SUMMARY
Kinesin-1 is a two-headed motor that takes processive 8-nm hand-over-hand steps and transports intracellular cargos towards the plus end of microtubules. Processive motility requires a gating mechanism to coordinate the mechanochemical cycles of the two heads. Kinesin gating involves the neck linker (NL), a short peptide that interconnects the heads, but it remains unclear whether gating is facilitated by the NL orientation or tension. Using optical trapping, we measured the force-dependent microtubule release rate of kinesin monomers under different nucleotide conditions and pulling geometries. We find that pulling NL in the backward direction inhibits nucleotide binding and subsequent release from the microtubule. This inhibition is independent from the magnitude of tension (2–8 pN) exerted on NL. Our results provide evidence that the front head of a kinesin dimer is gated by the backward orientation of its NL until the rear head releases from the microtubule.
INTRODUCTION
Kinesin-1 (herein referred to as kinesin) is a dimeric motor that carries membranous organelles and vesicles towards the synapse in neurons (Hirokawa et al., 2009). Kinesin moves processively, taking hundreds of 8 nm steps along microtubules (MTs) before dissociation (Svoboda et al., 1993; Toprak et al., 2009). The processivity of kinesin-1 results from a coordinated mechanochemical cycle between the two catalytic heads. Each mechanical step is associated with a single ATP hydrolysis (Hua et al., 1997; Schnitzer and Block, 1997), also suggesting that mechanochemical cycles of the heads are coordinated. The heads alternately take a step (a pattern termed hand-over-hand stepping), in which the front head remains bound to the MT as the rear head steps forward (Asbury et al., 2003; Kaseda et al., 2003; Yildiz et al., 2004). This coordinated movement is facilitated by a gating mechanism that keeps the heads out of phase such that chemical or structural transitions in one head are inhibited until the partner head proceeds through its mechanochemical cycle. Two competing models have been proposed to explain which head is gated during processive movement. According to the fronthead gating model (Klumpp et al., 2004; Rosenfeld et al., 2003) nucleotide binding to the front head is inhibited. In the rear-head gating model (Crevel et al., 2004; Schief et al., 2004), ATP hydrolysis or MT release in the rear head are accelerated relative to the front head. Both models are consistent with the stepping of the rear head as the front head remains bound to a MT during processive motility, and they are not mutually exclusive.
Studies of kinesin motility have revealed that interhead coordination is mediated through NL, a 14 amino acid peptide that connects each of the kinesin heads to the common stalk (Asenjo et al., 2006; Block, 2007; Rosenfeld et al., 2001; Sindelar and Downing, 2010). The principal conformational change that drives motility is the docking of the NL onto the catalytic core of the front head (Rice et al., 1999) upon ATP hydrolysis (Milic et al., 2014). In a two-heads-bound (2HB) state, the heads are separated by 8 nm and the NL of the front head points backward whereas the NL of the rear head points forward. As a consequence, intramolecular tension develops between the heads via NLs (Hyeon and Onuchic, 2007). It remains unclear whether it is tension on NL (Guydosh and Block, 2006; Shastry and Hancock, 2010; Yildiz et al., 2008) or the asymmetric orientations of the NLs (Clancy et al., 2011) that gate kinesin motility (Figure 1A, Figure S1). In this study, we used single-molecule optical trapping assays to determine which of these potential gating mechanisms is responsible for kinesin processivity.
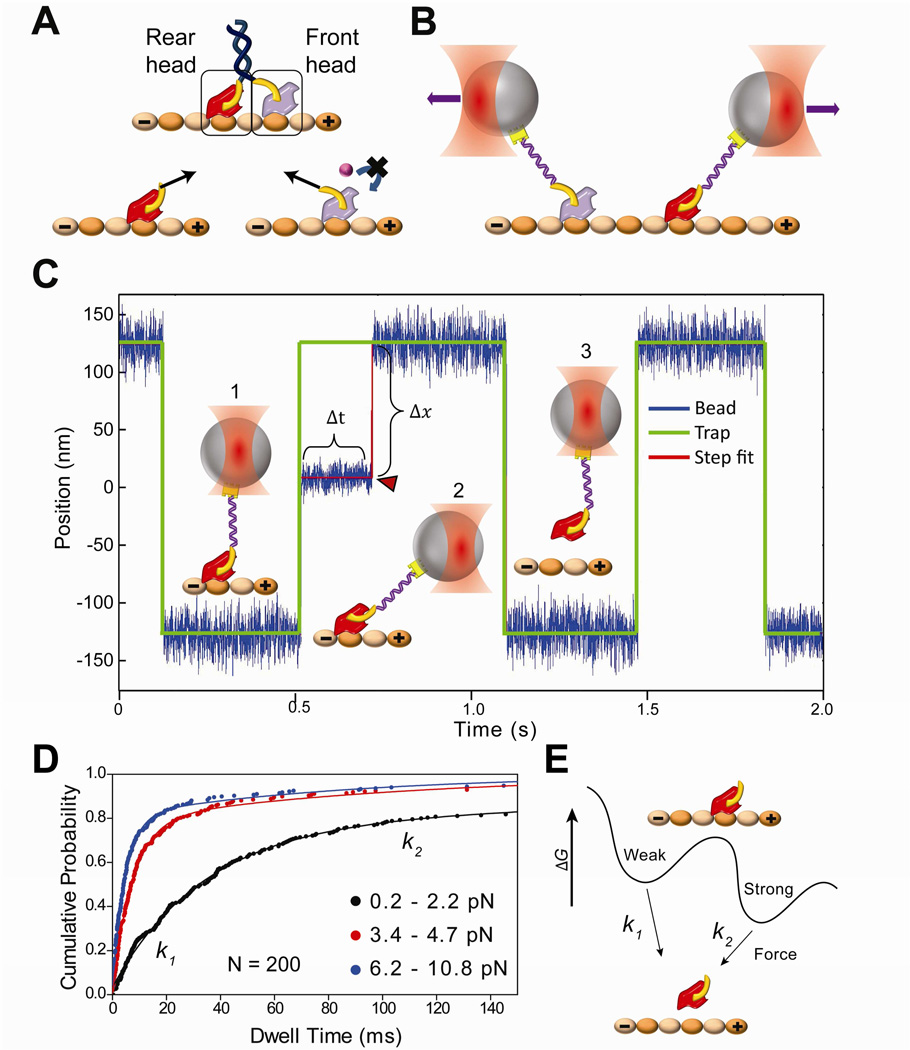
