Abstract
Survivin is a tumor-associated antigen (TAA) that inhibits apoptosis and is widely overexpressed in cancer cells; therefore, survivin has potential as a target for cancer immunotherapy. Application of HLA-A2–restricted survivin-specific T cell receptors (TCRs) isolated from allogeneic HLA–mismatched TCR repertoires has, however, been impeded by the inability of these TCRs to distinguish healthy cells expressing low levels of survivin from cancer cells with high survivin expression levels. Here, we identified an HLA-A2–restricted survivin-specific TCR isolated from autologous TCR repertoires that targets tumor cells in vitro and in vivo but does not cause fratricidal toxicity. Molecular modeling of the TCR-peptide-HLA ternary complexes and alanine scanning revealed that the autologously derived TCRs had tighter interactions with the survivin peptide than did fratricidal TCRs. Similar recognition patterns were observed among 7 additional TAA-specific TCRs isolated from allogeneic versus autologous repertoires. Together, the results from this study indicate that maximal peptide recognition is key for TCR selectivity and likely critical for reducing unwanted off-target toxicities. Moreover, isolating TCRs from autologous repertoires to maximize TCR selectivity has potential as a useful strategy to identify and select other shared tumor- and self-antigen–specific TCRs and ensure selective antitumor activity.
Introduction
Cancer-targeted adoptive T cell therapy with genetically engineered αβ T cell receptors (TCRs) has resulted in encouraging responses in some patients (1–3). Broadening this approach to a larger array of malignancies requires targeting more widely expressed tumor-associated antigens (TAAs). However, most TAAs are not exclusively tumor specific, but are also expressed at low levels in normal adult tissues, making TCR-mediated targeting of these important antigens a challenge. “On-target off-tumor” toxicity may occur when TCRs fail to discriminate levels of TAAs presented on normal versus tumor cells. For example, toxicity occurs when the antigen is expressed equally, or when the TCR not only recognizes low levels of the targeted TAA epitope, but also a cross-reactive epitope expressed on normal cells. Such combined target recognition may then lead to T cell activation, resulting in toxicity that apparently precludes safe targeting of the desired TAA. To explore this putative mechanism, we chose to use the TAA survivin as a model. The National Cancer Institute (NCI) prioritized survivin as a target for the development of immunotherapies (4) because of its ubiquitous overexpression in cancer and its crucial role in maintaining tumor cell phenotype and functions. Furthermore, compelling results from previous studies suggested that survivin is an excellent model antigen to study the problem of antigen threshold sensing and molecular discrimination. Autologous vaccination with survivin-derived peptides has proven safe (5) and effective in inducing survivin-specific T cell precursors (6), but objective clinical responses remain limited (6). Conversely, T cells expressing transgenic survivin–specific TCRs isolated from allorestricted TCR repertoires circumventing thymic selection have produced antitumor activity, but were incapable of discriminating self from tumor, causing severe fratricidal effects (7). This cytotoxic effect was considered on-target, off-tumor, as survivin mRNA was upregulated in activated T lymphocytes (7).
We hypothesized that selection of the TCR from an autologous repertoire leads to identification of survivin-specific clones with high affinity and selectivity capable of self-versus-tumor discrimination, since highly autoreactive and cross-reactive T cell clones have already undergone thymic selection, and surviving T cells should express TCRs tolerant to antigen thresholds present in healthy cells and tissues. Using an autologous repertoire selection strategy is in sharp contrast to other TCR-engineering approaches that aim at priming T cell responses from allogeneic or xenogenic repertoires devoid of human thymic selection (8) or generating TCRs with high or supraphysiologic avidities ex vivo (9). These methods have produced severe toxicities due to unrecognized cross-reactivities targeting epitopes from entirely unrelated proteins that can be expressed by healthy tissues (10, 11). We now report the successful cloning of a survivin-specific TCR from autologous cultures that has antitumor activity but lacks fratricidal effects or toxicity against normal hematopoietic stem/progenitor cells. To understand the mechanistic basis of the striking difference in molecular recognition of TCRs isolated from autologous versus allogeneic TCR repertoires, we performed structural modeling of the TCR-peptide-HLA ternary complexes combined with alanine substitution analysis of the survivin-specific TCRs. We then validated our observation in a set of additional TCRs targeting other TAAs. These studies provide critical insights into the determinants governing selective TCR molecular recognition.
Results
Generation of autologous survivin–specific T cell clones with selective antitumor effects.
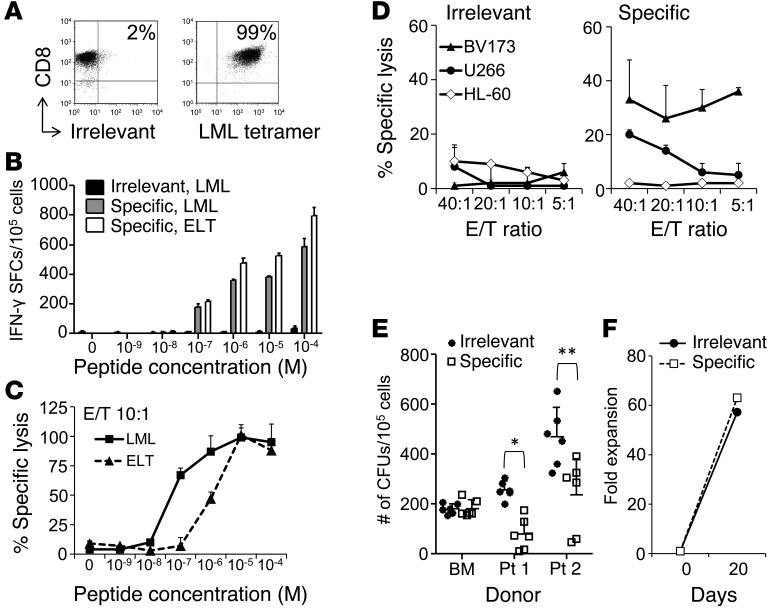
We used peripheral blood (PB) samples collected from HLA-A*02+ healthy donors to generate CD8+ cytotoxic T lymphocytes (CTLs) specific to the HLA-A*0201–restricted survivin95–104 (ELT) epitope, using its heteroclitic variant survivin96–10497M (LML) (12). As assessed by IFN-γ ELISpot assay, 3 of the 5 CTL lines (from donors 2, 4, and 5) were specifically reactive to both the LML (643 ± 5, 49 ± 1, and 96 ± 7 spot-forming cells [SFCs]/105 T cells) and the ELT peptides (662 ± 65, 45 ± 6, and 86 ± 9 SFCs/105 T cells) after 3 antigen-specific stimulations (data not shown). Single T cell clones were generated by limiting dilution from the most reactive donor (donor 2). Using multiple assays comparing survivin-specific and nonspecific (irrelevant) clones, we successfully identified one with optimal functional avidity. Specifically, we selected clone 24, which showed the highest specificity for the LML tetramer (>99%) (Figure 1A), the highest TCR avidity for both LML and ELT peptides (10–7 M when assessed by IFN-γ ELISpot assay [Figure 1B] and 5 × 10–8 M for LML and 10–6 M for ELT when measured by standard 51Cr-release assay [Figure 1C]). The functional avidity of clone 24 overlapped the broad range of previously described avidities of fratricidal TCRs (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI75876DS1). Importantly, clone 24 showed cytotoxic activity against the HLA-A*02+survivin+ tumor cell lines BV173 (leukemia) and U266 (myeloma) (Figure 1D) and inhibition of CFU of HLA-A*02+survivin+ leukemic progenitor cells (Figure 1E). By contrast, the same clone was not cytotoxic against the HLA-A*02–survivin+ cell line HL-60 or against HLA-A*02+ normal hematopoietic progenitor cells (Figure 1, D and E). This clone expanded effectively in vitro (>63-fold expansion after 3 weeks) (Figure 1F), indicating a lack of detectable T cell fratricidal effects.
Figure 1. Survivin-specific T cell clone with antitumor effects in the absence of toxicity.
