Abstract
FIB-SEM has become an essential tool for studying neural tissue at resolutions below 10×10×10 nm, producing datasets superior for automatic connectome tracing. We present a technical advance, ultrathick sectioning, which reliably subdivides embedded tissue samples into chunks (20 µm thick) optimally sized and mounted for efficient, parallel FIB-SEM imaging. These chunks are imaged separately and then ‘volume stitched’ back together, producing a final 3D dataset suitable for connectome tracing.
Today’s scanning electron microscopes (SEMs) achieve ~2nm spot diameters at landing energies low enough that only the top few nanometers of the block’s surface contributes significantly to the acquired image. In Focused Ion Beam SEM (FIB-SEM)1,2, this is combined with the FIB’s ability to polish (mill) away a few nanometers at a time. Repeating this cycle produces a sequence of images representing 3D ultrastructure and gives this technique a “section” (z) resolution significantly better3 than serial section techniques (e.g. ssTEM4,5 and ATUM-SEM6) and serial block EM (SBEM7).
Physically cutting ultrathin sections requires a knife sharpened (and positioned in height relative to the surface) to much less than the section thickness. In contrast, because ion milling is akin to sand blasting, FIB-SEM uses a “soft knife” whose diameter can be over a hundred times the desired surface removal thickness. The beam’s positioning accuracy is similarly less constrained and can be held constant relative to the surface simply by adjusting deflection voltages. In our lab, we have designed custom FIB-SEMs taking advantage of these characteristics, allowing us to operate our FIB-SEMs mostly unattended for months at a time.
However, the FIB-SEM technique has an “Achilles’ heel” –ion polishing accumulates artifacts when milling blocks longer than a few tens of microns in the direction of the ion beam. Artifacts take the form of streaks and waves of thickness variation. In our hands, imaging regions longer than ~100 µm results in unacceptable artifacts, thus representing a fundamental barrier to applying FIB-SEM to the field of connectomics8,9.
A solution would be a technique which can divide tissue into smaller chunks (Fig. 1a) –each large enough to be cut and handled reliably but small enough for FIB-SEM. Such subdivision must be virtually lossless so that imaged chunks can be stitched back together into a single dataset. We refer to such sections as “ultrathick”. An ultrathick series contains sections which are typically 20 µm or thicker so they are efficiently loadable into FIB-SEM (or SBEM), but whose surfaces are cut so cleanly that ultrastructural details remain traceable across the cuts.
Figure 1.
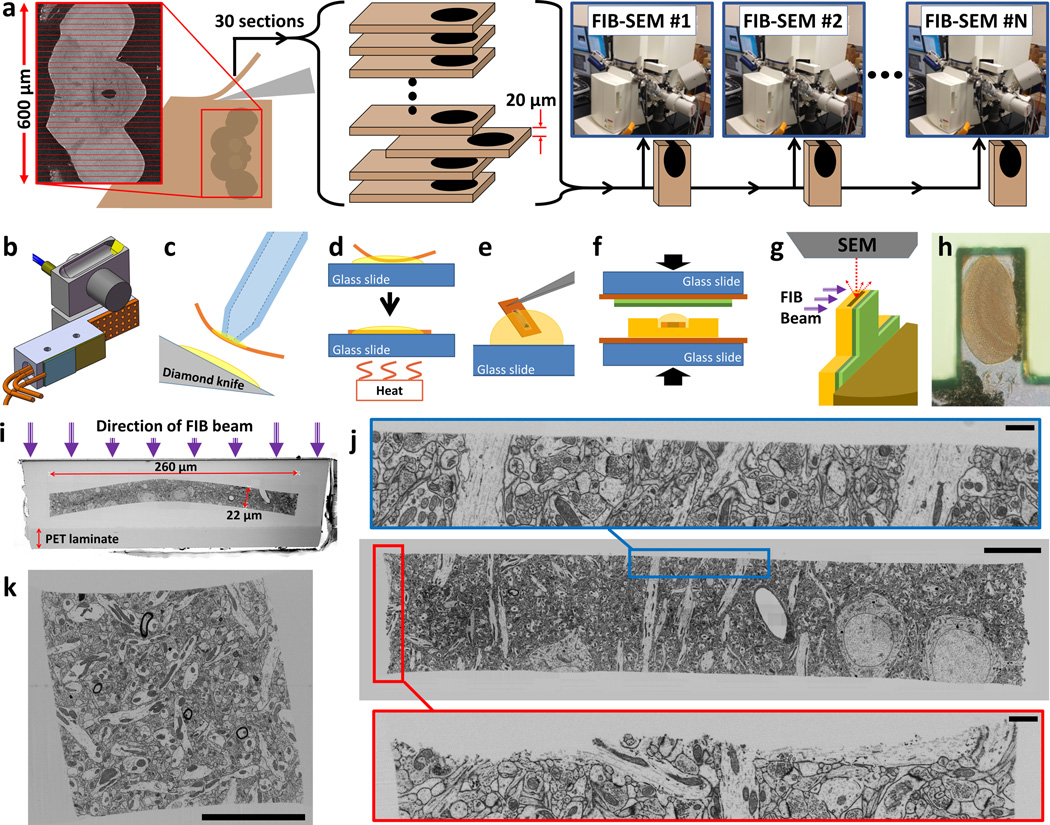
Overview of ultrathick sectioning. (a) Tissue volumes with dimensions too large for FIB-SEM can be “ultrathick” sectioned into optimally sized chunks and imaged across multiple FIB-SEMs. Depicted is our plan for imaging an entire adult Drosophila brain. (b) Heating jig designed for use with the Diatome ultrasonic knife. More typically we use our custom designed “Ultrathick Sectioning Testbed” (Supplementary Fig. 4). (c-g) Steps involved in preparing ultrathick sections for FIB-SEM imaging. Oil covered sections are retrieved using vacuum tweezers (c), and hot plate flattened (d). Oil is removed by dipping each section in a succession of Durcupan drops (e). Sections are flat embedded in Durcupan against a PET laminate (green layers) (f). Sections are cut out and adhered to metal studs. Each tab is UV laser and diamond trimmed and conductively coated