Figure 1. Force-dependent release of kinesin from MTs.
(A) (Top) Schematic of a kinesin dimer in a 2HB state. The NL (yellow) of the front head is oriented backwards and that of the rear head is oriented forward. (Bottom) Orientation of the NLs or tension between them (black arrows) may prevent ATP binding to the front head, or accelerate the nucleotide hydrolysis and subsequent MT release of the rear head to facilitate coordinated movement. (B) NL orientation of the front and rear heads can be mimicked by pulling a kinesin monomer from its NL via a short DNA tether using an optical trap (not to scale). (C) A trapped bead is oscillated between two positions 250 nm apart along the MT long axis. (1) When a monomer binds to the MT, (2) the movement of the bead to the next trap position is restricted. In this state, the trap exerts a constant force as a function of bead-trap separation (Δx) on the motor until it releases from the MT (Δt). (3) When the monomer releases from the MT (red arrowhead), the bead resumes following the trap. (D) Cumulative probability distributions (solid circles) represent the dwell time data for kinesin monomers pulled from the head towards the plus-end in the absence of nucleotide at different force ranges. N = 200 for each histogram. The release rates (k1 and k2) at a given force range were calculated by a two-exponential-decay fit (solid curves). (E) Model of kinesin-MT interaction shows two distinct binding modes in the apo state. k1 and k2 represent force-induced release rates from the weak and strong states, respectively.
See also Figures S1 and S2.
RESULTS
Force-dependent release rate of kinesin from MTs
To test the predictions of front- and rear-head gating models, we pulled on the NL to mimic its orientations in the front and rear head positions using an optical trap. To exert forces on the motor at a specific position, human kinesin monomers truncated at the C-terminus of the NL (hK349) were labeled with a 74 base pair DNA tether at the head (Guydosh and Block, 2009) or at NL (see Experimental Procedures). Labeling efficiency of the motors with a DNA tether was 15% (Figure S2). The other end of the DNA tether was functionalized with biotin and attached to streptavidin-coated polystyrene beads.
The MT release rate of a head was measured across a range of forces and nucleotide conditions (Figure 1B). Motor-coated beads were moved ±125 nm in a square wave pattern on polarity marked MTs (Figure 1C and S2) (Cleary et al., 2014). When a single kinesin monomer was bound to the MT, it restricted the movement of the bead to a new position of the trap. As a result, the trap exerted constant forces ranging from 0.5 pN to 10 pN on the motor, until it released from the MT. Previous kinesin rupture-force measurements were performed by moving a trapped bead along MTs under constantly increasing force (Kawaguchi and Ishiwata, 2001). Our assay better represents the situation in a kinesin dimer, where the heads are under constant tension in a 2HB state before MT release.
To calculate the force-dependent release rate of kinesin monomers, MT dwell-time data were sorted by applied force. We defined positive and negative forces as forces assisting and opposing kinesin’s natural direction of motion, respectively. Cumulative frequency distributions of ~200 dwells in a given force range were fit using two exponential decays (Figure 1D). The data could not be fitted well with a single exponential decay (p <0.001, F-test). The MT release rates were defined as the decay constants of the fit (k1 and k2). The results support a conventional model that kinesin has strong and weak binding modes to MT surface (Cross, 2004) and suggest that k1 and k2 represent force-induced exit from these weak and strong binding states, respectively (Figure 1E).