(A) FACS analysis of the survivin-specific T cell clone stained for CD8 and the LML-specific or irrelevant tetramer. (B) T cell avidity assessed by IFN-γ ELISpot assays of the irrelevant clone against the LML peptide (black bars) and the survivin-specific clone against the LML (gray bars) or the ELT peptides (white bars). SFCs per 105 cells. Data represent the mean ± SD of triplicate experiments. (C) T cell avidity assessed by 51Cr-release assay against LML- (squares, solid line) or ELT-pulsed T2 cells (triangles, dashed line). Data show the mean ± SD of triplicates of the specific lysis at a 10:1 E/T ratio. (D) Antitumor activity by 51Cr-release assay of an irrelevant (left panel) and a survivin-specific (right panel) clone derived from the same donor against the HLA-A*02+survivin+ target cell lines BV173 and U266 and the HLA-A*02–survivin+ target cell line HL-60. Data represent the mean ± SD of 3 technical replicates of 1 experiment. Two independent experiments were performed in triplicate. (E) Antileukemic activity and absence of toxicity to normal hematopoietic progenitor cells by CFU assay of the survivin-specific clone (white squares) and the irrelevant clone (black circles) against HLA-A*02+survivin+ primary leukemic blasts from 2 CML blast crisis patients and 1 HLA-A*02+ normal BM donor. Data represent the mean ± SD of 3 independent experiments performed in duplicate. *P < 0.001; **P = 0.001 by Student’s t test. (F) Absence of T cell fratricide, assessed as fold expansion over a 3-week culture period after superexpansion of the survivin-specific (white squares, dashed line) and irrelevant (black circles, solid line) clones.
Polyclonal T cells engineered to express the survivin-specific TCR are not fratricidal.
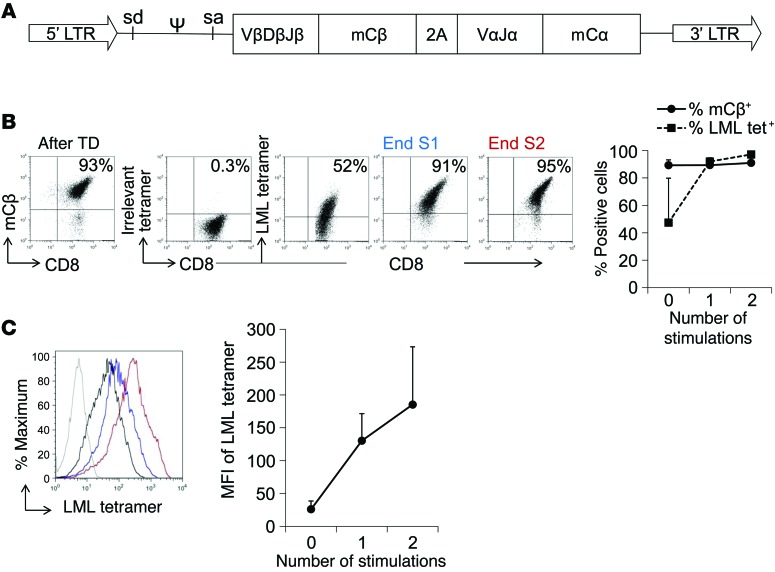
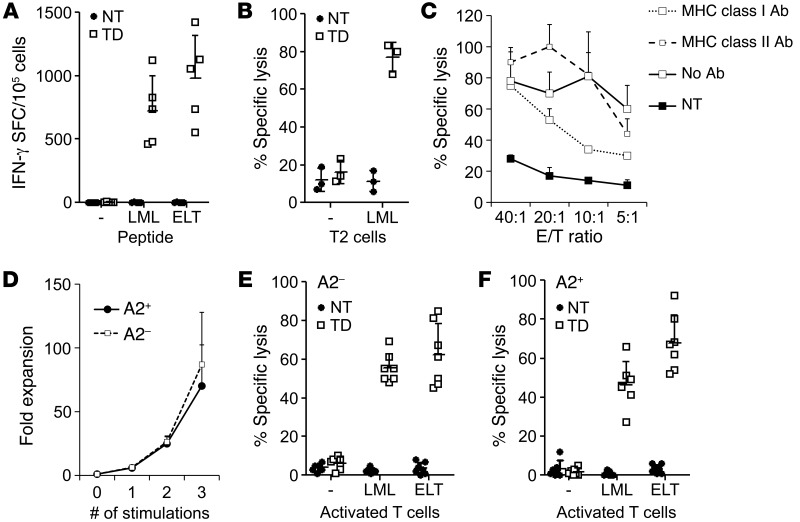
TCR α and β chains of clone 24 (referred to hereafter as s24-TCR) were cloned, codon optimized, and encoded into a retroviral vector after replacement of the constant regions with the corresponding murine regions (Figure 2A). TCR chain usage and complementarity-determining regions were completely distinct from the previously published fratricidal TCRs (Supplemental Tables 2 and 3). CD8+ T cells were transduced and expanded in the presence of LML peptide–pulsed artificial antigen–presenting cells (aAPCs) and IL-2. Immediately after transduction, 89% ± 4% of T cells stained for the murine constant β chain (mCβ+) and 47% ± 32% with the LML tetramer (Figure 2B). Although positivity for the LML tetramer was modest, with a mean fluorescence intensity (MFI) of 26 ± 12, after expansion in the presence of LML-pulsed aAPCs, we observed a significant enrichment in LML tetramer+ cells (97% ± 1%) (Figure 2, B and C). The ectopically expressed s24-TCR was functional, with s24-TCR+ T cells producing IFN-γ in response to both the LML (725 ± 274 SFCs/105 T cells) and ELT (978 ± 341 SFCs/105 T cells) peptides (Figure 3A). s24-TCR+ T cells also lysed LML peptide–pulsed T2 cells (77% ± 8% specific lysis, with an effector-to-target [E/T] ratio of 20:1) (Figure 3B) in an HLA-restricted fashion, as cytotoxic activity was significantly reduced by preincubation with MHC class I–blocking Abs (Figure 3C) (53% ± 10% specific lysis, E/T 20:1; P = 0.03). To confirm that s24-TCR+ T cells did not cause fratricide, we compared the phenotype, expansion, and cytotoxic activity of s24-TCR+ cells generated from both HLA-A*02+ and HLA-A*02– donors. The TCR was efficiently expressed in both (Supplemental Figure 1), and s24-TCR+ T cells expanded identically in response to LML-pulsed aAPCs and IL-2 (66- ± 38- vs. 76- ± 38-fold expansion after 3 stimulations for HLA-A*02+ and HLA-A*02– donors, respectively) (Figure 3D). Furthermore, we did not detect cytotoxic activity by s24-TCR+ T cells against HLA-A*0201+ T cells. As shown in Figure 3, E and F, lysis of activated T cells was negligible (2% ± 4% vs. 6% ± 3% specific lysis, E/T 20:1, HLA-A*02+ vs. HLA-A*02– donors), and these cells became targetable by s24-TCR+ T cells only after loading with the LML or ELT peptide (46% ± 12% vs. 55% ± 7% specific lysis for LML-loaded T cells; 68% ± 14% vs. 62% ± 16% for ELT-loaded T cells, E/T 20:1, HLA-A*02+ vs. HLA-A*02– donors). As expected, control T cells had no cytotoxic activity against activated T cells (Figure 3, E and F).
Figure 2. Efficient expression of the transgenic survivin-specific TCR by polyclonal CD8+ T cells.
(A) Scheme of the retroviral vector. sd, splice donor; sa, splice acceptor. (B) Transduction efficiency detected by staining for the mCβ and the LML tetramer. Enrichment of LML tetramer+ cells during T cell expansion in the presence of LML-pulsed aAPCs (2 weekly stimulations). Representative FACS plots immediately after transduction (After TD), and after 1 (End S1) or 2 (End S2) stimulations. Numbers indicate the percentage of positive cells in each quadrant. Graph shows the mean ± SD of 4 donors. (C) Increase in LML tetramer MFI after weekly antigen-specific stimulations. Representative histogram staining with irrelevant tetramer (gray), LML tetramer after TD (black), “End S1” (blue), and “End S2” (red). Graph shows the mean ± SD of 4 donors.
Figure 3. The ectopically expressed survivin-specific TCR is functional and specific, but not fratricidal, in vitro.
(A–C, E, and F) Symbols represent the mean of triplicates per donor; error bars represent the mean ± SD. (A) Production of IFN-γ (ELISpot) in response to LML and ELT peptides by nontransduced (NT, black circles) and transduced (TD, white squares) T cells (n = 5). (B) Killing of LML-pulsed T2 cells (51Cr release) by NT and TD T cells, with the percentage of specific lysis at an E/T ratio of 20:1 (n = 3). (C) Evaluation of HLA restriction of the killing of LML-pulsed T2 cells by preincubation with HLA class I–blocking Ab (dotted line) or HLA class II–blocking Ab (dashed line), or in the absence of Ab (solid line) by TD T cells (white squares) and NT T cells (black squares, solid line). Data are representative of 2 independent experiments. (D) Expansion of transgenic T cells with weekly antigen-specific stimulations generated from HLA-A*02+ (black circles, solid line) or HLA-A*02– (white squares, dashed line) donors. Mean ± SD, n = 7, P = NS. (E and F). 51Cr-release assays of NT (black circles) and TD T cells (white squares) against activated HLA-A*02+ target T cells loaded or not with LML or ELT peptide. Percentage of specific lysis at an E/T ratio of 20:1 of survivin-specific TCR expressed in (E) HLA-A*02– (n = 7) or (F) HLA-A*02+ donors (n = 7).
