Abstract
The development of high-throughput DNA sequencing technologies has enabled large-scale characterization of functional antibody repertoires, a new method of understanding protective and pathogenic immune responses. Important parameters to consider when sequencing antibody repertoires include the methodology, the B-cell population and clinical characteristics of the individuals analysed, and the bioinformatic analysis. Although focused sequencing of immunoglobulin heavy chains or complement determining regions can be utilized to monitor particular immune responses and B-cell malignancies, high-fidelity analysis of the full-length paired heavy and light chains expressed by individual B cells is critical for characterizing functional antibody repertoires. Bioinformatic identification of clonal antibody families and recombinant expression of representative members produces recombinant antibodies that can be used to identify the antigen targets of functional immune responses and to investigate the mechanisms of their protective or pathogenic functions. Integrated analysis of coexpressed functional genes provides the potential to further pinpoint the most important antibodies and clonal families generated during an immune response. Sequencing antibody repertoires is transforming our understanding of immune responses to autoimmunity, vaccination, infection and cancer. We anticipate that antibody repertoire sequencing will provide next-generation biomarkers, diagnostic tools and therapeutic antibodies for a spectrum of diseases, including rheumatic diseases.
Introduction
Antibodies are a major component of the adaptive immune system and have critical roles in protective and pathogenic immune responses. In response to microbial infection, vaccination, autoimmune disease or cancer, the immune system generates distinct antibody repertoires. Analysis of these antibody repertoires, particularly those contributing to functional immune responses, can provide important information on protective and pathogenic immunity. In autoimmune diseases, including autoimmune rheumatic diseases, antibody characterization has enabled the identification of autoantigens and has provided insights into the underlying mechanisms of disease; furthermore, detection of autoantibodies has become a cornerstone of modern diagnostics.1–3 In infectious diseases, in which antibody responses are usually protective, there is growing interest in isolating antibodies that could be developed as novel therapeutic agents4,5 and in using microbial antigens and epitopes targeted by antibody responses to develop vaccines.4,6,7 A challenge in understanding, as well as in diagnostically and therapeutically harnessing, antibody responses is the identification of antibodies that underlie functional immune responses, that is, antibodies that directly contribute to an immune outcome, such as neutralizing a microbial pathogen or mediating autoimmune tissue injury. This Review provides an overview of technologies for large-scale sequencing of antibody repertoires, and discusses how these technologies can be applied to characterize immune responses and identify antibodies of therapeutic, diagnostic or mechanistic relevance to autoimmune diseases, including rheumatic diseases.
Antibody responses
Antibodies are comprised of an immunoglobulin heavy chain (IgH) and light chain (IgL), each containing an antigen-binding domain that is generated by the recombination, junctional diversification and somatic hypermutation of variable (V), joining (J) and/or diversity (D) gene segments during B-cell development.8,9 The complementarity-determining regions (CDRs), CDR1, CDR2 and CDR3, as well as the surrounding framework regions, together form the antigen-binding site of the antibody.10 In the primary B-cell repertoire, substantial diversity in antibody specificity comes from the IgH CDR3 owing to its generation both from combinatorial gene segments and from N-region diversity.10 In an ongoing immune response to an antigen, B cells that produce antibodies specific for that antigen further diversify by undergoing clonal expansion and by somatic hypermutation of their antibody variable regions. In this process, termed affinity maturation, B cells that express antibodies with an increased affinity for the activating antigen as a result of somatic hypermutation are selected for further expansion (Figure 1).11–13 These antigen-activated, affinity-matured B cells are then released from germinal centers into the blood as short-lived or long-lived plasmablasts, which migrate to other secondary lymphoid organs and sites of tissue injury, and can differentiate into long-lived memory B cells and plasma cells.14,15
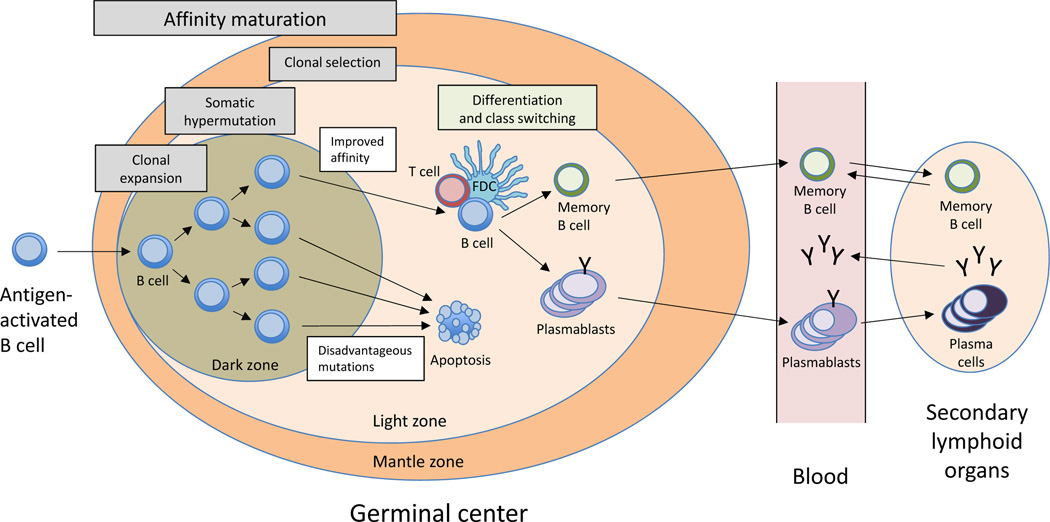
Figure 1.
The B-cell response. B cells undergo clonal expansion and affinity maturation after encountering antigen and T-cell help or co-stimulatory signals, a process that generally occurs in germinal centres within secondary lymphoid organs.12,13 Affinity maturation involves two processes, somatic hypermutation and clonal selection. Somatic hypermutation is a cytidine-deaminase-mediated process in which antibody CDRs are mutated ~1–2 times per cell division. Clonal selection involves competition of B cells for antigen and growth factors in germinal centres, resulting in B cells that express the highest-affinity antibodies being selected to expand and survive.12,13 After multiple rounds of somatic hypermutation and clonal selection, antibody-expressing B cells produce antibodies with increased affinity for their target antigen.10 B cells that express high-affinity antibodies respond further to growth factors and other signals that induce differentiation into plasmablasts and memory B cells.11,14,15 Plasmablasts transiently circulate in the blood and migrate to secondary lymphoid organs and tissues involved in the disease process, including tissues under autoimmune, infectious or malignant attack. Memory B cells circulate in the blood, and enable rapid recall responses upon re-exposure to their target antigen, whereas plasma cells localize primarily in the bone marrow and lamina propria, where they secrete antibodies. Abbreviation: CDRs, complementarity determining regions; FDC, follicular dendritic cell.
Identifying antigens
For most diseases, knowledge of the antigens and epitopes targeted by antibody responses is limited. For example, autoimmune diseases affect 3% of the global population, yet in most of these diseases the pathogenic autoantigens have not been identified.16 In infectious diseases, most of the critical microbial antigens and epitopes targeted by protective antibody responses are also unidentified,17 with the exception of HIV antigens, influenza and some other microbes.18 With regards to cancer, T-cell-mediated responses are still considered the primary form of immunity. Although immunotherapies are now revolutionizing cancer therapy,19,20 the specific targets of the immune response to these therapies are unclear, as is the contribution of antibody responses to effective anticancer immunity.
A variety of ‘array and display’ approaches have been developed for investigating antibody responses, including protein and peptide arrays,21 aptamer and peptoid arrays,22 random peptide arrays,23 large-scale peptide arrays,24 human protein arrays,25 and human proteomic display arrays.26 One major challenge of these approaches is the use of serum or plasma samples as the source of antibodies analysed. Serum (and plasma) samples contain a pool of all the antibodies produced by an individual, including antibodies that are heterogeneous, bind to many different antigens, and have differing affinities and avidities, thereby increasing background reactivity and diluting potential antibodies of interest. Furthermore, most serum antibodies are produced by plasma cells generated in prior immune responses, and are not produced by the plasmablasts or plasma cells responding to the immunogenic antigen of interest.