We first established baseline release rates by pulling kinesin monomers under the nucleotide-free (apo) state. A kinesin motor has a strong affinity for MT in the apo state. Figure 2A shows that NL-pulled kinesins have similar release properties to head-pulled kinesins in the apo state. k1 was an order of magnitude faster than k2, and represented ~60–80% of the release events under a wide range of applied forces (Figure S3). In head- and NL-pulled motors, k1 and k2 were less than 30 s−1 and 3 s−1, respectively, at low forces (±1.4 pN) and gradually increased with load in both forward (MT plus-end) and backward (MT minus-end) directions. At high forces, k1 and k2 were on the order of 100 s−1 and 10 s−1, respectively. Release under positive forces was ~20% faster through a wide range of applied forces, consistent with a weak net preference of kinesin to release towards its natural direction of motion under load (Uemura et al., 2002). This preference is in contrast to cytoplasmic dynein, which prefers to release towards the minus-end in a force-dependent manner, while the release towards the plus-end is slow and forceindependent (Cleary et al., 2014).
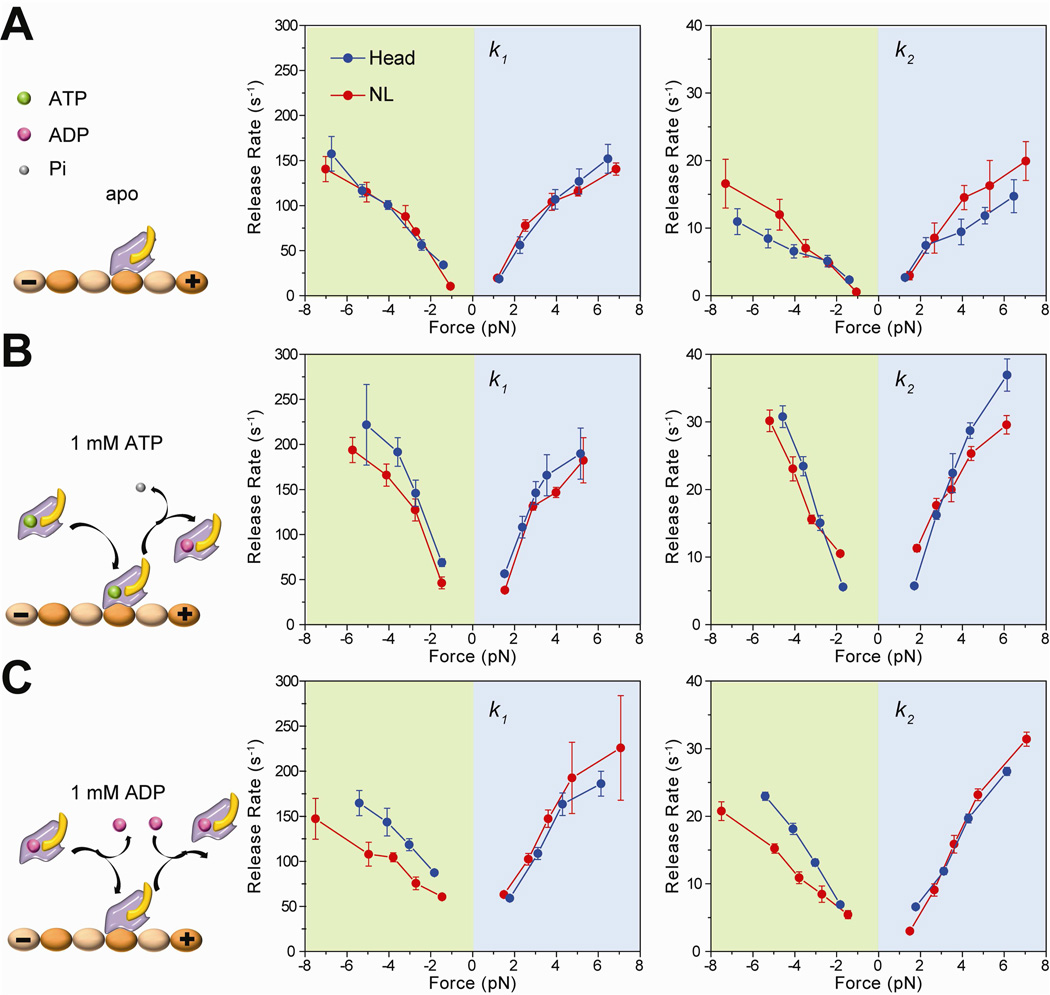
Figure 2. MT release rates of head- and NL-pulled kinesins under different nucleotide conditions.
(A) Without nucleotide, monomers release in response to external force (left). k1 (middle) and k2 (right) of kinesin monomers pulled from the head and NL increase with force in both forward (positive forces) and backward directions. (B) With 1 mM ATP in solution, kinesin can release by force or hydrolysis of the bound nucleotide (left). MT release rates of the headand NL-pulled kinesins increase compared to the apo condition. (C) At 1 mM ADP, kinesin can release from MTs by force or ADP binding (left). NL-pulled kinesins show slower release under backward forces. Error bars represent 95% confidence intervals.
