs16991615 in reproductive senescence has not been elucidated. Our study shows that MCM8, when inherited in a Mendelian fashion, is essential for normal gonadal development. Future studies and follow-up on these and additional individuals will be necessary to define the spectrum of human phenotypes associated with MCM8 variants. The role of the novel MCM8/MCM9 pathway in women with idiopathic POF needs to be explored further.
Methods
Further information can be found in the Supplemental Methods.
Genetic studies.
Regions of homozygosity within the family were mapped using the Affymetrix GeneChip Human Mapping 250K Nsp array (Affymetrix). Data were deposited in the NCBI’s Gene Expression Omnibus (GSE56043). Exons and splice sites were captured using the Agilent Haloplex All Exon Kit, and WES was performed on an Illumina HiSeq 2500. Raw data were deposited in the Sequence Read Archive (ID SRP046742). Sanger sequencing was used to confirm WES-discovered variants and to evaluate 200 fertile controls for putative damaging variants.
Functional analysis of chromosomal instability.
We assayed DNA repair capabilities in peripheral lymphocytes and patient fibroblasts exposed to diepoxybutane and MMC. We evaluated the ability of exogenous wild-type and mutant MCM8-GFP to form foci at locations of DNA damage in HEK293T cells.
DNA-binding assay.
We generated wild-type and mutant MCM8 proteins and compared their ability to bind DNA. MCM8 wild-type and mutant soluble proteins were expressed and purified from BL21(DE3) Rosetta 2 Competent cells (EMD Millipore). EMSAs were performed with a random 46-nt-long ssDNA.
Statistics.
In chromosomal breakage analyses, ANOVA for single-factor variation was used to determine both the effect of drug concentration within each cell line and the effect of the cell line within each drug concentration. Comparisons between cell lines at a single drug concentration and differences in GFP foci formed with MMC treatment were evaluated by 2-tailed t tests, assuming unequal variance. A P value of less than 0.01 was considered significant.
Study approval.
The study of the family was approved by the Ethical Review Committee of King Khalid University Hospital. The recruitment of fertile women from Magee-Womens Hospital was approved by the IRB of the University of Pittsburgh. Written informed consent was obtained from all participating subjects.
Supplementary Material
Acknowledgments
We thank the patients and their family for participating in this research study on the genetics of POF. We thank the College of Medicine Research Center, Deanship of Scientific Research, King Saud University. We also thank the Cytogenetics Laboratory at Magee-Womens Hospital for their assistance with the chromosomal breakage studies, Pittsburgh Clinical Genomics Laboratory for WES, and the Center for Biological Imaging at the University of Pittsburgh for access to microscopy resources. Funding was provided by the National Institute of Child Health and Human Development (R01HD070647 and R21HD074278, to A. Rajkovic), the Samuel and Emma Winters Biomedical Award (to M.A. Trakselis), and the Magee-Womens Research Institute Postdoctoral Fellowship (to M.A. Wood).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(1):258–262. doi:10.1172/JCI78473.
Michael A. Trakselis’s present address is: Department of Chemistry and Biochemistry, Baylor University, Waco, Texas, USA.
References
- 1.Nelson LM. Clinical practice. Primary ovarian insufficiency. N Engl J Med. 2009;360(6):606–614. doi: 10.1056/NEJMcp0808697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood MA, Rajkovic A. Genomic markers of ovarian reserve. Semin Reprod Med. 2013;31(6):399–415. doi: 10.1055/s-0033-1356476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vijayraghavan S, Schwacha A. The eukaryotic Mcm2-7 replicative helicase. Subcell Biochem. 2012;62:113–134. doi: 10.1007/978-94-007-4572-8_7. [DOI] [PubMed] [Google Scholar]
- 4.Gozuacik D, et al. Identification and functional characterization of a new member of the human Mcm protein family: hMcm8. Nucleic Acids Res. 2003;31(2):570–579. doi: 10.1093/nar/gkg136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lutzmann M, et al. MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination. Mol Cell. 2012;47(4):523–534. doi: 10.1016/j.molcel.2012.05.048. [DOI] [PubMed] [Google Scholar]
- 6.Oostra AB, Nieuwint AW, Joenje H, de Winter JP. Diagnosis of fanconi anemia: chromosomal breakage analysis. Anemia. 2012;2012:238731. doi: 10.1155/2012/238731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishimura K, et al. Mcm8 and Mcm9 form a complex that functions in homologous recombination repair induced by DNA interstrand crosslinks. Mol Cell. 2012;47(4):511–522. doi: 10.1016/j.molcel.2012.05.047. [DOI] [PubMed] [Google Scholar]
- 8.Park J, et al. The MCM8-MCM9 complex promotes RAD51 recruitment at DNA damage sites to facilitate homologous recombination. Mol Cell Biol. 2013;33(8):1632–1644. doi: 10.1128/MCB.01503-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giri N, Batista DL, Alter BP, Stratakis CA. Endocrine abnormalities in patients with Fanconi anemia. J Clin Endocrinol Metab. 2007;92(7):2624–2631. doi: 10.1210/jc.2007-0135. [DOI] [PubMed] [Google Scholar]
- 10.Stolk L, et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet. 2012;44(3):260–268. doi: 10.1038/ng.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuh-Huerta SM, Johnson NA, Rosen MP, Sternfeld B, Cedars MI, Reijo Pera RA. Genetic markers of ovarian follicle number and menopause in women of multiple ethnicities. Hum Genet. 2012;131(11):1709–1724. doi: 10.1007/s00439-012-1184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CT, et al. Replication of loci influencing ages at menarche and menopause in Hispanic women: the Women’s Health Initiative SHARe Study. Hum Mol Genet. 2012;21(6):1419–1432. doi: 10.1093/hmg/ddr570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He C, et al. A large-scale candidate gene association study of age at menarche and age at natural menopause. Hum Genet. 2010;128(5):515–527. doi: 10.1007/s00439-010-0878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carty CL, et al. Replication of genetic loci for ages at menarche and menopause in the multi-ethnic Population Architecture using Genomics and Epidemiology (PAGE) study. Hum Reprod. 2013;28(6):1695–1706. doi: 10.1093/humrep/det071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spencer KL, et al. Genetic Variation and Reproductive Timing: African American Women from the Population Architecture Using Genomics and Epidemiology (PAGE) Study. PLoS One. 2013;8(2):e55258. doi: 10.1371/journal.pone.0055258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.