Another major challenge is that most approaches involve screening the binding of blood antibodies against panels of preselected antigens. For example, important autoantigens in rheumatoid arthritis (RA) are post-translationally citrullinated,27,28 and in systemic lupus erythematosus (SLE) are post-translationally cleaved (or phosphorylated);29 therefore, preselecting antigens might result in the failure to detect pathogenic autoantibody clonal families. Antigen-agnostic approaches (Box 1) are therefore needed for identifying important functional antibodies and their targets. An ideal solution is to isolate, recombinantly express, and characterize antibodies produced by the specific subset of B cells participating in functional immune responses to vaccination, infection, cancer or other diseases, in humans or other animals. In this Review, I describe several DNA sequencing approaches that are currently used to achieve this end.
Box 1 | Antigen agnostic.
The term antigen agnostic means independent of an antigen. For the identification of novel autoantigens, analysis of immune responses in an antigen agnostic fashion can be important for identifying pathogenic B cells and T cells.
B-cell populations
Selection of B-cell populations is an important consideration for antibody sequencing. The best B cells for characterizing the functional antibody repertoire include plasmablasts, plasma cells, memory B cells and tissue-infiltrating B cells (Figure 1). Of these cells, plasmablasts and memory B cells are the two populations that are present in peripheral blood and are therefore most easily isolated from humans.
Plasmablasts and plasma cells
Plasmablasts and plasma cells are the antibody-producing cells of the immune system. During an immune response, plasmablasts differentiate from antigen-activated B cells and are released from germinal centers to transiently circulate in the bloodstream before migrating to secondary lymphoid organs or to inflamed tissues, where they can differentiate into longer-lived tissue-resident plasma cells.30–32 Plasmablasts in peripheral blood form not only from newly generated naive B cells, but also from reactivated memory B cells. Together with the relative ease of isolating plasmablasts, their origin from either naive or memory B cells make them the ideal B-cell population to analyse when searching for antibodies associated with any given immune response. For example, studies sequencing the antibody response to influenza vaccination have detected a transient increase in peripheral blood plasmablast numbers, >75% of which produced anti-influenza antibodies.33,34
Although autoimmune diseases are chronic, evidence exists that underlying autoantibody responses involve continuous activation and reactivation of new B-cell clones,35 such that peripheral blood plasmablasts could produce new pathogenic antibodies at any stage of the disease. Indeed, in patients with RA, the proportion of plasmablasts in the blood correlates positively with disease activity,27,36 and these cells produce anti-citrullinated protein antibodies (ACPAs)27,28 that are known to contribute to the pathology of RA.37,38 Tissue-resident plasma cells might also produce ACPAs and might even be generated in situ in the synovial tissue of patients with RA; however, these tissue-resident cells are not easily accessible.39–41
Memory B cells
Memory B cells form during germinal centre reactions and provide an accelerated antibody response upon re-exposure to a target antigen.42 Whereas >75% of peripheral blood plasmablasts are specific for the antigens targeted by the ongoing immune response,33,34,43 antigen-specific memory B cells are present at much lower frequencies; approximately 1 of 2,500–100,000 memory B cells in blood are specific for the inciting antigen.42,44,45 These memory B cells can be isolated by antigen-sorting (by fluorescence-activated cell sorting [FACS], with fluorescently-labelled antigen baits to sort the cells by the specificity of their membrane immunoglobulins) of memory B cells from peripheral blood; however, because of the low frequency of antigen-specific memory B cells, such sorting requires large numbers of peripheral blood mononuclear cells. The substantial differences in the antigen load, prior exposure history, route of exposure, and associated immunostimulatory molecules in different vaccines, microbial infections, and autoimmune diseases influence the magnitude and properties of antibody responses, and modifications of antibody repertoire sequencing methods might be needed to effectively characterize the antibody responses in these conditions.
Tissue-infiltrating B cells
Analysis of tissue-infiltrating B cells could also help to characterize functional antibody responses. Despite being more difficult to access than peripheral blood B cells, characterization of tissue-infiltrating B cells in both autoimmune disease46,47 and in cancer48 has identified disease-associated antibodies. Integrated analysis of the B-cell antibody repertoires in the synovium and blood of patients with RA, and the brain and blood of patients with multiple sclerosis,46,47 is providing insights into the sites of B-cell activation and affinity maturation in these diseases.
Small-scale sequencing
An approach used in the 1990s and 2000s for the analysis of antibody repertoires involves reverse transcriptase polymerase chain reaction (RT-PCR) and Sanger sequencing of antibodies expressed by individual B cells. Because the experimental steps in this technique require manual handling, in practice this approach can only characterize tens to hundreds of B cells per experiment. Despite low throughput, the technique has been used to identify antibodies that neutralize clinically important pathogens such as HIV-1 and influenzae viruses,5,33,49–52 or autoantibodies that contribute to autoimmune disease.46,53
A variety of approaches have been used for single-cell RNA sequencing,54–57 several of which enable sequencing of coexpressed genes and use next-generation technology. However, these assay platforms and short-read sequencing technologies, for example, HiSeq™ platforms (Illumina, USA), read considerably fewer base pairs than are necessary (>500) for sequencing the entire IgL and IgH variable regions.58
Large-scale sequencing
A variety of high-throughput next-generation sequencing technologies have now been developed59,60 and are transforming the analysis of B cells and their antibody repertoires.4,58,61–63 These technologies are applied in a variety of ways, and important parameters for characterization of functional antibody responses include the methodological approach, the B-cell populations characterized, the clinical characteristics of the individuals analysed, the approach to bioinformatic analysis and the objective of the experiment.
Monitoring specific B cells
A number of research groups have developed approaches for deep-sequencing IgH (or in some cases the IgL), or solely the CDR3, in genomic DNA or cDNA generated from bulk RNA isolated from individuals with autoimmune disease,47,64 infections65 or cancer,66,67 or from vaccine recipients.68 In some instances, molecular ‘barcodes’ are used to enhance single-chain or CDR3 sequencing, that is, random hexamers are used as primers10,69,70 or adapters with a unique molecular identifier54,55 to tag each cDNA. Molecular barcoding enables the correction of base-calling errors inherent to the sequencing platform or of PCR bias, although not the correction of RT-PCR errors or accurate identification of the clonal proportions of the B cell pool.
Sequencing immunoglobulin genes in genomic DNA is challenging because as many as 50% of B cells have multiple rearranged immunoglobulin gene loci, a result of nonproductive rearrangements and receptor editing.71,72 Furthermore, receptor editing, an important feature in B-cell maturation that helps to prevent autoimmunity, can lead to allelic inclusion at the immunoglobulin IgL loci and the development of B cells that coexpress two different immunoglobulin IgLs, thereby confounding the sequencing of IgH and IgL cDNA from individual B cells.71,72 Moreover, although rapid, these methods usually involve the sequencing of only one immunoglobulin chain and, therefore, provide limited information about the antibodies generated during an immune response. Although both IgHs and IgLs can be sequenced simultaneously in bulk mRNA, pairing of the IgH and IgL sequences obtained in this way can only be attempted according to similarities in sequence frequency;73 this pairing is not always accurate. Without the accurate pairing of the cognate IgHs and IgLs, the information generated about an antibody response is incomplete.
Although not optimal for analysis of antibody repertoires, single-chain sequencing from bulk RNA can be used to track malignant or clonal populations of B cells by detecting their unique IgH, IgL or CDR3 sequences.74,75 This method is proving to be powerful for detecting and monitoring the recurrence of B-cell malignancies and has better sensitivity for detecting residual disease or disease recurrence than flow cytometry or other conventional approaches.66,67
IgH CDR3 sequencing is also used in the search for biomarkers of autoimmune diseases and in characterizing B-cell responses to infection and vaccination, and to immunomodulation in patients with cancer. Results from bulk IgH sequencing of unsorted B cells suggest that direct analysis of isolated B-cell subsets might be necessary for obtaining the most informative data regarding diseases of immune function.76 Additional data are needed to assess whether sequencing of a single chain or a single-chain CDR3 will provide biomarkers with clinical utility.77
The functional antibody repertoire
Many challenges must be overcome to understand and identify functional antibody repertoires (Table 1). In addition to high throughput analysis, a comprehensive survey of the B-cell repertoire requires sequences that enable accurate recreation of endogenous, bioinformatically selected antibodies, so that their antigen specificity, binding and functional properties can be characterized. To accurately recreate recombinant versions of endogenous antibodies, one must sequence and correctly pair the cognate IgHs and IgLs expressed by individual B cells, ensure that the sequencing covers the entire variable regions encoding the CDR1, CDR2, CDR3 and framework regions of both the IgHs and IgLs, and ensure that the sequences are error-free.10,61,78,79 High-process fidelity and quality are major challenges for the robust analysis of antibody repertoires, and are further discussed in this article. Furthermore, to enable bioinformatic identification of important functional antibodies, antibody repertoire sequencing must be applied to the appropriate B-cell subsets derived from individuals with an immunological phenotype of interest (Table 1).