Survivin-specific TCR–redirected T cells exert antitumor activity without toxicity to normal hematopoietic progenitor cells.
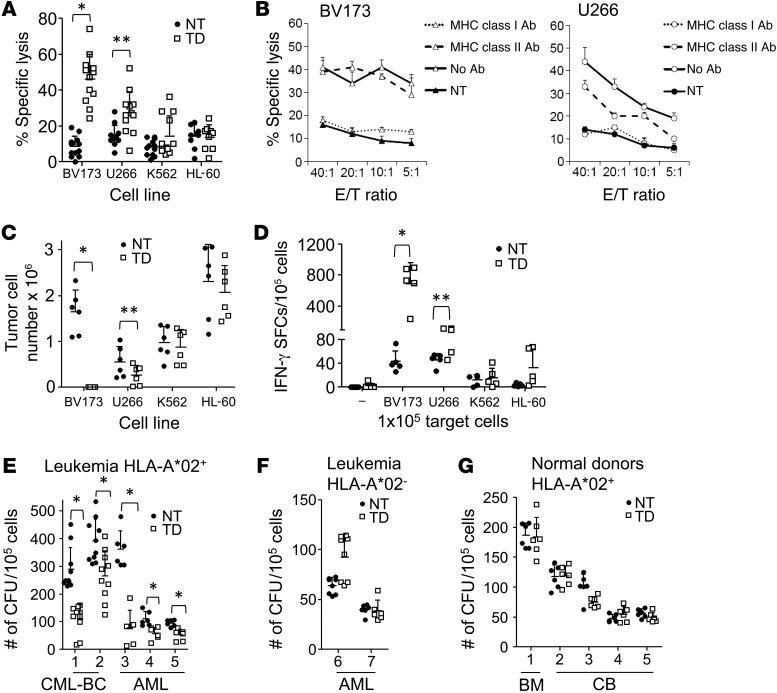
To ensure that the lack of fratricide was not at the expense of reduced antitumor activity, we evaluated the cytotoxic activity of s24-TCR+ T cells against survivin+ hematological malignancies. We demonstrated that s24-TCR+ T cells produced significantly greater lysis of the HLA-A*02+survivin+ leukemia cell line BV173 (46% ± 14% specific lysis at an E/T ratio of 20:1) and the HLA-A*02+survivin+ multiple myeloma–derived cell line U266 (27% ± 12%) than was observed in control T cells (8% ± 6 % and 14% ± 6%, respectively) (P < 0.001 for BV173; P = 0.003 for U266) (Figure 4A). In contrast, we observed negligible killing for both transduced and control T cells against the control target HLA-A*02–survivin+ leukemia cell lines K562 and HL-60 (Figure 4A). Cytotoxic activity of s24-TCR+ T cells was MHC class I restricted, as preincubation of target cells with HLA class I–blocking Abs abrogated the cytotoxic activity against BV173 and U266 cells (Figure 4B). In longer-term assays in which we cocultured control or s24-TCR+ T cells with HLA-A*02+survivin+ tumor cells for 5 days, we found a significant reduction of both BV173 and U266 tumor cells only in the presence of s24-TCR+ T cells (Figure 4C and Supplemental Figure 2). These cytotoxic effects were paralleled with IFN-γ production by s24-TCR+ T cells against the BV173 and U266 cell lines as assessed by ELISpot assays (Figure 4D) and by the release of Th1 cytokines as assessed by cytometric bead arrays (Supplemental Figure 3). We also confirmed antitumor effects of s24-TCR+ T cells in CFU assays against primary leukemic samples. As shown in Figure 4E, leukemic CFU formation was significantly reduced in all 5 HLA-A*02+ leukemia samples incubated with s24-TCR+ T cells as compared with that seen in control T cells, with a median 48% reduction of CFU formation in the presence of s24-TCR+ T cells (range, 32%–78%; P = 0.03). In addition, we observed no cytotoxic effects against 2 HLA-A*0201– leukemia samples (Figure 4F). In sharp contrast, we found that CFU formation of hematopoietic stem/progenitor cells from HLA-A*0201+ healthy donors was unaffected by incubation with s24-TCR+ T cells, with a median 3% reduction of CFU in the presence of s24-TCR+ T cells compared with that detected in cultures with control T cells (Figure 4G).
Figure 4. Survivin-specific TCR–redirected T cells have antitumor activity in vitro, while lacking toxicity against normal hematopoietic stem/progenitor cells.
(A) 51Cr-release assays of survivin-specific TCR+ TD (white squares) and NT control T cells (black circles) against HLA-A*02+survivin+ cancer cell lines BV173 and U266 and the HLA-A*02–survivin+ targets HL-60 and K562. Symbols indicate the mean of triplicates per donor; error bars represent the mean ± SD of the percentage of specific lysis at an E/T ratio of 20:1 (n = 12 donors). *P < 0.001; **P = 0.003 by Student’s t test. (B) HLA restriction of TCR+ TD (white symbols) and NT T cells (black symbols) assessed by preincubation of BV173 (left panel, triangles) or U266 (right panel, circles) with HLA class I–blocking Ab (dotted lines), HLA class II–blocking Ab (dashed lines), or in the absence of Ab (solid lines). Data represent the mean ± SD of 3 technical replicates from 1 donor. Cells from 2 donors were analyzed. (C) Quantification of residual tumor cells in cocultures on day 5 of TCR+ TD (open squares) and NT (black circles) T cells cultured with BV173, U266, K562, and HL-60 cells (E/T 5:1). Mean ± SD of residual tumor cells (n = 6). *P < 0.001 and ** P = 0.02 by Student’s t test. (D) IFN-γ production by TCR+ TD (white squares) and NT (black circles) T cells against BV173, U266, K562, and HL-60 cells by ELISpot assay. Symbols represent the mean of triplicates per donor; error bars indicate the mean ± SD (n = 5). *P < 0.001 and **P = 0.01 by Student’s t test. (E–G) Assessment of leukemic colony formation by TCR+ TD (white squares) and NT (black circles) T cells against HLA-A*02+ CML blast crisis (n = 2) and AML (n = 3) (E), HLA-A*02– leukemic blasts (F), and HLA-A*02+ healthy donor–derived BM (n = 1) or CB (n = 4) progenitor cells (G). Data represent the mean ± SD of CFU plated in duplicate for 5 donors. *P < 0.001 by Student’s t test.
Survivin-specific TCR transgenic T cells have antitumor activity in vivo and improve survival.
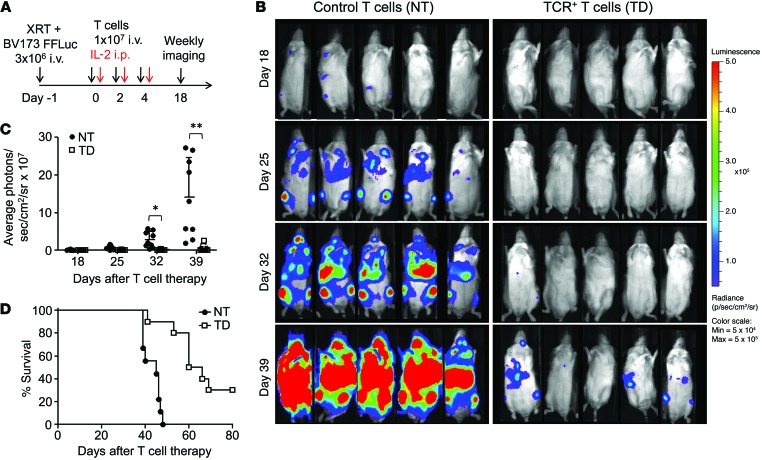
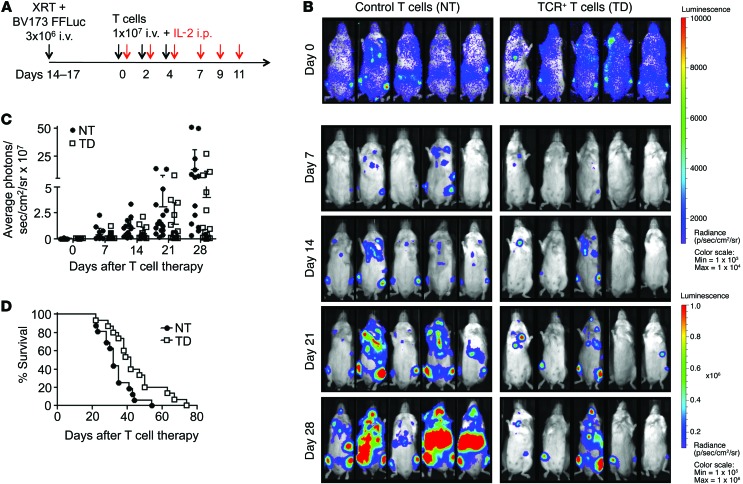
To confirm the in vivo antitumor function of s24-TCR+ T cells, we used a xenogeneic NSG mouse model systemically engrafted with BV173 cells genetically modified with firefly luciferase (FFLuc) and used bioluminescent imaging (BLI) to monitor tumor growth. In conditions that mimicked residual leukemia, mice underwent adoptive T cell transfer with either control or s24-TCR+ T cells the day after leukemia infusion (Figure 5A). On day 40 after infusion, mice treated with s24-TCR+ T cells had significantly better control of leukemia growth than did mice receiving control T cells (5.4 × 106 ± 7.7 × 106 vs. 141 × 106 ± 105 × 106 photons/second; P < 0.0001) (Figure 5, B and C). This translated into an improved overall survival of s24-TCR+–treated mice by day 80 (P < 0.001) (Figure 5D), with 3 of 10 s24-TCR+ T cell–treated mice found to be tumor free. To measure antitumor activity in mice with a high leukemic burden, we infused T cells 2 weeks after leukemia inoculation when disease dissemination and burden were documented by BLI (Figure 6A). Mice receiving s24-TCR+ T cells had a slower leukemia progression compared with that observed in control mice, resulting in a lower bioluminescent signal by day 28 (Figure 6, B and C) (40 × 106 ± 73 × 106 vs. 128 × 106 ± 178 × 106 photons/second; P = 0.006). This TCR-mediated antileukemic activity translated into significantly improved survival of the mice (P = 0.01) (Figure 6D).