readying it for FIB-SEM imaging (g). (h) Light micrograph of a tab containing a 20 µm ultrathick section through the fly medulla. (i) SEM image of top surface of a sample tab. (j) FIB-SEM image of a 20 µm ultrathick section of mouse cortex cut from an original 100 µm vibratome section (scale bar 10 µm for main image, 1 µm for insets). Note smooth hot knife cut edge (blue inset) relative to original vibratome cut (red inset). (k) Ultrathick sectioning is not limited to one dimension. Here a 20 µm ultrathick section of mouse cortex has been re-embedded and sliced again orthogonal to the original cut plane. This creates a ‘pillar’ with a square cross section. All edges are smooth including sharp corners of the square, in principle allowing for volume stitching with neighboring pillars (scale bar 10 µm).
We first considered vibratomy of unembedded tissue but this results in unacceptable damage to the cut surfaces. To avoid this damage, ultrathin sectioning relies on tissue infiltrated with hard resin10 to hold the ultrastructure in place. However, when attempting to section a hard block at 20 µm, sections crumble. As long ago as 1967, McGee-Russell and Gosztonyi11,12 and separately West13 developed a solution. By superficially heating the block’s surface, it is locally softened so sections thicker than 10 µm can be cut. These researchers showed that such sections can be re-sectioned for serial electron microscopy; however, they did not address surface damage which, since they used metal blades, was likely considerable.
To assess what quality of surface could be obtained if “hot knife microtomy” was upgraded with a diamond knife, we built a knife heater and cut 20–30 µm sections. We found 60°C worked best. At colder temperatures curling occurs, while sections distort at higher temperatures. An immediate problem was that water, which provides lubrication in traditional ultrathin sectioning, was ineffective for thick sectioning. SEM images of cut faces showed considerable damage (Supplementary Fig. 1). Using oil for knife lubrication dramatically reduced stick-slip, however it did not eliminate it. Ultrasonic vibration has been shown to reduce compression14, and we thought it might help maintain lubrication, so we switched to using a heated ultrasonic knife (Fig. 1b). This eliminated stick-slip and distortions. We acquired SEM images of the surfaces of two matched 20 µm thick sections (Supplementary Figs. 2 and 3). None of the previous damage was visible and matching neuronal processes were readily traced across the cut faces. We found the (Fig. 1b) jig inadequate for in-depth research, so we built a custom “Ultrathick Sectioning Testbed” (Supplementary Fig. 4) which includes the ability to record top and side video microscope views during sectioning (Supplementary Video 1).
Additional steps are required to prepare ultrathick sections for FIB-SEM imaging (Fig. 1c-i). We acquired 3D FIB-SEM stacks of ultrathick sections to evaluate cut quality (Fig. 1j,k and upplementary Videos 2, 3 and 7). From these videos it is clear that the cut surfaces are exceptionally smooth but not perfectly flat. Small undulations remain due to local differences in rigidity. We developed algorithms to computationally flatten these surfaces so that sequential sections could be “volume stitched” (Supplementary Figs. 5–7). First, coordinates of the cut edge are found in each image individually, then a 3D surface is fit representing the cut boundary. This is used to “flatten” the edge in each image up to the same vertical position. This results in a stack where the cut surface resides in a single plane.