Table 1.
Challenges and solutions to sequencing the functional antibody repertoire
| Challenge | Details of challenge | Solution |
|---|---|---|
| Analyse the functional repertoire | Identify clinical phenotype Analyse B cells participating in an immune response of interest Perform integrated analysis of coexpressed functional genes |
Focus analysis on individuals with immune phenotypes of interest Focus analysis on functional B cell populations, such as plasmablasts, antigen-specific memory B cells, plasma cells or tissue-infiltrating B cells Cell barcode to enable analysis of coexpressed functional genes in B cell subsets |
| Overcome the scale necessary for comprehensive repertoire analysis | Analyse thousands of B cells per experiment | Use high-throughput approaches |
| Demonstrate function | Accurately pair IgH + IgL expressed by individual B cells Sequence full-length variable regions Have error-free sequences (correction of both RT-PCR and sequencing errors) |
Use cell barcoding and linkage PCR methods Sequence entire variable regions Error correct, using cell barcodes to enable correction for RT-PCR and sequencing errors, and molecule barcodes to enable correction for sequencing but not RT-PCR errors |
| Overcome poor fidelity and quality | V-gene primers fail to sequence some antibodies PCR bias distorts clonal proportions RT-PCR and sequencing errors yield artefact clonal families PCR contamination produces artefacts |
Use template switching or other non-V-gene primer approach to add 5′ barcode Use cell barcodes Use cell barcodes to enable correction of both types of error Use cell or molecule barcodes |
Abbreviations: IgH, immunoglobulin heavy chain; IgL, immunoglobulin light chain; PCR, polymerase chain reaction; RT-PCR, reverse transcriptase polymerase chain reaction.
Several new single-cell sequencing methods can preserve the endogenous pairing of IgHs and IgLs (Figure 2). These methods include linkage-PCR based, barcode-based, bead-based, microwell-based and droplet-based methods, with most approaches integrating more than one of these methods.
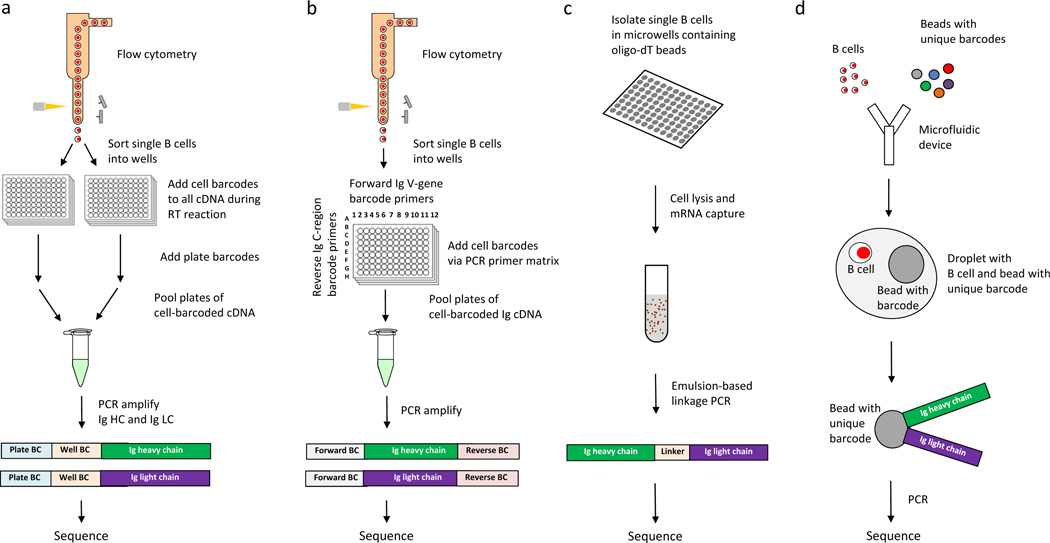
Figure 2.
Approaches for high-throughput sequencing of functional antibody repertoires. Comprehensive characterization of the functional antibody repertoire necessitates high-throughput, full-length and error-free sequencing of IgH and IgL pairs expressed by individual B cells. a | Cell barcoding via linkage PCR. Single B cells are isolated and lysed, then their RNA is captured by poly-T beads. cDNAs of the IgH and IgL expressed by individual B cells are then linked by emulsion to RT-PCR, and then pooled and sequenced.82 b | Cell barcoding via template switching. Single B cells are sorted, the template switching activity of RT adds a unique cell-specific barcode to all cDNAs generated from an individual B cell. Plate-specific barcodes are then added, resulting in cDNAs that have compound cell barcodes. Finally, the compound cell-barcoded IgH and IgL genes are amplified by PCR, pooled and sequenced.27,34,43 c | Cell barcoding by forward and reverse primer matrix. Single B cells are sorted, then V-gene forward primers and C-region reverse primers are used to add cell-specific barcodes to, and amplify by PCR, IgH and IgL cDNA generated from an individual B cell. Single-barcoded immunoglobulin genes are then pooled and sequenced.85 d | Microfluidic combination of beads with unique barcodes and single B cells into individual droplets. Using microfluidics, single B cells and beads with unique barcodes are combined in individual droplets, followed by lysis of the B cell, PCR and sequencing.61,83 Abbreviations: C, constant; IgH, immunoglobulin heavy chain; IgL, immunoglobulin light chain; PCR, polymerase chain reaction; RT, reverse transcriptase; V, variable.
Linkage-PCR
A method for physically linking and thereby sequencing IgH and IgL pairs from individual B cells is overlap-extension, also known as linkage PCR.80,81 Several groups have adapted this technique for high-throughput sequencing of IgH and IgL pairs.82,83 One such method involves depositing single B cells in high-density microwell plates, capturing their mRNA with oligo-dT beads, and then using emulsion-based linkage PCR to physically link and amplify IgH and IgL pairs before sequencing their CDR3 regions (Figure 2a).82 Integrating long-read sequencing technologies, cell barcodes and universal 5′ priming into linkage-PCR-based approaches should yield robust antibody repertoire datasets.