Figure 5. Survivin-specific TCR–redirected T cells have in vivo antileukemic activity.
(A) Experimental plan. Intravenous administration of 3 × 106 BV173-FFluc cells to NSG mice after sublethal irradiation (120 cGy), followed by T cell infusions, IL-2, and weekly BLI starting on day 18. (B) Time course of BLI in representative individual mice from both treatment groups. Scale: 5 × 104 to 5 × 105 photons/second/cm2/sr. (C) Average photons/second/cm2/sr per mouse, determined by BLI, comparing mice treated with control T cells (NT, n = 9, black circles) or survivin-specific TCR+ T cells (TD, n = 10, white squares). Data represent the mean ± SD. *P = 0.01 at day 32 and **P = 0.009 at day 39, by Student’s t test with Bonferroni’s correction for multiple comparisons. The intensity signals were also log transformed, and the response profiles over time were analyzed by the robust generalized estimating equations method (P < 0.0001). Data represent 2 independent experiments. (D) Kaplan-Meier survival curve of mice treated with survivin-specific TCR+ T cells (TD) or control T cells (NT). P < 0.001 by generalized Wilcoxon test.
Figure 6. Survivin-specific TCR+ T cells prolong survival of mice with high leukemia burden.
(A) Experimental plan. Intravenous administration of 3 × 106 BV173 FFluc cells to NSG mice after sublethal irradiation (120 cGy). T cells were infused 14–17 days later, when leukemia was disseminated and established in multiple organs as detected by BLI. T cell infusions, IL-2, and weekly BLI. (B) Time course of BLI in representative individual mice from both treatment groups. Scale: 1 × 103 to 1 × 104 photons/second/cm2/sr (day 0); 1 × 105 to 1 × 106 photons/second/cm2/sr (days 7–28). (C) Average photons/second/cm2/sr/mouse comparing mice treated with control T cells (NT, n = 16) or survivin-specific TCR+ T cells (TD, n = 15). The intensity signals were also log transformed, and the response profiles over time were analyzed by the robust generalized estimating equations method (P = 0.006). Data represent the mean ± SD of 3 independent experiments. (D) Kaplan-Meier survival curve of mice treated with survivin-specific TCR+ T cells (TD) or control T cells (NT). P = 0.01 by generalized Wilcoxon test.
Computational molecular modeling and alanine scanning reveal a TCR-binding mode optimized for recognition of the survivin epitope.
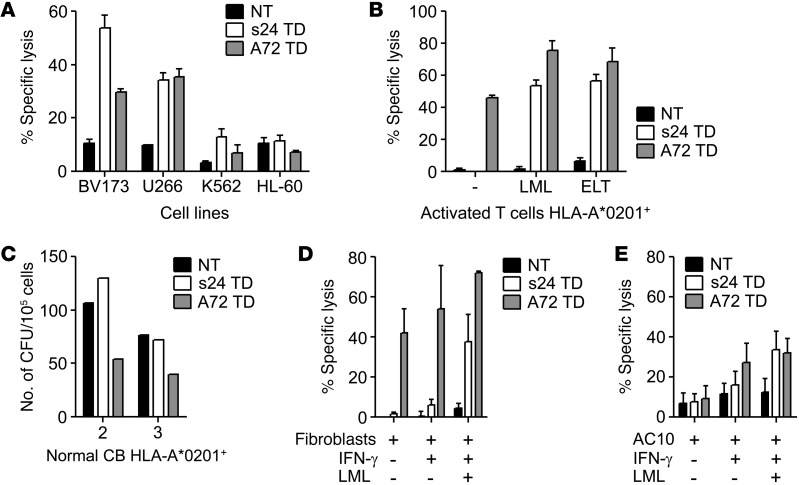
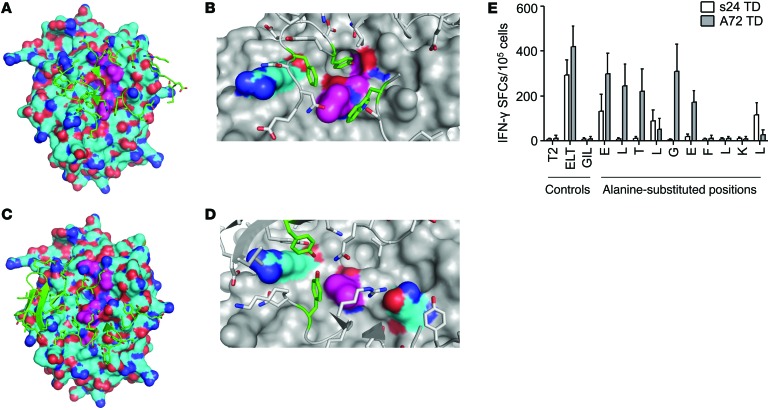
To understand the mechanism by which the s24-TCR produced antitumor activity without fratricide or toxicity to normal hematopoietic cells, we first compared T cells expressing either s24-TCR or the reported fratricidal TCR (A72-TCR) (7) in side-by-side experiments. While we observed no significant differences between s24-TCR+ and A72-TCR+ T cells in terms of antitumor activity (Figure 7A), only T cells expressing A72-TCR showed autoreactivity (Figure 7B) and toxicity against normal hematopoietic stem/progenitor cells (Figure 7C). Furthermore, A72-TCR+ T cells, but not s24-TCR+ cells, also showed cytotoxic activity against nonhematopoietic cells such as fibroblasts (Figure 7D) and cardiomyocytes (Figure 7E). Importantly, the safer profile of s24-TCR+ T cells was retained even in conditions mimicking an inflammatory insult, such as when targets were preincubated with IFN-γ, which modulates HLA-A*0201 expression (Supplemental Figure 4). This favorable toxicity profile was not due to reduced antitumor activity in vivo in the BV173 tumor model, as s24-TCR+ T cells mediated superior tumor control compared with A72-TCR+ T cells (P < 0.0001; Supplemental Figure 5). Structural models of the TCRs were generated using Rosetta software (13–15) and were docked onto models of the HLA-survivin peptide complex to predict the optimal bound structures of the TCR-HLA-survivin ternary complex. The overall binding energies of both s24-TCR and A72-TCR with the HLA-survivin complex were similar (Figure 8, A–D, and Supplemental Table 4). In contrast, the contribution of the binding interface derived from contacts between the TCR and the survivin peptide was significantly higher for s24-TCR compared with that of the A72-TCR (Supplemental Table 4). As shown in Figure 8, A and C, s24-TCR made extensive contact with most of the accessible survivin peptide residues, while A72-TCR interacted mostly with the HLA-A*02 groove. Specifically, s24-TCR created a network of highly optimized physical interactions involving numerous aromatic residues with the local region of the survivin peptide including Leu4, Gly5, and Phe7 (Figure 8B). While the A72-TCR still created strong interactions with Leu4 of the survivin peptide, most of its optimal physical interactions were established with polar residues of the HLA-A*02 groove (Figure 8D). The structural analysis was then corroborated by functional analysis of the survivin peptide performed by alanine substitution experiments. As shown in Figure 8E, every single residue (10 of 10) of the survivin peptide appeared to be crucial for s24-TCR functional activation, since 7 of 10 single substitutions completely abrogated IFN-γ release and 3 of 10 significantly reduced it. By contrast, only 3 of 10 substitutions were critical for the complete functional loss of A72-TCR activation (Figure 8E), suggesting a smaller and less optimal TCR-peptide–binding interface. Based on both the prediction model and the alanine substitution analysis, we queried the UniProtKB/Swiss-Prot database sequence for protein sequences containing the motifs XXXLGXFXXX, XLTXGEFLKX, and XXXLXXFLKL to identify potential cross-reactive epitopes. We selected 8 peptides associated with cells of the hematopoietic or immune systems, including T lymphocytes. IFN-γ ELISpot assays showed that A72-TCR reacted against T2 cells pulsed with several of these peptides, but s24-TCR did not (Supplemental Table 5).