See also Figures S3 and S4.
Tension on the NL is not critical for nucleotide hydrolysis
We next tested gating models based on the NL orientation and tension by pulling monomers from the NL and the head under different nucleotide conditions. To test the rear-head gating model, we measured release rates at saturating (1 mM) ATP. Under this condition, kinesin can release from MT due to tension exerted by the trap either in the apo state or in different nucleotide states following the hydrolysis of the bound ATP. If ATP hydrolysis at the rear head were accelerated in a 2HB state (Hancock and Howard, 1999), kinesin monomers would release faster when NL is pulled in the forward direction. Figure 2B shows that k1 and k2 were nearly symmetric between positive and negative forces. Increase in k1 and k2 was ~1.5-fold steeper in 1 mM ATP compared to the apo condition, consistent with faster detachment of kinesin monomers from MT in the presence of nucleotide (Figure S4). NL and head-pulled kinesin have similar release rates, indicating that nucleotide hydrolysis of the rear head is not accelerated when NL is pulled forward. These results disfavor the rear-head gating model.
k1 and k2 were significantly accelerated by ATP addition at low (±1.4 pN) forces. Unlike the apo condition, in which k1 and k2 were approaching to near 0 s−1, in 1 mM ATP k1 and k2 were 60 s−1 and 5 s−1 for head-pulled and 42 s−1 and 7 s−1 for NL-pulled motors, respectively. While k1 values at low forces agree well with rapid (50 s−1 in ATP and 70 s−1 in ADP) MT detachment measured in bulk (Hancock and Howard, 1999), k2 is more consistent with slow (3–5 s−1) release of the monomer-coated beads from MT (Hackney, 2002) in unloaded conditions. The discrepancy between the results of the previous measurements could be explained by the fact that bulk measurements reflect the average release rate, whereas bead-release assays are unable to detect fast MT-bead interactions due to limited temporal resolution. Unlike apo conditions, probability of fast and slow release events was nearly equal under a wide range of forces (Figure S3), presumably due to the changes in kinesin’s affinity to MT as a function of its nucleotide state.
ADP binding is inhibited by backward orientation of the NL
We next tested force-dependent MT release of kinesin monomers from MT in ADP. Kinesin interacts weakly with the MT in the ADP state (Uemura et al., 2002) and an unbound head must release its ADP before MT attachment (Hackney, 1994). Therefore, MT-bound monomers release from the MT either by external tension in the apo state or in the ADP-bound state. Previous unbinding force measurements on a kinesin dimer in ADP conditions indicated that kinesin’s affinity for ADP is enhanced by external load exerted on kinesin along the direction of motility and weakened by backward load (Uemura and Ishiwata, 2003). However, nucleotide binding rate, not the dissociation constant, is critical to the front-head gating mechanism, because kinesin is gated at both limited and saturating nucleotide concentrations (Guydosh and Block, 2006; Toprak et al., 2009). In addition, these experiments do not distinguish between ADP binding to the front and rear heads, because the dimers sample both 1HB and 2HB states in ADP.
The front-head gating model predicts that a head is unable to bind to a nucleotide when a kinesin monomer is pulled backward from its NL. We measured the effect of nucleotide binding to a kinesin head in both front and rear head orientations of the NL using the trap assay at saturating (1 mM) ADP. In head-pulled kinesins, addition of 1 mM ADP resulted in a ~1.5-fold increase in k1 and k2 under both positive and negative forces compared to the apo condition (Figure 2C). k1 and k2 were 72 s−1 and 7 s−1, respectively, at ±1.8 pN. The increase in both rates as a function of applied force was comparable to that of 1 mM ATP (Figure 2B). Remarkably, we observed a clear asymmetry in the release rates when kinesin was pulled from its NL. Under positive forces, both k1 and k2 were similar to those of head-pulled motors. However, k1 and k2 of NL-pulled motors were significantly lower than the corresponding rates of head-pulled motors over a wide range of negative forces.
To estimate the degree to which negative forces exerted on NL slow down the release rate upon ADP binding, we subtracted k1 and k2 values in 1 mM ADP from those of the apo condition (see Experimental Procedures, Figure 3). In contrast to the previously proposed enhancement of ADP binding affinity to kinesin under forward load (Uemura and Ishiwata, 2003), we observed that k1-ADP − k1-apo and k2-ADP − k2-apo were within 20% to those of the head-pulled condition when NL is pulled forward (Figure 3A and 3B). Therefore, increase in k1-ADP − k1-apo and k2-ADP − k2-apo under positive forces is due to faster release of the motors in the ADP-bound state under increased load, not to increased ADP binding affinity in the forward orientation of NL.