Cell barcoding
In my laboratory, we developed a cell-barcoding method for antibody repertoire sequencing.27,34,43,84 In this approach, all cDNA generated from each individual B cell was labelled with a unique cell barcode by using a universal 5′ adapter (Figure 2b, Figure 3a), which was added via the template-switching activity of reverse transcriptase, and enabled full-length and unbiased sequencing of the antibody variable regions. Because each B cell has multiple copies of mRNA encoding its IgH and IgL genes, cell barcoding creates consensus IgH and IgL sequences for each cell and thereby enables correction not only of errors inherent to the sequencing platform, but also of those arising from RT-PCR (Figure 3b). Furthermore, cell-barcoding enables accurate calculation of B-cell clonal proportions (Figure 3c), providing error-corrected sequences of the full-length V regions of natively paired IgHs and IgLs and enabling direct gene synthesis of the variable regions of the antibodies for recombinant expression. This approach has been used to understand human plasmablast antibody responses to RA,27 Staphylococcus aureus infection,43 and influenza vaccination.34 Bioinformatic analysis of sequence datasets can lead to the development of phylogenetic trees to guide selection and recombinant expression of plasmablast antibodies, including ACPAs implicated in the pathogenesis of RA,27 or antibodies that mediate opsonophagocytosis of Staphylococcus aureus43 or that neutralize influenza.34 With this method, rapid and comprehensive characterization of antibody responses is possible, as is the generation of affinity-matured, monoclonal human antibodies with high efficiency relative to hybridoma and other classical approaches for generating monoclonal antibodies. Wardemann and colleagues developed a 2D barcode primer matrix PCR that utilizes distinct sets of barcoded V-gene forward primers and barcoded constant-region reverse PCR primers for multiplex cell-based barcoding of single-cell-sorted B cells (Figure 2c).61,85
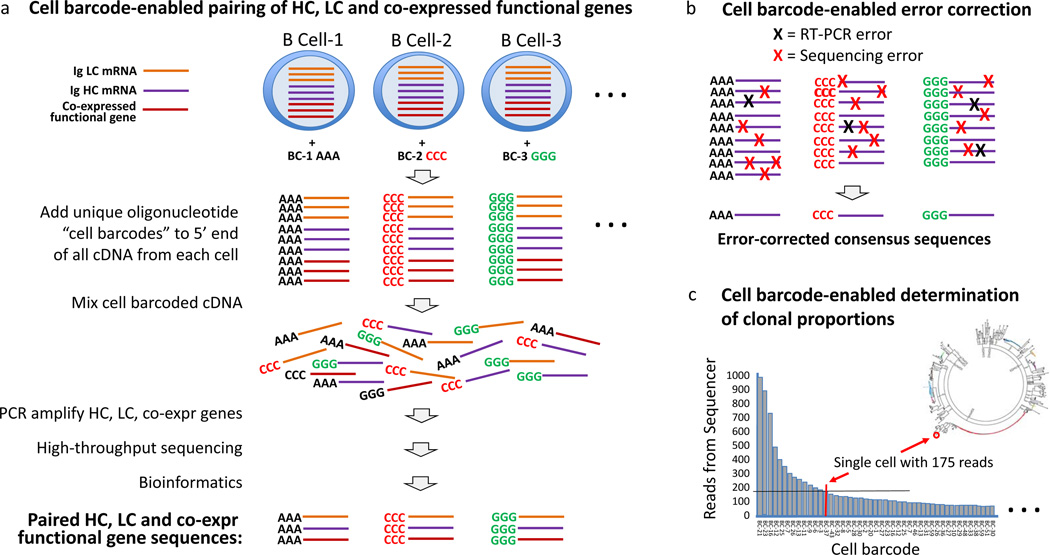
Figure 3.
Cell barcodes enable robust antibody repertoire sequencing. Cell barcodes (represented by AAA, CCC and GGG) added with the template switching activity of RT27,34,43 have many advantages, including preservation of the IgH and IgL pairs expressed by individual B cells, enabling accurate determination of the error-corrected sequence and of clonal proportions. a | In the approach presented, all cDNA generated from each cell receives the same unique barcode; therefore, coexpressed functional genes indicative of functional subsets of B cells (or T cells) can also be characterized.27,34,43 Both cell and molecular barcodes enable quantitative analysis of mRNA in individual cells. b | Multiple copies of each mRNA are expressed by each cell, and for each cell all mRNA receives the same cell barcode; therefore, cell barcoding enables correction of both PCR and high-throughput sequencing errors. c | Cell barcodes enable accurate determination of clonal proportions. Cell barcodes enable binning of the IgH and IgL chain reads from each B cell, thereby providing accurate determination of clonal proportions. By contrast, molecular barcodes do not enable RT-PCR error correction, IgH and IgL pairing, analysis of coexpressed genes or accurate determination of clonal proportions.
Droplet PCR
Several groups are working to develop droplet-based approaches that utilize beads, linkage-PCR or barcodes to enable the sequencing of IgH and IgL pairs expressed by individual B cells (Figure 2d).61,83 These techniques offer the potential to increase throughput analysis and reduce costs. Furthermore, droplet PCR, also termed digital PCR, isolates and amplifies individual DNA molecules in emulsion droplets and thereby reduces PCR bias.86
Bioinformatic analysis of clonal families
Sampling depth
An important requirement of next-generation single-cell sequencing technologies is that they generate datasets with IgH and IgL pairs from sufficient numbers of B cells to enable bioinformatic generation of phylogenetic trees and the identification of clonal antibody families. The sampling depth necessary to characterize a functional antibody response depends on the population of B cells analysed. For example, antigen-specific memory B cells are present in blood at frequencies of 1:2,500–100,000 cells;42 therefore, a relatively large number of cells must be obtained from the blood (or in some cases by leukapharesis) to isolate memory B cells with a specific antigen specificity, or to identify clonal families of memory B cells. By contrast, plasmablasts produced during an active antibody response have extensive redundancy in clonal families, and a number of groups have calculated that >75% of peripheral blood plasmablasts express antibodies specific to the antigenic target of an ongoing immune response.33,34,43,87 As a result, when sequencing the plasmablast antibody repertoire of an individual with a functional immune response, analysis of hundreds to thousands of plasmablasts (obtained from a little as 15 mL blood, or just 1–3 mL from infants) is sufficient for robustly identifying and characterizing the clonal families.27,34,43
Bioinformatic methods
A number of bioinformatics methods have been developed for V(D)J gene assignment and antibody repertoire analysis,88,89 and clonal families can be defined as B cells that share IgH VJ and IgL VJ usage. In stricter definitions of clonal families, the IgH VJs and IgL VJs must have identical CDR3 lengths or the amino acid sequences must have a certain degree of homology, suggesting that the B cells that express these antibodies arose from the same B cell progenitor.90 Clonal families can be generated by expansion of a common progenitor B cell, by independent de novo recombination events, or by convergent evolution of B-cell responses, that is, accumulation of mutations in B cells that express IgH V(D)J and IgL VJ of different genomic origins, resulting in convergence of encoded antibody sequences.91,92 Regardless of their particular ontology, clonal families are defined by the generation of B cells from a common progenitor selected and expanded by the ongoing immune response. In other words, B cells express high-affinity antibodies that enable them to ‘out-compete’ other B cells in the germinal centre for antigen and growth factors during affinity maturation.14,15 Furthermore, cross-patient analysis of antibody repertoires of individuals with a common immune phenotype has proven a powerful method of identifying shared IgH V(D)J and IgL VJ that encode antibodies with functional activity at an increased frequency as compared to identification of clonal families alone.51,65,93
Analysis of coexpressed B-cell functional genes
An important direction for antibody repertoire sequencing is the integration of analysis of coexpressed genes, in an approach analogous to the simultaneous sequencing of T-cell receptors and functional genes that demarcate T-cell subsets.94,95 Investigators in my laboratory are now simultaneously sequencing the IgH and IgL genes along with a set of functional B-cell subset genes (Figure 3), a strategy that will integrate information about functional B-cell subsets into our antibody repertoire data and thereby enhance our ability to identify functional antibodies. For B cells, coexpressed functional genes include transcription factors and effector molecules indicative of differentiation into plasma cells, memory B cells, long-lived plasmablasts or other maturated B-cell subsets. For quantitative analysis of coexpressed functional genes that are indicative of distinct functional B-cell (or T-cell94) subsets, both cellular and molecular barcoding are needed54,55 to measure the expression of the coding mRNAs in each cell.
Need for high-fidelity high-quality datasets
Obtaining large-scale, high-fidelity and high-quality sequencing data is a critical challenge in sequencing antibody repertoires, and is essential for biomarker analysis and for the generation of diagnostic and therapeutic antibodies. To achieve these ends, accurate determination of B-cell clonal proportions is needed, as is error correction for provision of high-fidelity sequences, comprehensive analysis of all variable regions expressed in the chosen B-cell subset for capturing the entire repertoire, and the ability to detect PCR contamination (Table 1).