Figure 7. Fratricidal activity of allogeneic repertoire–derived survivin-specific TCR.
(A–E) Comparison of TCR+ T cells transduced with the s24 survivin-specific TCR (s24 TD, white bars) with the A72 survivin-specific TCR (A72 TD, gray bars) or NT control T cells (black bars). Data represent the mean ± SD of triplicate experiments for 2 to 4 donors. (A) 51Cr-release assay against HLA-A2+survivin+ (BV173 and U266) and HLA-A2–survivin+ (HL-60 and K562) cancer cell lines. Data represent the mean ± SD of triplicates for the percentage of specific lysis at an E/T ratio of 20:1. (B) 51Cr-release assay against activated HLA-A*0201+ target T cells in the absence of exogenous peptide (–) or pulsed with LML or ELT peptide. Data represent the mean ± SD of triplicates for the percentage of specific lysis at an E/T ratio of 20:1. (C) CFU assay with normal HLA-A2+ CB donors. Data represent the mean of duplicates at an E/T ratio of 10:1. 51Cr-release assay against HLA-A*0201+ fibroblasts (D) and the HLA-A*0201+ cardiomyocyte cell line AC10 (E) with IFN-γ pretreatment or IFN-γ pretreatment and pulsed with LML peptide. Data represent the mean ± SD of the percentage of specific lysis at an E/T ratio of 20:1 for 4 donors.
Figure 8. Different molecular recognition patterns of autologous versus allogeneic repertoire–derived survivin-specific TCRs.
(A–D) Structural modeling reveals a highly epitope-specific binding interface of the novel autologous survivin-specific TCR s24. (A and C) HLA-A*0201 (cyan) and survivin ELT peptide (magenta) are represented at the surface; TCR regions directly in contact with the HLA-peptide complex are represented in green. (B and D) Residues with the most energetically favorable contacts across the binding interface are in green (TCR), cyan (HLA) or magenta (ELT peptide). (B) Trp 289, Tyr 352, and Phe 356 from the s24-TCR, Leu 4, Gly 5, and Phe 7 from the survivin peptide, and Arg 65 from the HLA. (D) Trp 244, Tyr 290 from the A72-TCR, Leu 4 from the survivin peptide, and Arg 65 and Gln 155 from the HLA. (E) Alanine substitution analysis testing s24 TD (white bars) or A72 TD (gray bars) T cells for recognition of peptide-pulsed T2 cells by IFN-γ ELISpot assay. Data represent the mean ± SD (n = 4 donors).
TCRs isolated from allorestricted repertoires have a high potential for cross-reactivity.
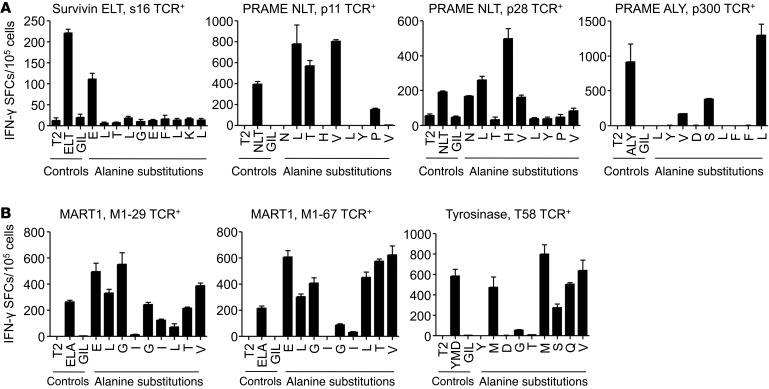
To test whether the highly optimized molecular recognition patterns by autorestricted TCRs that we found could be extended to a broader range of TCRs and TAAs, we performed comparative functional analyses on a set of TCRs derived from either autologous or allogeneic repertoires. Using the same methodology described for the identification of the s24 TCR, we isolated additional TCRs targeting the survivin ELT epitope (s16), the preferentially expressed antigen of melanoma (PRAME) NLT epitope (p11 and p28 TCRs), and the PRAME ALY epitope (p300 TCR). For comparison, we also selected 3 TCRs derived from allogeneic repertoires generated with the same approach for the identification of the A72 TCR. These allogeneic TCRs targeted the MART-1 ELA epitope (M1-29 and M1-67 TCRs) and the tyrosinase YMD epitope (T58 TCR), respectively (16). Each TCR was expressed within the same retroviral vector cassette as that of the s24 TCR (Figure 2A), and TCR-transgenic T cells were generated by retroviral transduction. We achieved TCR expression comparable to that of the s24 TCR. Alanine substitution analysis by IFN-γ ELISpot assay revealed that all (4 of 4) TCRs derived from autologous repertoires maintained a highly conserved molecular recognition pattern (Figure 9A). Indeed, all epitope motifs obtained by IFN-γ ELISpot assay, when queried with the ExPASy UniProtKB/Swiss-Prot database, failed to identify significant alternative epitopes for each TCR (Supplemental Table 6). By contrast, all (3 of 3) TCRs derived from allogeneic repertoires displayed reduced epitope specificity with a highly promiscuous recognition pattern by IFN-γ ELISpot assay (Figure 9B). Database queries with the obtained motifs yielded significant potential for cross-reactivity by recognition of 111 to more than 4,000 alternative sequences (Supplemental Table 6).
Figure 9. TCRs derived from autologous repertoires have a lower potential for cross-reactivity.
Alanine substitution analysis of T cells engineered to express transgenic autologous TCRs (A) and allogeneic TCRs (B) for recognition of peptide-pulsed T2 cells by IFN-γ ELISpot assay when targeting different TAAs. Data represent the mean ± SD of triplicates, with 2 donors tested for each TCR.
Discussion
We have isolated from an autologous TCR repertoire what we believe to be a novel survivin-specific s24-TCR that, when engrafted in polyclonal T cells, shows sufficient functional avidity to eliminate a variety of tumor cells both in vitro and in vivo, without producing autotoxicities. This novel TCR is capable of discriminating survivin on self-tissues from tumor-associated survivin expression and selectively mediates antitumor reactivity without on-target, off-tumor toxicity. Structural modeling and functional data revealed that the selective tumor specificity of the s24-TCR relies on a tight and extended TCR-survivin-MHC complex binding interface particularly enriched in TCR-survivin peptide contacts. Thus, the optimal recognition of a self-peptide by the TCR confers its selectivity, minimizing cross-reactivity and hence autoreactivity. Our findings challenge the previous conclusion that functional survivin-specific TCRs are not suitable for clinical use due to their toxicity, as well as the claim that the fratricidal effect mediated by such TCRs is exclusively due to on-target recognition of activated T cells (7). Our observation was corroborated and generalized in a comparative analysis of a set of 7 additional TCRs derived from autologous or allogeneic repertoires targeting several different TAAs, suggesting that the thymic selection step is key for minimizing TCR cross-reactivity.