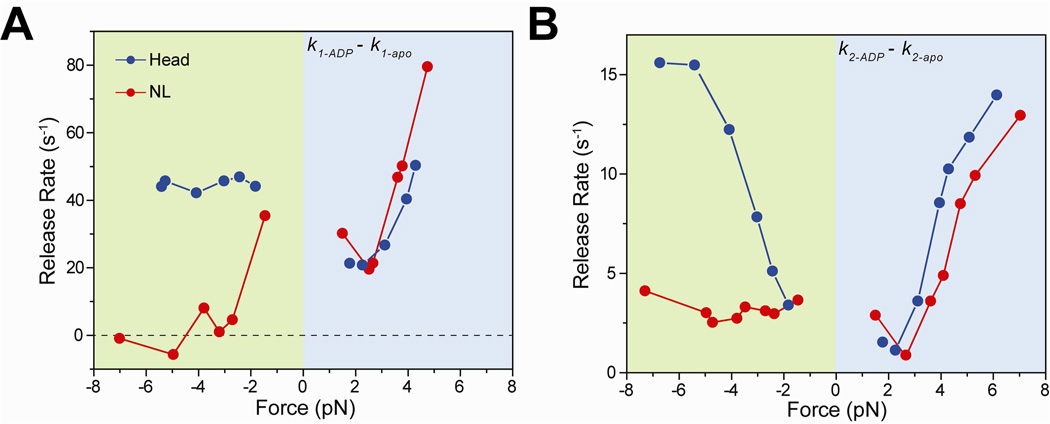
Figure 3. Nucleotide binding to a kinesin head is inhibited when the NL is oriented backward.
(A) k1 values of the apo condition were subtracted from that of 1 mM ADP to calculate the nucleotide-binding induced MT release rate from the weakly-bound state. k1-ADP − k1-apo of NL-pulled kinesins was 35 s−1 at −1.5 pN, and decreased to ~0 s−1 at higher negative forces. (B) k2 values of the apo condition were subtracted from that of 1 mM ADP to calculate the nucleotide-binding induced MT release rate from the strongly-bound state. k2-ADP − k2-apo of NL-pulled kinesins remained nearly constant at 3.0 s−1 under negative forces, whereas k2-ADP − k2-apo of head-pulled kinesins increased from 3.4 s−1 at −1.8 pN to 14.2 s−1 at −6.7 pN.
When the NL is pulled backward, nucleotide-dependent release of kinesin occurs at a significantly slower rate than when it is pulled from the head (Figure 3A and 3B). k1-ADP − k1-apo of NL-pulled kinesins was 35 s−1 at −1.5 pN and decreased abruptly to 0 ± 5 s−1 (mean ± sd) at higher negative forces. k1-ADP − k1-apo of head-pulled kinesins remained constant at 45 ± 2 s−1 under all negative forces. Furthermore, k2-ADP − k2-apo of NL-pulled kinesins remained largely constant at 3.0 ± 0.5 s−1 from −1.5 pN to −7.3 pN, whereas k2-ADP − k2-apo of head-pulled kinesins increased from 3.4 s−1 at −1.8 pN to 14.2 s−1 at −6.7 pN. We concluded that reduction in nucleotide-dependent MT release of NL-pulled kinesins is independent on the magnitude of tension from −1.5 to −7 pN.
DISCUSSION
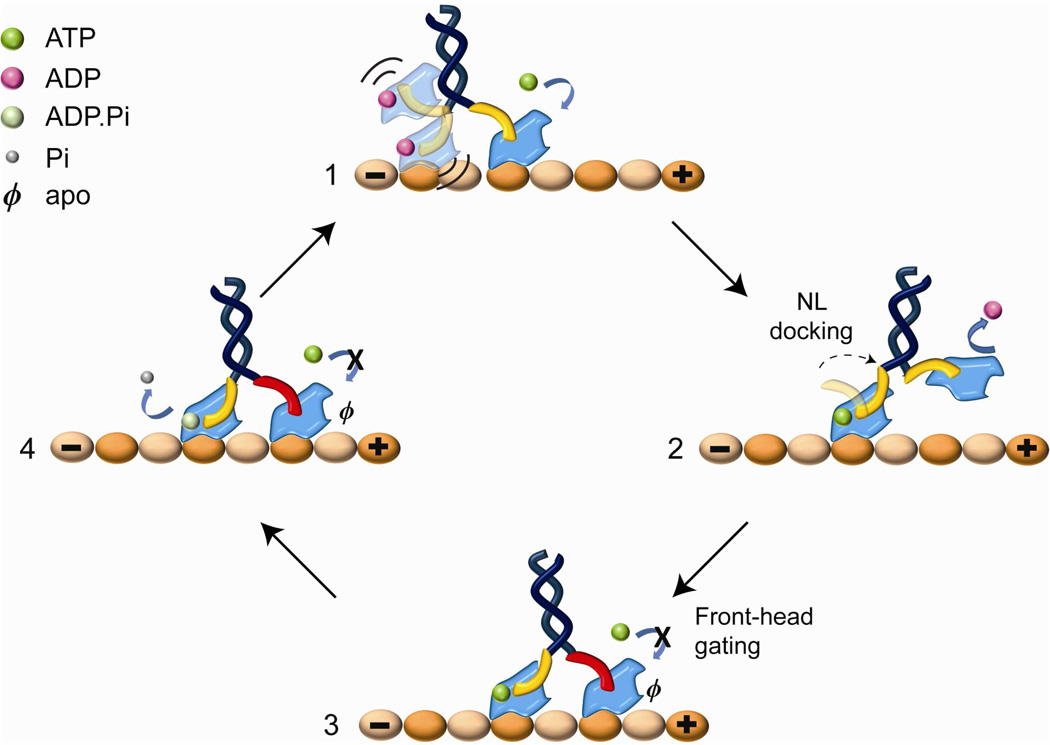
Our results strongly support the front-head gating model (Figure 4) for coordination of the processive motility of a kinesin dimer. When kinesin waits for an ATP molecule, the front head remains tightly attached and the rear head is either weakly interacting with or unbound from the MT. ATP binding to the front head triggers NL docking, moving the rear head towards its next tubulin binding site in the plus-end direction. Kinesin motility is gated when both heads are attached to the MT. In this state, the NL of the rear head orients forward and is free to dock, but the NL of the front head is restricted from docking because it is oriented backward by the trailing head. The rear head remains attached to the MT until it releases the phosphate, which triggers subsequent MT release (Milic et al., 2014). This is the rate-limiting step in kinesin’s ATPase cycle (Ma and Taylor, 1997) and the processivity would end prematurely if the front head were to bind and hydrolyze ATP during this process. As a result, the rear head hydrolyzes ATP and releases the inorganic phosphate while the front head remains strongly attached to the MT. Consistent with this scheme, our results showed that pulling NL in the backward direction greatly reduces the nucleotide-binding induced detachment rate of a head from MT. At high negative forces, k1-ADP − k1-apo of head-pulled motors were 4 fold faster than NL-pulled motors. In addition, k2-ADP − k2-apo of head-pulled kinesins was at 45 ± 2 s−1, while that of NL-pulled kinesins remained constant at 0 ± 5 s−1. The observed reduction in k1 agrees with the estimation that ATP unbinding to the front head is six fold faster than to the rear head of a walking kinesin dimer (Clancy et al., 2011).