Analysis of clonal proportions
Accurately measuring the size of clonal families is important for estimating the degree to which the immune system has selected and expanded a B cell expressing a particular antibody. Selection by the immune system is indicative of a high affinity of B-cell membrane immunoglobulins for cognate antigens, and thereby out-competing other B cells for antigen and growth factors in the germinal centre (Figure 1). In studying the antibody response to influenza vaccination with a cell-barcoding approach (Figure 2b), investigators in my laboratory found that large clonal families encode antibodies that bind to influenza hemagglutinin with higher affinities, and in certain cases are more effective at neutralizing influenza virus than antibodies that are not part of large clonal families.34 PCR bias, in which certain templates amplify more efficiently than other templates, can distort clonal proportions.96 In addition, activated B cells, such as plasmablasts, express as much as 100-fold more RNA than memory B cells and other resting B cells,97 which in the absence of cell-based barcoding27 or amplification within emulsion droplets98 might suggest artificial differences in clonal proportions. Analysis of antibodies expressed by individual cells, with cell barcodes27,34,43,54,55 (Figure 3c) and emulsion droplets,98 can correct or minimize PCR bias to enable accurate determination of clonal proportions.
Biological variation versus error
Robust analysis of datasets is highly dependent on obtaining high-fidelity, error-free sequences. Sequence errors arise from both RT-PCR and the sequencing platform.78 Several groups have demonstrated, by sequencing DNA plasmids that encode single antibodies (or single T-cell receptors), that the error from next-generation sequencing alone results in artefactual identification of clonal families.10,79,99 A variety of approaches have been developed for error correction, two of which use molecule10,54,55,70,100 or cell barcodes.27,34,43,54,55 Cell barcodes provide multiple advantages over molecular barcodes, including error correction of both RT-PCR and base-calling sequencing errors as well as accurate assessment of clonal proportions (Figure 3).
Analysis of antibody variable regions
Most approaches for sequencing antibody repertoires use 5′ V-gene primers, which might fail to amplify variable regions containing V genes encoded by germline sequences with insufficient homology or that have acquired somatic hypermutations in the residues to which such primers hybridize. Indeed, in my laboratory’s antibody repertoire analysis of plasmablasts from influenza vaccine recipients,34 we found that ~10% of all antibodies analysed had an IgH or IgL variable region that would not have been amplified by state-of-the-art V-gene-specific primers. Capturing such highly mutated antibodies is important because they might have acquired variable region mutations that confer binding and functional activity.
Ability to detect PCR contamination
Most next-generation sequencing approaches for antibody repertoire analysis utilize PCR to amplify DNA isolated or cDNA generated from B cells. PCR exponentially amplifies sequences, and is highly prone to contamination by amplicons from prior experiments.101 As the sequences isolated by antibody repertoire analysis have a high degree of nucleotide sequence homology, detecting PCR contamination can be difficult.27 Both cell and molecule barcodes provide a means to bioinformatically monitor and detect PCR contamination.
Applications
Generation of recombinant antibodies
Monoclonal antibodies are a mainstay of diagnostic tests, therapeutics and research tools. High-throughput sequencing of antibody repertoires is a powerful new approach to generate recombinant monoclonal antibodies directly from humans or other animals during a functional immune response. Therapeutic monoclonal antibodies have revolutionized care for patients with autoimmune diseases (anti-TNF, anti-IL-6, anti-IL-1 and anti-IL-12p40 monoclonal antibodies), infection (anti-respiratory syncytial virus antibody) and cancer (anti-CD20, anti-CTLA4, and anti-programmed-death 1 [PD-1] antibody). Investigators in my laboratory used barcode-enabled antibody repertoire sequencing (Figure 2b) of plasmablasts isolated from Staphylococcus aureus-infected humans to generate recombinant antibodies that bind and mediate killing of the bacteria.43 Others have isolated individual B cells in microwells, then performed linkage RT-PCR and sequencing (Figure 2a) to generate tetanus toxoid (TT) reactive recombinant antibodies from TT-sorted peripheral blood plasmablasts isolated from a TT-vaccinated human.82
Monitoring immune responses
More effective approaches are needed to effectively monitor immune responses in health, autoimmunity, microbial infection, biothreat detection, vaccination and cancer. Monitoring immune responses provides opportunities for early diagnosis, predicting flare, monitoring remission and assessing responses to immunomodulatory therapies and vaccines.
Autoimmunity
Investigators in my laboratory used barcode-enabled antibody repertoire sequencing (Figure 2b) of peripheral blood plasmablasts derived from patients with RA to identify sequences encoding autoantibodies that target citrullinated fibrinogen and citrullinated enolase.27 Others have used IgH (single-chain) sequencing of bulk RNA isolated from synovium of patients with RA to identify dominant clones utilizing the IGHV4–34 gene segment and having CDR3s longer than those of antibodies expressed by naive B cells.102 Sequences encoding pathogenic autoantibodies have the potential to be used as predictive biomarkers to identify individuals likely to develop disease, experience an autoimmune disease flare or respond to immunomodulatory therapy.77 A leading hypothesis is that mucosal sites, including the lung and oral cavity, might be critical in the initiation of ACPA responses that lead to the development of RA.103 Some antibody repertoire sequencing technologies27 generate sequencing reads that extend sufficiently far into the antibody constant region to identify the antibody isotype and subclasses, thereby enabling IgA ACPA-expressing B cells to be pinpointed to gain insight into the potential environmental exposures and microbial infections that initiate their production in RA. Finally, these sequences also have the potential to be used as pharmacodynamic biomarkers to monitor the response to an immunomodulatory drug, or to expedite clinical development by demonstrating the activity of these drugs in proof-of-concept studies.77
Vaccine development
A great need exists for the development of vaccines for a variety of pathogens. Antibody repertoire sequencing is used to identify the microbial antigens and epitopes targeted by effective antimicrobial antibody responses that naturally control infection in humans and other animals,4,6,43,104,105 thereby enabling the development of vaccines based on these antigens. In addition, antibody repertoire sequencing is used in clinical proof-of-concept trials to demonstrate that a candidate vaccine, adjuvant or vaccination regimen can induce protective immune responses.106
Immunomodulatory drug development
Immunomodulatory ‘checkpoint inhibitors’ are revolutionizing the care of patients with autoimmune diseases and cancer. Abatacept (CTLA4-Ig) blocks CD28-mediated activation of T cells and is efficacious in the treatment of RA.107 In patients with metastatic melanoma and other cancers, ipilimumab (anti-CTLA4 antibody) and pembrolizumab (anti-PD-1 antibody) are therapeutically effective by blocking inhibition of T cells.19,20,108 Antibody repertoire analysis and sequencing provides an approach to monitor and characterize the immune responses induced by candidate immunomodulatory drugs109 or vaccines.110 Checkpoint inhibitor therapy in cancer results in the induction of autoimmunity in a subset of patients, and antibody repertoire sequencing has the potential to be used to identify individuals at risk for developing autoimmune disease and to characterize the autoimmune response in individuals that do.19,20,108
B-cell repertoire development
By analysing the sequences of antibody repertoires in an individual and comparing them to the sequences of the corresponding germline antibody genes, the lineage development of B cells and evolution of antibody responses can be tracked.42,58,61,62 Using this analysis, one can model and investigate the evolution of functional antibody responses in health and disease, reveal mechanisms underlying the development of native B-cell repertoires111,112 and the activation, development and trafficking of B cells in immune responses of interest.47,113,114 Several academic laboratories are in the process of utilizing antibody repertoire sequencing to investigate the development of anti-citrullinated-protein-reactive B cells in RA and anti-nuclear-antigen-reactive B cells in SLE. Such studies might identify autoantigens and provide insight into disease initiation and progression in these and other diseases.
Antigen and epitope discovery
Identification of the critical antigens and epitopes targeted by functional antibody responses is needed. Most approaches require prior knowledge of the target antigen, which is used for screening vast numbers of hybridomas, B cells or panels of candidate antibodies.115 Antibody repertoire sequencing, by contrast, does not require such knowledge. When applied to plasmablasts derived from a human or other animal with an immune phenotype of interest, this method enables the bioinformatic identification of clonal families of antibodies within a functional B-cell response without having to express or screen the antibodies. Such clonal antibody families define important antigens, as determined by the immune response. Recombinant antibodies generated from plasmablast clonal families can be used to identify the antigenic specificity of these cells. Such antigen discovery can uncover not only the targets of antibodies but also those of T cells, because B cells and T cells coordinate responses against the same macromolecular antigen complexes.116 Comprehensive datasets generated by large-scale sequencing of antibody repertoires, in combination with identification of the corresponding antigen targets of important antibodies, is anticipated to enable bioinformatic modelling of antibody–antigen structure-binding relationships that will ultimately lead to antibody variable region sequences being used to predict antibody specificity.