Numerous studies have indicated that survivin is upregulated in most cancers, but is also functional in normal cells such as CD34+ hematopoietic stem cells, T lymphocytes (17), and cardiomyocytes (18, 19). For instance, conditional knockout mice show that loss of survivin at an early stage blocks T cell transition from the double-negative to double-positive cells, while at late stages, this loss decreases their numbers in the circulation (20). Survivin was also found to be upregulated upon TCR cross-linking in human T cells (7, 21) and is involved in the proliferation of cardiac myocyte remodeling during congestive cardiac failure (18) and in the ischemic/reperfused heart (19). Despite these functional activities of survivin, no cardiac toxicities and no delays in stem cell engraftment or T cell reconstitution have been reported in patients receiving survivin-based vaccines after autologous stem cell transplantation, even with elicitation of survivin-specific CTLs (5). In line with this clinical experience, we observed no impairment in the growth of normal hematopoietic progenitor cells or any negative impact on T cell expansion in the presence of our s24-TCR–redirected T cells. The safety profile occurred without compromising the antitumor effects. Polyclonal T cells expressing the s24-TCR significantly eliminated leukemic progenitors and tumor cells both in vitro and in vivo. We found that toxic effects, however, were consistently induced by polyclonal T cells expressing the A72-TCR that was isolated from allogeneic HLA–mismatched TCR repertoires.
Our conclusion that s24-TCR selectively recognizes tumor cells is supported by the modeling studies we performed to analyze the structure of the TCR-epitope-MHC complex and by functional alanine substitution analyses of the survivin epitope. These studies demonstrate that the s24-TCR is not fratricidal or autoreactive, because it establishes most of its strongest interactions with the survivin peptide, while the known fratricidal A72-TCR contacts the HLA molecule more strongly than it does the peptide, thus favoring peptide cross-reactivity. Our findings are in line with previous studies showing that native cross-reactivity appears to be focused on a limited number of hot-spot residues in any given peptide-MHC complex. (22, 23). Murine studies previously showed that peptide cross-reactivity only occurs in an allogeneic setting, as negative selection occurring in physiological conditions severely limits the number of distinct ligands recognized by a TCR (24). Indeed, cross-reactive TCRs capable of “accepting” amino acid substitutions within the targeted peptide, and thus a restricted number of binding hot spots, can be found only in mice in which negative selection has been experimentally limited (25). By comparing the molecular recognition patterns of the autologous versus allorestricted survivin-specific TCRs, we identified the molecular determinants explaining the mechanism for the capability of the s24-TCR to discriminate native from tumor survivin expression levels. We therefore consider that the fratricidal effect reported with the survivin-specific A72-TCR generated from an allogeneic TCR repertoire may be due not only to a lower threshold of on-target recognition on activated T cells, but also to an off-target recognition of cross-reactive peptides resulting from a suboptimal peptide-TCR interaction. As a consequence, the in vivo antitumor function of A72-TCR+ T cells may be limited in comparison with our s24 TCR, as we indeed observed in the BV173 mouse model. By validating our findings with an additional set of autologous and allogeneic TCRs targeting different TAAs, our study suggests that cross-reactivity with potential for off-target toxicity is a general problem of TCRs isolated from allogeneic repertoires. Our findings have broader implications for therapeutic tumor targeting by means of transgenic TCRs. While TCRs derived from allogeneic or xenogenic repertoires or TCRs with experimentally enhanced affinities have been widely used as means to attain high-affinity TCRs, autologous repertoire selection still remains a valid strategy. Although it may seem more challenging to identify TCRs with high functional avidity within the autologous repertoire, the task is worthwhile if a better safety profile is warranted.
In conclusion, we have reestablished the validity of survivin as a target in cancer immunotherapy by means of the ectopic expression of a TCR that fulfills the requirements of epitope specificity, antitumor activity, and lack of autoreactivity. This TCR relies on the optimal and selective recognition of the MHC-epitope complex and is capable of sensing survivin antigen levels on self-tissue versus tumor targets. This approach may be adapted for the identification of additional TCRs targeting other shared tumor/self-antigens, while reducing the risk of generating TCR-mediated autoreactivity.
Methods
Cell lines.
The tumor cell line BV173 (B cell acute lymphoblastic leukemia) was obtained from the German Cell Culture Collection (DSMZ), and the tumor cell lines U266B1 (multiple myeloma), K562 (erythroleukemia), HL-60 (acute myelomonocytic leukemia), CEM-T2 (TAP transporter deficient), 293T, and the cardiomyocyte cell line AC10 were obtained from the American Type Culture Collection (ATCC). Cells were maintained in culture with RPMI 1640 (HyClone; Thermo Scientific), IMDM (Invitrogen) for 293T cells, or DMEM/F12 medium (Invitrogen) for AC10 cells, containing 10% or 20% FBS (HyClone), according to the manufacturers’ recommendations, 1% L-glutamine, and 1% penicillin-streptomycin (Invitrogen) in a humidified atmosphere containing 5% CO2 at 37°C. The BV173 cell line was transduced with a retroviral vector encoding the FFluc and neomycin resistance genes as previously described (26). The K562 cell line was engineered to express the HLA-A*0201 molecule and CD40L, CD80, and OX40L as costimulatory molecules and used as aAPCs for T cell expansion (27). Cell lines were authenticated by the University of Texas MD Anderson Cancer Center Characterized Cell Line Core Facility. AC10 cells were confirmed to be HLA-A*0201+ by high-resolution sequence-based typing (Houston Methodist Hospital, Houston, Texas, USA).
Samples from healthy donors and leukemia patients.
Buffy coats from healthy volunteer blood donors were obtained through the Gulf Coast Regional Blood Center (Houston, Texas, USA). Deidentified cord blood (CB) units were obtained from the MD Anderson Cord Blood Bank (University of Texas, Houston, Texas, USA). PB and BM samples from deidentified patients with acute myeloid leukemia (AML) or chronic myeloid leukemia (CML) were provided by the Texas Children’s Cancer Center Tissue Bank. Dermal fibroblasts were collected from HLA-A*0201+ healthy donors (confirmed by high-resolution sequence-based typing) and generated as previously reported (28).
Peptides and alanine substitution experiments.
The native 10-mer survivin peptide ELT (ELTLGEFLKL), its heteroclitic 9-mer variant LML (LMLGEFLKL), PRAME P435 (NLTHVLYPV), PRAME P300 (ALYVDSLFFL), MART1 ELA (ELAGIGILTV), tyrosinase YMD (YMDGTMSQV), and alanine substitution variants for each amino acid position of all peptides were synthesized by Genemed Synthesis. All peptides were reconstituted in DMSO and used at a concentration of 5 μM unless otherwise indicated. Influenza matrix protein58–66 (flu) (GILGFVFTL) was used as an irrelevant control (27). Recognition of the HLA-peptide complex by transgenic T cells was analyzed by IFN-γ ELISpot assay using peptide-pulsed T2 cells as targets.
Generation and expansion of survivin-specific T cell lines and clones.
PB mononuclear cells (PBMCs) were isolated by Lymphoprep (Accurate Chemical and Scientific Corporation) density gradient centrifugation. HLA-A2 status was assessed by FACS, and survivin-specific T cell lines were generated from HLA-A2+ donors as previously described (27). Briefly, DCs were generated from CD14-selected monocytes (using CD14+ beads and manual MACS columns from Miltenyi Biotec) and, after maturation, pulsed with 5 μM of the specific peptide for 2 hours at 37°C. DCs were then used to stimulate autologous CD8+ T cells (obtained by immunomagnetic selection) (Miltenyi Biotec) at an E/T ratio of 20:1 in complete CTL media containing 45% Click’s medium (Irvine Scientific), 45% RPMI 1640, 5% heat-inactivated human AB serum (Valley Biomedical), 1% L-glutamine, and 1% penicillin-streptomycin (Invitrogen) in the presence of a previously validated combination of cytokines IL-7 (10 ng/ml), IL-12 (1 ng/ml), and IL-15 (2 ng/ml) (from PeproTech or R&D Systems). At days 9 and 16 of culture, T cells were restimulated with peptide-pulsed aAPCs at an E/T ratio of 10:1 in media containing IL-7, IL-12, and IL-15. IL-2 (50 U/ml) (Teceleukin; Hoffmann La-Roche) was added to the culture from day 16, as previously described (27).
Single-cell survivin-specific T cell clones were generated from LML- and ELT-reactive T cell lines by limiting dilution as previously described (29). Growing cells were screened for survivin-specific reactivity in IFN-γ ELISpot assays and were further expanded in the presence of allogeneic feeder cells, IL-2, and OKT3 (Orthoclone). In parallel, nonspecific (irrelevant) clones were expanded from the same donors and served as controls. The expanded clones were confirmed to be HLA-A*0201+ by high-resolution sequence-based typing. PRAME-specific clones were generated following the same methodology.
Immunophenotyping.
Cells were stained with FITC-, phycoerythrin- (PE-), peridinin chlorophyll protein– (PerCP-), or allophycocyanin-conjugated (APC-conjugated) Abs against HLA-A2, CD3, CD4, CD8, CD33, CD34, CD38, CD56, and CD45 from BD Biosciences or Beckman Coulter; PE-conjugated Abs against survivin (R&D Systems), APC-conjugated Abs against murine TCR constant β chain (eBioscience), or PE-conjugated LML or ELT survivin–specific tetramers were prepared by the Baylor College of Medicine MHC Tetramer Production Facility. Data acquisition was performed on a BD FACSCalibur using CellQuest software. Data analysis was performed with FlowJo software (Tree Star Inc.).