Figure 4. Front-head gating model for kinesin.
(1) In the ATP waiting state, the rear head is ADP-bound and weakly interacting with MT. (2) ATP binding to the front head triggers NL docking that pulls the rear head forward. (3) The unbound head releases ADP and rebinds MT ahead of its partner head. (3–4) In 2HB state, the NL of the front head (red) is oriented backward and ATP binding to this head is inhibited until the rear head hydrolyzes ATP and releases from MT.
Kinesin heads experience up to 15 pN tension in a 2HB state (Hyeon and Onuchic, 2007). When tension between the heads is reduced by extending NLs, the rear head strongly binds to a MT in the ATP-waiting state (Clancy et al., 2011), and as a result, kinesin loses its ability to convert ATP hydrolysis to a mechanical step and undergoes futile cycles of ATP hydrolysis (Yildiz et al., 2008). While high intramolecular tension is crucial for the energetic efficiency of the kinesin motor, our results show that it is not critical for the interhead coordination. Nucleotide binding to a head can be inhibited at as low as 2 pN backward tension and independent of the magnitude of tension exerted on the NL (Figure 3). Based on this result, we propose that kinesin gating is facilitated by the backward orientation of NL of the front head. This gating mechanism does not require substantial tension between the heads and is mainly facilitated by restricting NL of the front head to orient backwards by the rear head in a 2HB state. Consistent with our model, kinesin maintains its gating mechanism with reduced tension on NLs (Clancy et al., 2011).
A possible clue for how ATP binding to the front head may be suppressed by the inability of its NL to dock onto the catalytic core comes from structural studies (Kikkawa et al., 2001; Rice et al., 1999; Sindelar and Downing, 2010). The kinesin motor domain contains two hydrophobic pockets on opposite sides, the “switch pocket” and the “docking pocket” that facilitate nucleotide binding and NL docking, respectively (Sindelar, 2011). ATP binding to the switch pocket triggers opening of the docking pocket and leads to NL docking. When the rear head is strongly attached to the MT, it prevents forward extension of the NL of the front head and occupation of its docking pocket. Under this conformation, the switch pocket remains closed and ATP binding to the front head is disfavored, because the nucleotide sensing loops cannot interact with γ-phosphate of ATP (Sindelar, 2011).