Conclusions
Effective characterization of the antibody repertoire requires high-fidelity analysis of the full-length IgH and IgL pairs expressed by individual B cells, to enable robust bioinformatic analysis of the repertoires and accurate recombinant generation of selected antibodies that can be used to identify target antigens and to investigate the mechanisms of their protective and pathogenic functions. Technologies that utilize cell-barcodes and analyse full-length paired IgH and IgL are anticipated to provide the greatest utility in terms of identifying biomarkers and gaining new insights into the pathogenesis of RA, SLE, other autoimmune rheumatic diseases, and other diseases. Antibody repertoire analysis will transform our understanding of immune responses to autoimmunity, vaccination, infection and cancer, providing new biomarkers and diagnostic tools, and enabling efficient generation of therapeutic antibodies.
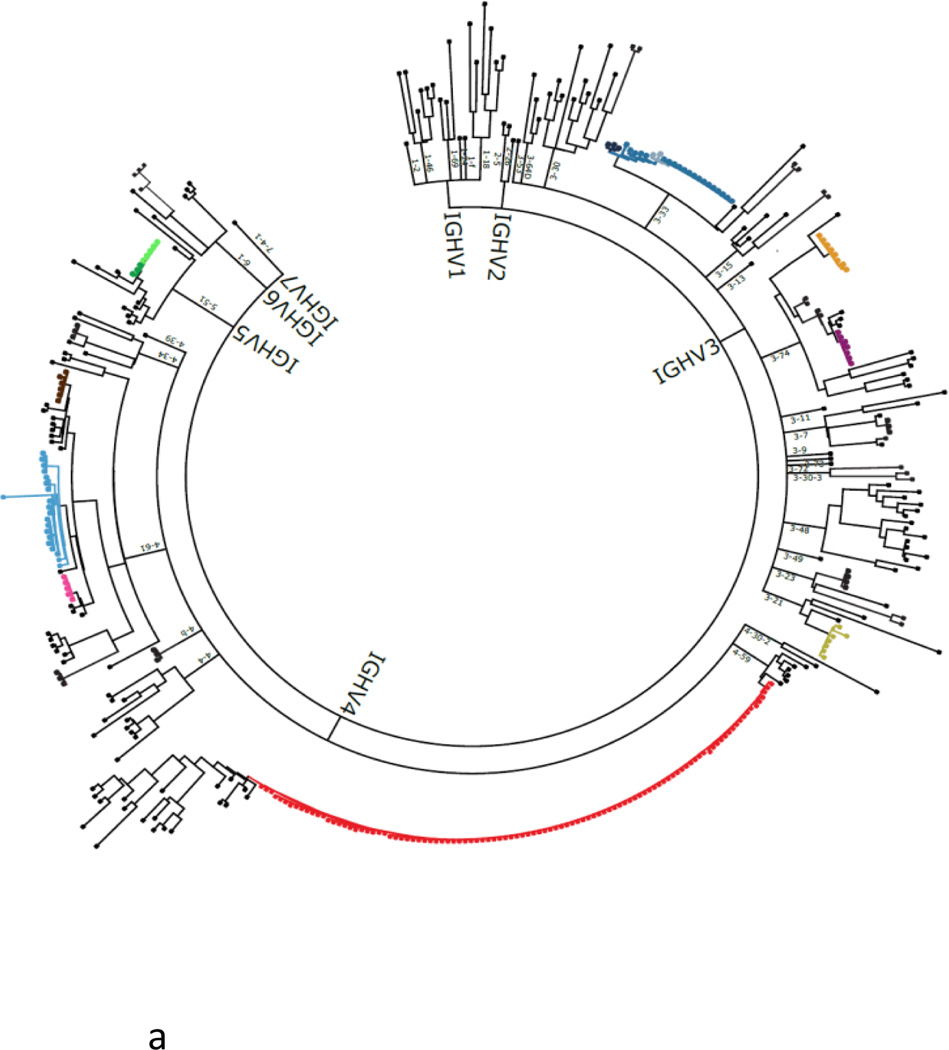
Figure 4.
Phylogenetic tree of the antibody repertoire. Example of a phylogenetic tree of a human plasmablast response generated using cell barcoding. Phylogenetic trees reveal clonal families and guide the expression and screening of the identified antibodies. Each major branch represents a single IgH V(D)J, outer branching is determined by the IgL VJ sequence, and each terminal leaf represents the error-corrected, full-length, paired IgH and IgL sequences expressed by an individual B cell. Clonal families (colour coded) contain antibodies that share IgH VJ and IgL VJ sequences. Immunodominant (red) and rare (other colours) families are identified. Single nucleotide substitutions can substantially alter antibody binding specificity and affinity117, and in order to evaluate the spectrum of antibodies encoded within each clonal family recombinant antibodies representing the range of variants represented within each family should be expressed and characterized. Abbreviations: D, diversity; IgH, immunoglobulin heavy chain; IgL, immunoglobulin light chain; J, joining; RT-PCR, reverse transcriptase polymerase chain reaction, V, variable.
Key Points.
The development of high-throughput DNA sequencing has enabled large-scale analysis of antibody repertoires and responses
Critical parameters for characterization of functional antibody repertoires include the methodology, the B-cell population characterized, clinical characteristics of the individuals analysed and the bioinformatic analysis
High-fidelity analysis of full-length paired heavy and light chains expressed by individual B cells is critical for the characterization of functional antibody repertoires
Analysis of coexpressed functional genes can identify antibodies that contribute to functional immune responses
Sequencing antibody repertoires is transforming our understanding of pathogenic and protective immune responses in autoimmunity, infection, vaccination and cancer
Antibody repertoire sequencing should lead to the generation of biomarkers, diagnostic tools and therapeutic antibodies for various diseases, including rheumatic diseases
Acknowledgements
I thank Drs. Tito Serafini, Yann-Chong Tan, Mark Davis, Chia-Hsin Ju, Lisa Blum, Jeremy Sokolove, Wayne Volkmuth, Daniel Emerling, Sohail Ahmed, Larry Steinman, Lynda Stuart and Chris Karp for their scientific insights and discussion. I thank Daniel Lu and Sarah Kongpachith for their scientific insights, and Dr Tamsin Lindstrom for her scientific and editorial input. My research is supported by NIH NHLBI Proteomics Center N01-HV-00242, NIH NIAMS R01 AR063676, NIH NIAID U01 AI101981, NIH NIAID U01 AI057229, NIH NIAID U19 AI110491, NIH NCI R33 CA183659, NIH NIAMS/NIAID/FNIH AMP Program UH2 AR067681, the Brennan Family, the Northern California Chapter of the Arthritis Foundation (NCCAF) Center of Excellence, and the Bill and Melinda Gates Foundation.
Footnotes
Competing interests
The author declares that he is a member of the Board of Directors, consultant for, and owner of equity in Atreca, Inc.