ELISpot assay.
The IFN-γ ELISpot assay was performed as previously described (27). In brief, 1 × 105 T cells per well were plated in triplicate and then stimulated with 5 μM or the indicated concentration (Figure 1B) of the specific peptides or with 1 × 105 cells of the respective target cell lines or media alone. As a positive control, T cells were stimulated with 25 ng/ml PMA and 1 μg/ml ionomycin (both from Sigma-Aldrich). The IFN-γ SFCs were enumerated (ZellNet).
51Cr-release assay.
The cytotoxic activity of T cells was evaluated using a standard 4-hour (for determination of TCR avidity using peptide-pulsed T2 cells) to 6-hour (for assessing killing of tumor cell lines, activated T cells, fibroblasts, and cardiomyocytes) 51Cr-release assay as previously described (27). Target cells were incubated in medium alone or in 1% Triton X-100 (Sigma-Aldrich) to determine spontaneous and maximum 51Cr release, respectively. The mean percentage of specific lysis of triplicate wells was calculated as follows: [(test counts – spontaneous counts)/(maximum counts – spontaneous counts)] × 100%. For blocking experiments, target cells were preincubated with anti-HLA class I– or class II–blocking Abs (Dako) as previously described (27). In selected experiments, fibroblasts and cardiomyocytes were preincubated with IFN-γ (100 U/ml; PeproTech) for 48 hours before being used as targets in the absence or presence of the LML peptide (28).
Cocultures and cytometric bead arrays.
Transduced or nontransduced T cells (1 × 106/well) were cocultured with tumor cell lines (2 × 105/well) at an E/T ratio of 5:1 in 24-well plates, in the absence of cytokines. After day 5 of culture, cells were harvested and stained for CD3 and specific tumor markers (CD19 for BV173, CD138 for U266, CD33 for HL-60 and K562). Residual tumor cells in cultures were enumerated by FACS using CountBright beads (Invitrogen). Coculture supernatant was harvested after 24 hours of culture and cytokines measured using specific cytometric bead arrays (BD) according to the manufacturer’s instructions.
CFU assay of leukemic and normal hematopoietic progenitors.
Mononuclear cells (MNCs) from BM, CB, or PB of healthy donors or leukemia patients were coincubated with survivin-specific or nonspecific T cell clones or with survivin-specific TCR–transduced or –nontransduced T cells at an E/T ratio of 10:1 for 6 hours and then plated in duplicate in methylcellulose-based medium supplemented with recombinant cytokines (MethoCult H4434 Classic; STEMCELL Technologies), as previously described (27). Granulocyte-macrophage CFU and erythrocyte CFU were scored using a high-quality inverted microscope after 2 weeks of culture.
Isolation of TCR genes and generation of a retroviral vector.
Total RNA was isolated from the survivin-specific clones 24 (s24) and 16 (s16), or from the PRAME-specific clones p11, p28, and p300 using the RNeasy kit from QIAGEN and TCR cDNAs cloned by 5′ RACE PCR (GeneRacer Kit; Invitrogen) according to the manufacturer’s instructions. The PCR products were cloned into the pCR4 TOPO vector (Invitrogen) and transformed into One Shot TOP10 Competent Cells (Invitrogen). Plasmid DNAs were prepared from 40 individual colonies, 20 containing the TCR α chain cDNA and 20 containing the TCR β chain cDNA. Full-length inserts from 10 plasmids per TCR chain were sequenced (SeqWright) to determine the TCR usage of the CTL clones. After identification, TCR sequences were modified by replacing the human TCR constant regions with murine TCR constant regions, linked by a 2A sequence, codon optimized by GeneArt (Invitrogen), and finally introduced into the SFG retroviral vector (Figure 2A). Native α and β chain TCR sequences of the A72-TCR (7) were codon optimized and synthesized by GeneArt (Invitrogen) and introduced separately into the SFG retroviral vector without further modification. MART1 (M1-29 and M1-67) and tyrosinase (T58) TCR sequences (16) were codon optimized, linked by a 2A sequence, synthesized by GeneArt, and introduced into the SFG retroviral vector.
Generation of retroviral supernatant, T cell transduction, and expansion.
Transient retroviral supernatant was prepared by transfection of 293T cells as previously described (30) and used to transduce CD8+ T cells isolated from PBMCs from healthy donors using magnetic beads (Miltenyi Biotec). Transduced cells were expanded in CTL media containing 10% FBS and by weekly stimulations with IL-2 (50 U/ml) and survivin LML peptide–loaded γ-irradiated (80 Gy) aAPCs at an E/T ratio of 4:1. Nontransduced T cells were maintained in CTL media containing 10% FBS and IL-2 (50 U/ml) and restimulated with immobilized OKT3 and anti-CD28 Abs (BD).
BV173 leukemia xenograft model and in vivo BLI.
NSG mice (8–10 weeks of age) were purchased from the Jackson Laboratory and maintained at the Baylor College of Medicine Animal Facility. Sublethally irradiated (120 cGy) NSG mice were infused i.v. via the tail vein with 3 × 106 FFluc-labeled BV173 cells. Leukemia burden was monitored by BLI (photons/second/cm2/sr) using the Xenogen in vivo imaging system (IVIS) (Caliper Life Sciences). A total of 3 T cell infusions (2 days apart) of transduced or nontransduced T cells (10 × 106/mouse) were injected retro-orbitally either 24 hours (low tumor burden model; Figure 5A) or 14 to 17 days (high tumor burden model; Figure 6A) after BV173 inoculation. Recombinant human IL-2 (1,000 U/mouse) was administered i.p. during the T cell infusions and in the following week, for a total of 6 doses. Leukemia growth was monitored weekly by imaging and survival recorded. Sick mice were sacrificed, and organs (spleen, blood, BM, lymph nodes, and liver) were analyzed by FACS for the presence of leukemia and T cells.
Computational modeling of TCR-peptide-HLA interactions.
Models of survivin peptide were built by threading the sequence of survivin (ELTLGEFLKL) onto the backbone structure of one of the HLA-bound 10-mer peptide homolog structures (MART1, PDBID: 3hg1). Using the Rosetta FlexPepDock server (14, 31), models of survivin were docked onto the HLA-A*0201 crystal structure, and the models of the survivin-HLA-A*0201 complex with the lowest energy interface score were selected, refined using Rosetta’s FastRelax protocol, and clustered. The center of the most populated cluster exhibiting the lowest overall energy was selected as the predicted structure of the HLA-survivin complex for the subsequent modeling steps.
A72 and s24 TCR models were generated by homology modeling using Rosetta. The closest structural homologs for each target TCR were identified using the HHpred sequence and structure alignment technique (32). The Protein Data Bank (PDB) IDs of the best homolog template for A72 α and β chains and s24 α and β chains were 4g9f, 3hg1, 2oi9, and 2oi9, respectively. Since target and template sequences were very similar except in the CDR3 regions, the target TCR sequence was threaded onto the template TCR backbone, with the exception of the CDR3 loops, which were reconstructed de novo using fragment insertion techniques and refined at all-atom (13, 33). Then, the initial models were clustered based on the structure similarity of the CDR3 loops. The center of the most populated clusters representing a different backbone conformation of the CDR3 loops was further relaxed with constraints to the starting pose to generate low-energy final models. A total of 16 models representing different backbone conformations of CDR3 loops for each TCR were used in subsequent docking of the TCRs to the HLA-survivin-peptide complex.
The Rosetta docking perturbation protocol (15) was used to dock different TCR models onto the survivin-HLA-A*0201 model. This protocol finds low-energy docking poses by randomly perturbing the starting docking poses (15) defined by users. The starting poses were taken from the templates and other available structures of the TCR-peptide-HLA-A*0201 complexes. Using this approach, about 5,000 docking decoys were generated for each starting TCR model, resulting in a total of ~80,000 decoys for the A72 TCR-survivin-HLA-A*0201 complex and ~144,000 docking decoys for the s24 TCR-survivin-HLA-A*0201 complex. Based on the lowest total energy and interface energy scores, ~1,000 docking decoys were selected and clustered based on structure similarity. The lowest energy models of the largest cluster exhibiting general features of the native TCR-peptide-HLA complex (with the α chains of TCRs located above the N-terminal of the peptide and the β chains located above the C-terminal of the peptide) (34) were selected and analyzed for binding energy contributions of the TCR HLA versus TCR survivin peptides to the total interface.