Our results have broader implications for understanding the communication of the heads of a dimeric motor during processive motility. The nucleotide hydrolysis of the catalytic core drives the conformational change of a mechanical element (referred to as a lever arm in myosins, a linker in dyneins, and an NL in kinesins). These mechanical elements sense intramolecular tension in a 2HB state. A tension sensing mechanism has been shown to affect nucleotide binding to myosin-V heads (Dunn et al., 2010) and inhibit ATP-dependent MT release of cytoplasmic dynein (Cleary et al., 2014). These observations suggest that tension sensing and asymmetric conformations of these structures in the front and rear heads of a walking dimer play a major role in achieving processivity.
EXPERIMENTAL PROCEDURES
Preparation and labeling of kinesin constructs
The hK349 construct contains the entire motor domain, NL and a short region of the neck coiled coil. To label the kinesin head with biotin maleimide, E215C mutation was inserted to a cysteine-light kinesin construct (hK349-E215C-CLM). To label the distal end of the NL, HaloTag (HT, a 26 kDa protein tag) was inserted to the C-terminus (hK349-HT). Kinesin monomers were expressed in E.coli, and purified by affinity chromatography.
To label hK349-E215C-CLM with DNA at the head, we used a 74 bp DNA tether, modified with biotin and a free amine at opposite ends. 40× excess amine to sulfyhydryl crosslinker (Sulfo-SMCC) was incubated with the DNA solution for 90 minutes at 37 °C. DNA-SMCC was reacted with hK349-E215C-CLM in 1:1 ratio for 2 hours at 4 °C. This surface-exposed residue was chosen for DNA attachment point because it is a solvent-exposed residue located at the back side of the motor domain and distal from regions known to be critical for motility, such as the NL, nucleotide binding cleft and MT binding surface. E215C mutation has no detectable effect on kinesin motility and force production (Mori et al., 2007; Tomishige et al., 2006; Yildiz et al., 2004, 2008). To label K349-HT monomers with DNA, the free amine at the 5’-end of the DNA tether was conjugated with a 40× excess HT-succinimidyl ester ligand at room temperature for 6 hours. The DNA-HT ligand was reacted with K349-HT for 5 hours at 4°C. Motors were purified by MT bind and release assay to remove excess DNA.
Optical trapping assay
DNA-labeled kinesin monomers were diluted in BRBC (BRB80 with 2.5 mg/ml casein, BRB80 = 80 mM PIPES pH 6.8, 1 mM MgCl2, 1 mM EGTA, 2 mM DTT) and incubated with 860 nm streptavidin-coated polystyrene beads. The residual ATP in kinesin solution was removed by pelleting and resuspending the beads in BRBC. To deplete the residual ATP, 0.5 U/ml apyrase was added for the apo condition, and 2 U/ml hexokinase and 0.4% glucose was added for the ADP condition.
Trapped beads were positioned over a Cy5-labeled axoneme, oscillated ±125 nm along the axoneme in a square wave pattern and held for 0.375 seconds in each position. Microtubule polarity was determined by Alexa488-labeled S. cerevisiae cytoplasmic dyneins, which decorate the minus-end of MTs. The traces of the bead and trap centers are recorded at 5 kHz. Because the bead was moved within the linear range of the trap (±150 nm), the force exerted on a motor during a binding event was calculated by multiplying trap stiffness (0.045 pN/nm) with bead-trap separation.
Data Analysis
MT binding events were determined by a custom step finding algorithm written in MATLAB (MathWorks) (Cleary et al., 2014). Rare (~10%) multiple-step release events and events showing a dwell time of less than 2.5 ms were discarded from the data analysis. Data consisting of applied force and dwell time were sorted by force and binned every 150–300 data points under different pulling geometries and nucleotide conditions. Cumulative distribution of force-induced MT release data were fitted to the sum of two exponentials in MATLAB. Because the forces in apo and ADP conditions do not perfectly match, subtraction of the rates at each force value was performed by linear interpolation between the adjacent data points in Origin (Origin Labs).
A complete and comprehensive list of the methods, reagents, and statistics are described in Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank V. Belyy, S. Wichner and A. Majumdar for helpful discussions and R. D. Vale for providing plasmids. This work has been supported by NIH (GM094522 (AY)), NSF CAREER Award (MCB-1055017 (AY)) and NSF Graduate Research Fellowship (DGE 1106400 (FBC)).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes supplemental experimental procedures and four figures, and can be found with this article online.
AUTHOR CONTRIBUTIONS
Y.S.D., S.C., F.B.C. and A.Y. designed experiments, V.P. cloned and purified the protein, Y.S.D. and S.C. performed optical trapping assays, and Y.S.D., S.C. and A.Y. wrote the manuscript.