References
- 1.Leadbetter EA, et al. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 2.Rosen A, Casciola-Rosen L. Autoantigens as substrates for apoptotic proteases: implications for the pathogenesis of systemic autoimmune disease. Cell Death Differ. 1999;6:6–12. doi: 10.1038/sj.cdd.4400460. [DOI] [PubMed] [Google Scholar]
- 3.Tan EM. Autoantibodies, autoimmune disease, and the birth of immune diagnostics. J. Clin. Invest. 2012;122:3835–3836. doi: 10.1172/JCI66510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson PC, Andrews SF. Tools to therapeutically harness the human antibody response. Nat. Rev. Immunol. 2012;10:709–719. doi: 10.1038/nri3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein F, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton DR, et al. A blueprint for HIV vaccine discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotech. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 9.Alt FW, Blackwell TK, Yancopoulos GD. Development of the primary antibody repertoire. Science. 1987;238:1079–1087. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- 10.Shugay M, et al. Towards error-free profiling of immune repertoires. Nat. Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 11.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 12.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509:637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Victora GD, Nussenzweig MC. Germinal centers. Ann. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 14.Kuppers R. Somatic hypermutation and B cell receptor selection in normal and transformed human B cells. Ann. NY Acad. Sci. 2003;987:173–179. doi: 10.1111/j.1749-6632.2003.tb06046.x. [DOI] [PubMed] [Google Scholar]
- 15.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat. Rev. Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 16.Hueber W, Robinson WH. Proteomic biomarkers for autoimmune disease. Proteomics. 2006;6:4100–4105. doi: 10.1002/pmic.200600017. [DOI] [PubMed] [Google Scholar]
- 17.Koff WC, et al. Accelerating next-generation vaccine development for global disease prevention. Science. 2013;340:1232910. doi: 10.1126/science.1232910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couzin-Frankel J. Breakthrough of the year cancer immunotherapy. Science. 2013;342:1432–1433. doi: 10.1126/science.342.6165.1432. [DOI] [PubMed] [Google Scholar]
- 20.Riley JL. Combination checkpoint blockade—taking melanoma immunotherapy to the next level. N. Engl. J. Med. 2013;369:187–189. doi: 10.1056/NEJMe1305484. [DOI] [PubMed] [Google Scholar]
- 21.Robinson WH, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat. Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 22.Reddy MM, et al. Identification of candidate IgG biomarkers for Alzheimer’s disease via combinatorial library screening. Cell. 2011;144:132–142. doi: 10.1016/j.cell.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legutki JB, et al. Scalable high-density peptide arrays for comprehensive health monitoring. Nat Commun. 2014;4785 doi: 10.1038/ncomms5785. [DOI] [PubMed] [Google Scholar]
- 24.Price JV, et al. On silico peptide microarrays for high-resolution mapping of antibody epitopes and diverse protein-protein interactions. Nat. Med. 2012;18:1434–1440. doi: 10.1038/nm.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phizicky E, Bastiaens PI, Zhu H, Snyder M, Fields S. Protein analysis on a proteomic scale. Nature. 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- 26.Larman HB, et al. Autoantigen discovery with a synthetic human peptidome. Nat. Biotech. 2011;29:535–541. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan YC, et al. Barcode-enabled sequencing of plasmablast antibody repertoires in rheumatoid arthritis. Arthritis Rheumatol. 2014;66:2706–2715. doi: 10.1002/art.38754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerkman PF, et al. Circulating plasmablasts/plasmacells as a source of anticitrullinated protein antibodies in patients with rheumatoid arthritis. Ann. Rheum. Dis. 2013;72:1259–1263. doi: 10.1136/annrheumdis-2012-202893. [DOI] [PubMed] [Google Scholar]
- 29.Utz P, Anderson P. Posttranslational modifications, apoptosis, and bypass of tolerance to autoantigens. Arthritis Rheum. 1998;41:1152–1160. doi: 10.1002/1529-0131(199807)41:7<1152::AID-ART3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 30.Fooksman DR, et al. Development and migration of plasma cells in the mouse lymph node. Immunity. 2010;33:118–127. doi: 10.1016/j.immuni.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiepe F, et al. A. Long-lived autoreactive plasma cells drive persistent autoimmune inflammation. Nat. Rev. Rheumatol. 2011;7:170–178. doi: 10.1038/nrrheum.2011.1. [DOI] [PubMed] [Google Scholar]
- 32.Fink K. Origin and function of circulating plasmablasts during acute viral infections. Front. Immunol. 2012;3:78. doi: 10.3389/fimmu.2012.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan YC, et al. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin. Immunol. 2014;151:55–65. doi: 10.1016/j.clim.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verpoort KN, et al. Isotype distribution of anti-cyclic citrullinated peptide antibodies in undifferentiated arthritis and rheumatoid arthritis reflects an ongoing immune response. Arthritis Rheum. 2006;54:3799–3808. doi: 10.1002/art.22279. [DOI] [PubMed] [Google Scholar]
- 36.Owczarczyk K, et al. A plasmablast biomarker for nonresponse to antibody therapy to CD20 in rheumatoid arthritis. Sci. Transl. Med. 2011;3:101ra192. doi: 10.1126/scitranslmed.3002432. [DOI] [PubMed] [Google Scholar]
- 37.Kuhn KA, et al. Antibodies against citrullinated proteins enhance tissue injury in experimental autoimmune arthritis. J. Clin. Invest. 2006;116:961–973. doi: 10.1172/JCI25422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokolove J, Zhao X, Chandra PE, Robinson WH. Immune complexes containing citrullinated fibrinogen costimulate macrophages via Toll-like receptor 4 and Fcγ receptor. Arthritis Rheum. 2011;63:53–62. doi: 10.1002/art.30081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantaert T, et al. B lymphocyte autoimmunity in rheumatoid synovitis is independent of ectopic lymphoid neogenesis. J. Immunol. 2008;181:785–794. doi: 10.4049/jimmunol.181.1.785. [DOI] [PubMed] [Google Scholar]
- 40.Humby F, et al. Ectopic lymphoid structures support ongoing production of class-switched autoantibodies in rheumatoid synovium. PLoS Med. 2009;6:e1. doi: 10.1371/journal.pmed.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toes RE, Huizinga TW. Autoimmune responses in the rheumatoid synovium. PLoS Med. 2009;6:e9. doi: 10.1371/journal.pmed.1000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McHeyzer-Williams M, Okitsu S, Wang N, McHeyzer-Williams L. Molecular programming of B cell memory. Nat. Rev. Immunol. 2012;12:24–34. doi: 10.1038/nri3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu DR, et al. Identification of functional anti-Staphylococcus aureus antibodies by sequencing patient plasmablast antibody repertoires. Clin. Immunol. 2014;152:77–89. doi: 10.1016/j.clim.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franz B, May KF, Jr, Dranoff G, Wucherpfennig K. Ex vivo characterization and isolation of rare memory B cells with antigen tetramers. Blood. 2011;118:348–357. doi: 10.1182/blood-2011-03-341917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doria-Rose NA, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J. Virol. 2009;83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amara K, et al. Monoclonal IgG antibodies generated from joint-derived B cells of RA patients have a strong bias toward citrullinated autoantigen recognition. J. Exp. Med. 2013;210:445–455. doi: 10.1084/jem.20121486. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Stern JN, et al. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci. Transl. Med. 2014;6:248ra107. doi: 10.1126/scitranslmed.3008879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shigematsu Y, et al. Malignant mesothelioma-associated antigens recognized by tumor-infiltrating B cells and the clinical significance of the antibody titers. Cancer Sci. 2009;100:1326–1334. doi: 10.1111/j.1349-7006.2009.01181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 50.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 51.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Traggiai E, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Niro R, et al. High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nat. Med. 2012;18:441–445. doi: 10.1038/nm.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 56.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn JA, Crowe JE., Jr Impact of new sequencing technologies on studies of the human B cell repertoire. Curr. Opin. Immunol. 2013;25:613–618. doi: 10.1016/j.coi.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotech. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 60.Metzker ML. Sequencing technologies—the next generation. Nat Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 61.Georgiou G, et al. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotech. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathonet P, Ullman CG. The application of next generation sequencing to the understanding of antibody repertoires. Front. Immunol. 2013;4:265. doi: 10.3389/fimmu.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robins H. Immunosequencing: applications of immune repertoire deep sequencing. Curr. Opin. Immunol. 2013;25:646–652. doi: 10.1016/j.coi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 64.Palanichamy A, et al. Immunoglobulin class-switched B cells form an active immune axis between CNS and periphery in multiple sclerosis. Sci. Transl. Med. 2014;6:248ra106. doi: 10.1126/scitranslmed.3008930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parameswaran P, et al. Convergent antibody signatures in human dengue. Cell Host Microbe. 2013;13:691–700. doi: 10.1016/j.chom.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu D, et al. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci. Transl. Med. 2012;4:134ra163. doi: 10.1126/scitranslmed.3003656. [DOI] [PubMed] [Google Scholar]