ExPASy prosite motif search for cross-reactive peptides.
The ExPASy ScanProsite tool (http://prosite.expasy.org/scanprosite/) (SIB Swiss Institute of Bioinformatics) was used to search for peptide motifs within all UniProtKB/Swiss-Prot data entries (release 2013_10 of 16-Oct-2013 with 541,561 entries for comparison of s24 and A72 TCRs; release 2014_07 of 09-Jul-2014 with 546,000 entries, for all TCRs). Filters were set for Homo sapiens.
Statistics.
Data were summarized as the mean ± SD. Student’s t test was used to determine statistically significant differences between treatment groups, with Bonferroni’s correction for multiple comparisons when appropriate. To compare the leukemia growth trend in mice over time, bioluminescent signal intensity at every time point was log transformed and analyzed by robust generalized estimating equations for repeated measurements. Survival analysis was performed using the Kaplan-Meier method (Prism 4.0; GraphPad Software). The generalized Wilcoxon test was used to assess statistically significant differences between groups of mice. All P values less than 0.05 were considered statistically significant.
Study approval.
The protocols for collection of deidentified samples and cord blood units were approved by the IRB of Baylor College of Medicine. All animal studies were reviewed and approved by the IACUC of Baylor College of Medicine.
Supplementary Material
Acknowledgments
C. Arber is supported by Oncosuisse (BIL KFS 02506-08-2009), the Swiss National Science Foundation, the NIH (T32 HL092332, T32 DK060445, and P50CA126752), a 2012 American Society for Blood and Marrow Transplantation (ASBMT)/Celgene New Investigator Award, and is a 2013 American Society of Hematology (ASH) Research Scholar. H.E. Heslop is supported by a Dan L. Duncan Chair, NIH grants (P50CA126752 and PO1CA94237), and a SCOR grant from the Leukemia and Lymphoma Society. P. Barth is supported by NIH grants (R01GM097207 and P50CA126752) and a supercomputer allocation from XSEDE (MCB120101). G. Dotti is supported by a grant from the NIH (R01CA142636), a Leukemia and Lymphoma Society Translational Research grant, and a Department of Defense, Technology/Therapeutic Development Award (W81XWH-10-10425). B. Savoldo is supported by NIH grants (R01CA131027 and R01HL114564), a Leukemia and Lymphoma Society Translational Research grant, and a Cancer Prevention and Research Institute of Texas (CPRIT) award (RP120298). We also appreciate the support of shared resources from the Dan L. Duncan Cancer Center grant (P30CA125123). We are grateful to M.K. Brenner for helpful discussions and C. Gillespie for comments on the manuscript.
Footnotes
Conflict of interest: Helen E. Heslop, Gianpietro Dotti, and Barbara Savoldo are investigators on a collaborative research grant from the Center for Cell and Gene Therapy and Celgene to develop genetically modified T cells.
Reference information:J Clin Invest. 2015;125(1):157–168. doi:10.1172/JCI75876.
References
- 1.Morgan RA, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314(5796):126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson LA, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114(3):535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robbins PF, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29(7):917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheever MA, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15(17):5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rapoport AP, et al. Combination immunotherapy using adoptive T-cell transfer and tumor antigen vaccination on the basis of hTERT and survivin after ASCT for myeloma. Blood. 2011;117(3):788–797. doi: 10.1182/blood-2010-08-299396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker JC, et al. Survivin-specific T-cell reactivity correlates with tumor response and patient survival: a phase-II peptide vaccination trial in metastatic melanoma. Cancer Immunol Immunother. 2012;61(11):2091–2103. doi: 10.1007/s00262-012-1266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leisegang M, et al. MHC-restricted fratricide of human lymphocytes expressing survivin-specific transgenic T cell receptors. J Clin Invest. 2010;120(11):3869–3877. doi: 10.1172/JCI43437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spranger S, et al. TCR-transgenic lymphocytes specific for HMMR/Rhamm limit tumor outgrowth in vivo. Blood. 2012;119(15):3440–3449. doi: 10.1182/blood-2011-06-357939. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, et al. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat Biotechnol. 2005;23(3):349–354. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- 10.Cameron BJ, et al. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci Transl Med. 2013;5(197):197ra103. doi: 10.1126/scitranslmed.3006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linette GP, et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood. 2013;122(6):863–871. doi: 10.1182/blood-2013-03-490565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen MH, Pedersen LO, Capeller B, Brocker EB, Becker JC, thor Straten P. Spontaneous cytotoxic T-cell responses against survivin-derived MHC class I-restricted T-cell epitopes in situ as well as ex vivo in cancer patients. Cancer Res. 2001;61(16):5964–5968. [PubMed] [Google Scholar]
- 13.Qian B, et al. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450(7167):259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raveh B, London N, Schueler-Furman O. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins. 2010;78(9):2029–2040. doi: 10.1002/prot.22716. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhury S, Berrondo M, Weitzner BD, Muthu P, Bergman H, Gray JJ. Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS One. 2011;6(8):e22477. doi: 10.1371/journal.pone.0022477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilde S, et al. Dendritic cells pulsed with RNA encoding allogeneic MHC and antigen induce T cells with superior antitumor activity and higher TCR functional avidity. Blood. 2009;114(10):2131–2139. doi: 10.1182/blood-2009-03-209387. [DOI] [PubMed] [Google Scholar]
- 17.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3(1):46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 18.Wohlschlaeger J, et al. Cardiomyocyte survivin protein expression is associated with cell size and DNA content in the failing human heart and is reversibly regulated after ventricular unloading. J Heart Lung Transplant. 2010;29(11):1286–1292. doi: 10.1016/j.healun.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 19.Levkau B. Survivin signalling in the heart. J Mol Cell Cardiol. 2011;50(1):6–8. doi: 10.1016/j.yjmcc.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Xing Z, Conway EM, Kang C, Winoto A. Essential role of survivin, an inhibitor of apoptosis protein, in T cell development, maturation, and homeostasis. J Exp Med. 2004;199(1):69–80. doi: 10.1084/jem.20031588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornacker M, Verneris MR, Kornacker B, Scheffold C, Negrin RS. Survivin expression correlates with apoptosis resistance after lymphocyte activation and is found preferentially in memory T cells. Immunol Lett. 2001;76(3):169–173. doi: 10.1016/S0165-2478(01)00186-9. [DOI] [PubMed] [Google Scholar]
- 22.Tynan FE, et al. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat Immunol. 2005;6(11):1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 23.Birnbaum ME, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157(5):1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huseby ES, Crawford F, White J, Kappler J, Marrack P. Negative selection imparts peptide specificity to the mature T cell repertoire. Proc Natl Acad Sci U S A. 2003;100(20):11565–11570. doi: 10.1073/pnas.1934636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huseby ES, Crawford F, White J, Marrack P, Kappler JW. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat Immunol. 2006;7(11):1191–1199. doi: 10.1038/ni1401. [DOI] [PubMed] [Google Scholar]
- 26.Hoyos V, et al. Engineering CD19-specific T lymphocytes with interleukin-15 and a suicide gene to enhance their anti-lymphoma/leukemia effects and safety. Leukemia. 2010;24(6):1160–1170. doi: 10.1038/leu.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quintarelli C, et al. Cytotoxic T lymphocytes directed to the preferentially expressed antigen of melanoma (PRAME) target chronic myeloid leukemia. Blood. 2008;112(5):1876–1885. doi: 10.1182/blood-2008-04-150045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leen AM, et al. Fiber-modified adenoviruses generate subgroup cross-reactive, adenovirus-specific cytotoxic T lymphocytes for therapeutic applications. Blood. 2004;103(3):1011–1019. doi: 10.1182/blood-2003-07-2449. [DOI] [PubMed] [Google Scholar]
- 29.Perna SK, et al. Interleukin 15 provides relief to CTLs from regulatory T cell-mediated inhibition: implications for adoptive T cell-based therapies for lymphoma. Clin Cancer Res. 2013;19(1):106–117. doi: 10.1158/1078-0432.CCR-12-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintarelli C, et al. Co-expression of cytokine and suicide genes to enhance the activity and safety of tumor-specific cytotoxic T lymphocytes. Blood. 2007;110(8):2793–2802. doi: 10.1182/blood-2007-02-072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.London N, Raveh B, Cohen E, Fathi G, Schueler-Furman O. Rosetta FlexPepDock web server — high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011;39(Web Server issue):W249–W253. doi: 10.1093/nar/gkr431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barth P, Wallner B, Baker D. Prediction of membrane protein structures with complex topologies using limited constraints. Proc Natl Acad Sci U S A. 2009;106(5):1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.