REFERENCES
- Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asenjo AB, Weinberg Y, Sosa H. Nucleotide binding and hydrolysis induces a disorder-order transition in the kinesin neck-linker region. Nat Struct Mol Biol. 2006;13:648–654. doi: 10.1038/nsmb1109. [DOI] [PubMed] [Google Scholar]
- Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy BE, Behnke-Parks WM, Andreasson JO, Rosenfeld SS, Block SM. A universal pathway for kinesin stepping. Nat Struct Mol Biol. 2011;18:1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary FB, Dewitt MA, Bilyard T, Htet ZM, Belyy V, Chan DD, Chang AY, Yildiz A. Tension on the linker gates the ATP-dependent release of dynein from microtubules. Nat Commun. 2014;5(4587) doi: 10.1038/ncomms5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevel IM, Nyitrai M, Alonso MC, Weiss S, Geeves MA, Cross RA. What kinesin does at roadblocks: the coordination mechanism for molecular walking. The EMBO journal. 2004;23:23–32. doi: 10.1038/sj.emboj.7600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AR, Chuan P, Bryant Z, Spudich JA. Contribution of the myosin VI tail domain to processive stepping and intramolecular tension sensing. Proc Natl Acad Sci U S A. 2010;107:7746–7750. doi: 10.1073/pnas.1002430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Block SM. Backsteps induced by nucleotide analogs suggest the front head of kinesin is gated by strain. Proc Natl Acad Sci U S A. 2006;103:8054–8059. doi: 10.1073/pnas.0600931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Block SM. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009;461:125–128. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD. The rate-limiting step in microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains occurs while bound to the microtubule. J Biol Chem. 1994;269:16508–16511. [PubMed] [Google Scholar]
- Hancock WO, Howard J. Kinesin's processivity results from mechanical and chemical coordination between the ATP hydrolysis cycles of the two motor domains. Proc Natl Acad Sci U S A. 1999;96:13147–13152. doi: 10.1073/pnas.96.23.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Hua W, Young EC, Fleming ML, Gelles J. Coupling of kinesin steps to ATP hydrolysis. Nature. 1997;388:390–393. doi: 10.1038/41118. [DOI] [PubMed] [Google Scholar]
- Hyeon C, Onuchic JN. Internal strain regulates the nucleotide binding site of the kinesin leading head. Proc Natl Acad Sci U S A. 2007;104:2175–2180. doi: 10.1073/pnas.0610939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaseda K, Higuchi H, Hirose K. Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat Cell Biol. 2003;5:1079–1082. doi: 10.1038/ncb1067. [DOI] [PubMed] [Google Scholar]
- Kawaguchi K, Ishiwata S. Nucleotide-dependent single- to double-headed binding of kinesin. Science. 2001;291:667–669. doi: 10.1126/science.291.5504.667. [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Sablin EP, Okada Y, Yajima H, Fletterick RJ, Hirokawa N. Switch-based mechanism of kinesin motors. Nature. 2001;411:439–445. doi: 10.1038/35078000. [DOI] [PubMed] [Google Scholar]
- Klumpp LM, Hoenger A, Gilbert SP. Kinesin's second step. Proc Natl Acad Sci U S A. 2004;101:3444–3449. doi: 10.1073/pnas.0307691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YZ, Taylor EW. Interacting head mechanism of microtubule-kinesin ATPase. J Biol Chem. 1997;272:724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- Milic B, Andreasson JO, Hancock WO, Block SM. Kinesin processivity is gated by phosphate release. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1410943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM. Stepping and stretching. How kinesin uses internal strain to walk processively. J Biol Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld SS, Jefferson GM, King PH. ATP reorients the neck linker of kinesin in two sequential steps. J Biol Chem. 2001;276:40167–40174. doi: 10.1074/jbc.M103899200. [DOI] [PubMed] [Google Scholar]
- Schief WR, Clark RH, Crevenna AH, Howard J. Inhibition of kinesin motility by ADP and phosphate supports a hand-over-hand mechanism. Proc Natl Acad Sci U S A. 2004;101:1183–1188. doi: 10.1073/pnas.0304369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- Shastry S, Hancock WO. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr Biol. 2010;20:939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar CV. A seesaw model for intermolecular gating in the kinesin motor protein. Biophysical reviews. 2011;3:85–100. doi: 10.1007/s12551-011-0049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar CV, Downing KH. An atomic-level mechanism for activation of the kinesin molecular motors. Proc Natl Acad Sci U S A. 2010;107:4111–4116. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- Toprak E, Yildiz A, Hoffman MT, Rosenfeld SS, Selvin PR. Why kinesin is so processive. Proc Natl Acad Sci U S A. 2009;106:12717–12722. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Ishiwata S. Loading direction regulates the affinity of ADP for kinesin. Nat Struct Biol. 2003;10:308–311. doi: 10.1038/nsb911. [DOI] [PubMed] [Google Scholar]
- Uemura S, Kawaguchi K, Yajima J, Edamatsu M, Toyoshima YY, Ishiwata S. Kinesin-microtubule binding depends on both nucleotide state and loading direction. Proc Natl Acad Sci U S A. 2002;99:5977–5981. doi: 10.1073/pnas.092546199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.