- 67.Faham M, et al. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2012;120:5173–5180. doi: 10.1182/blood-2012-07-444042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boyd SD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci. Transl. Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Vlaminck I, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155:1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vollmers C, Sit RV, Weinstein JA, Dekker CL, Quake SR. Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proc. Natl Acad. Sci. USA. 2013;110:13463–13468. doi: 10.1073/pnas.1312146110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meffre E, Wardemann H. B-cell tolerance checkpoints in health and autoimmunity. Curr. Opin. Immunol. 2008;20:632–638. doi: 10.1016/j.coi.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 72.Halverson R, Torres RM, Pelanda R. Receptor editing is the main mechanism of B cell tolerance toward membrane antigens. Nat. Immunol. 2004;5:645–650. doi: 10.1038/ni1076. [DOI] [PubMed] [Google Scholar]
- 73.Reddy ST, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat. Biotech. 2010;28:965–969. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]
- 74.Lu DR, Robinson WH. Street-experienced peripheral B cells traffic to the brain. Sci. Transl. Med. 2014;6:248fs231. doi: 10.1126/scitranslmed.3009919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Logan AC, et al. High-throughput VDJ sequencing for quantification of minimal residual disease in chronic lymphocytic leukemia and immune reconstitution assessment. Proc. Natl Acad. Sci. USA. 2011;108:21194–21199. doi: 10.1073/pnas.1118357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mroczek ES, et al. Differences in the composition of the human antibody repertoire by B cell subsets in the blood. Front. Immunol. 2014;5:96. doi: 10.3389/fimmu.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson WH, Lindstrom TM, Cheung RK, Sokolove J. Mechanistic biomarkers for clinical decision making in rheumatic diseases. Nat. Rev. Rheumatol. 2013;9:267–276. doi: 10.1038/nrrheum.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loman NJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotech. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 79.Nguyen P, et al. Identification of errors introduced during high throughput sequencing of the T cell receptor repertoire. BMC Genomics. 2011;12:106. doi: 10.1186/1471-2164-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andersen PS, Hansen MH, Nielsen LS, Oleksiewicz MB. Method for linking sequences of interest. WO2005042774 A2005042772. US Patent. 2005
- 81.Meijer PJ, et al. Isolation of human antibody repertoires with preservation of the natural heavy and light chain pairing. J. Mol. Biol. 2006;358:764–772. doi: 10.1016/j.jmb.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 82.DeKosky BJ, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat. Biotech. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Church GM, Vigneault F, Laserson U, Bachelet I. High-throughput immune sequencing. US20130296535. US Patent. 2013
- 84.Robinson WH, Tan YC, Sokolove J. Identification of polynucleotides associated with a sample. WO2012148497 A2012148493. US Patent 2013. 2013
- 85.Busse CE, Czogiel I, Braun P, Arndt PF, Wardemann H. Single-cell based high-throughput sequencing of full-length immunoglobulin heavy and light chain genes. Eur. J. Immunol. 2014;44:597–603. doi: 10.1002/eji.201343917. [DOI] [PubMed] [Google Scholar]
- 86.Rubelt F, et al. Onset of immune senescence defined by unbiased pyrosequencing of human immunoglobulin mRNA repertoires. PLoS ONE. 2012;7:e49774. doi: 10.1371/journal.pone.0049774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wrammert J, et al. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J. Virol. 2012;86:2911–2918. doi: 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mehr R, Sternberg-Simon M, Michaeli M, Pickman Y. Models and methods for analysis of lymphocyte repertoire generation, development, selection and evolution. Immunol. Lett. 2012;148:11–22. doi: 10.1016/j.imlet.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 89.Kidd BA, Peters LA, Schadt EE, Dudley JT. Unifying immunology with informatics and multiscale biology. Nat. Immunol. 2014;15:118–127. doi: 10.1038/ni.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kepler TB, et al. Reconstructing a B-Cell clonal lineage. II. Mutation, selection, and affinity maturation. Front. Immunol. 2014;5:170. doi: 10.3389/fimmu.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu X, Zhou T, Zhu J, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krause JC, et al. Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J. Immunol. 2011;187:3704–3711. doi: 10.4049/jimmunol.1101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jackson KJ, et al. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe. 2014;16:105–114. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotech. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Steinman L. A brief history of TH17, the first major revision in the TH1/TH2 hypothesis of T cell-mediated tissue damage. Nat. Med. 2007;13:139–145. doi: 10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- 96.van Dijk EL, Jaszczyszyn Y, Thermes C. Library preparation methods for next-generation sequencing: tone down the bias. Exp. Cell Res. 2014;322:12–20. doi: 10.1016/j.yexcr.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 97.Kelley DE, Perry RP. Transcriptional and posttranscriptional control of immunoglobulin mRNA production during B lymphocyte development. Nucleic Acids Res. 1986;14:5431–5447. doi: 10.1093/nar/14.13.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zagnoni M, Cooper JM. Droplet microfluidics for high-throughput analysis of cells and particles. Methods Cell Biol. 2011;102:25–48. doi: 10.1016/B978-0-12-374912-3.00002-X. [DOI] [PubMed] [Google Scholar]
- 99.Zhu J, et al. Somatic populations of PGT135–137 HIV-1-neutralizing antibodies identified by 454 pyrosequencing and bioinformatics. Front. Microbiol. 2012;3:315. doi: 10.3389/fmicb.2012.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jiang N, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci. Transl. Med. 2013;5:171ra119–171ra119. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Longo MC, Berninger MS, Hartley JL. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 102.Doorenspleet ME, et al. Rheumatoid arthritis synovial tissue harbours dominant B-cell and plasma-cell clones associated with autoreactivity. Ann. Rheum. Dis. 2014;73:756–762. doi: 10.1136/annrheumdis-2012-202861. [DOI] [PubMed] [Google Scholar]
- 103.Demoruelle MK, Deane KD, Holers VM. When and where does inflammation begin in rheumatoid arthritis? Curr. Opin. Rheum. 2014;26:64–71. doi: 10.1097/BOR.0000000000000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- 105.Laserson U, et al. High-resolution antibody dynamics of vaccine-induced immune responses. Proc. Natl Acad. Sci. USA. 2014;111:4928–4933. doi: 10.1073/pnas.1323862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wiley SR, et al. Targeting TLRs expands the antibody repertoire in response to a malaria vaccine. Sci. Transl. Med. 2011;3:93ra69. doi: 10.1126/scitranslmed.3002135. [DOI] [PubMed] [Google Scholar]
- 107.Genovese MC, et al. Abatacept for rheumatoid arthritis refractory to tumor necrosis factor α inhibition. N. Engl. J. Med. 2005;353:1114–1123. doi: 10.1056/NEJMoa050524. [DOI] [PubMed] [Google Scholar]
- 108.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cha E, et al. Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Sci. Transl. Med. 2014;6:238ra270. doi: 10.1126/scitranslmed.3008211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Garren H, et al. Phase 2 trial of a DNA vaccine encoding myelin basic protein for multiple sclerosis. Ann. Neurol. 2008;63:611–620. doi: 10.1002/ana.21370. [DOI] [PubMed] [Google Scholar]
- 111.Boyd SD, et al. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J. Immunol. 2010;184:6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Watson CT, et al. Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am. J. Hum. Genet. 2013;92:530–546. doi: 10.1016/j.ajhg.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.von Budingen H, et al. Frontline: epitope recognition on the myelin/oligodendrocyte glycoprotein differentially influences disease phenotype and antibody effector functions in autoimmune demyelination. Eur. J. Immunol. 2004;34:2072–2083. doi: 10.1002/eji.200425050. [DOI] [PubMed] [Google Scholar]
- 114.Wu YC, et al. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116:1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bradbury AR, Sidhu S, Dubel S, McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat. Biotech. 2011;29:245–254. doi: 10.1038/nbt.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Robinson WH, Steinman L, Utz PJ. Protein arrays for autoantibody profiling and fine-specificity mapping. Proteomics. 2003;3:2077–2084. doi: 10.1002/pmic.200300583. [DOI] [PubMed] [Google Scholar]
- 117.Diamond B, Scharff MD. Somatic mutation of the T15 heavy chain gives rise to an antibody with autoantibody specificity. Proc. Natl Acad. Sci. USA. 1984;81:5841–5844. doi: 10.1073/pnas.81.18.5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]