The flattened surface images from two sequential ultrathick sections should match since they are two sides of the same physical cut. However these images are expected to be distorted relative to one another due to distortions during sectioning and embedding. An image transformation is found that will align one to the other. The two stacks can now simply be joined at their flattened surfaces resulting in a final 3D volume ready for tracing.
To test the full procedure, two sequential 20 µm ultrathick sections, cut from a Drosophila brain prepared via high-pressure freezing/freeze-substitution (HPF-FS), were FIB-SEM imaged and volume stitched (Supplementary Fig. 8). Each was imaged with 8×8×8nm voxels with a field of view encompassing the entire section (~120×32 µm) over a milling depth sufficient to encompass the entire lobula region (~80 µm). These large datasets (>1 TB) provided us the opportunity to survey thousands of square microns of matched cut area for significant damage. Since this volume of data overwhelms our current volume stitching tools, the dataset was collapsed to 32×32×32 nm voxels then volume stitched (Supplementary Videos 4 and 5). A smaller dataset was stitched at full resolution (Supplementary Video 6).
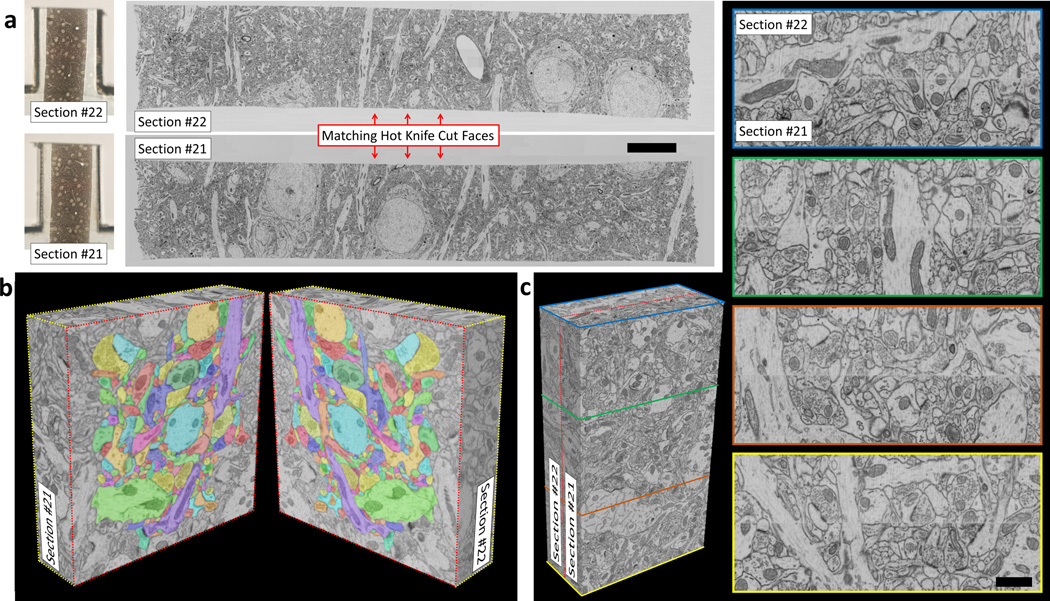
In a separate test, two sequential 20 µm ultrathick sections cut from a 100 µm thick vibratome slice of mouse cortex were FIB-SEM imaged and volume stitched (Fig. 2a). Datasets were acquired (10×10×10 nm voxels) for matching edge regions of the two sections. A volume stitch over a 9×16 µm cropped region was performed (Fig. 2c). Matching neuronal processes are easily identified across the stitch as was verified by tracing 200 densely-packed processes across the stitch (Fig. 2b and Supplementary Videos 7–9).
Figure 2.
Volume stitching results on mouse cortex tissue. (a) FIB-SEM images of two sequential 20 µm ultrathick sections (#21, #22) cut from a 100 µm thick vibratome slice. Images show quality of cut surfaces (see Supplementary Video 7) (scale bar 10 µm). Also shown are light micrographs of each corresponding sample tab. (b) Graphical depiction of an 8×8 µm cropped region which has been split open along the stitch plane. All processes in the densely packed central region (total of 200) were traced across the gap (denoted by colored outlines, Supplementary Video 9). (c) Volume stitching results over a 9×16 µm cropped region. The dashed red line on the 3D volume shows the stitch plane. Images with colored borders correspond to cut planes through this 3D stitched volume (Supplementary Video 8) (scale bar 1 µm).
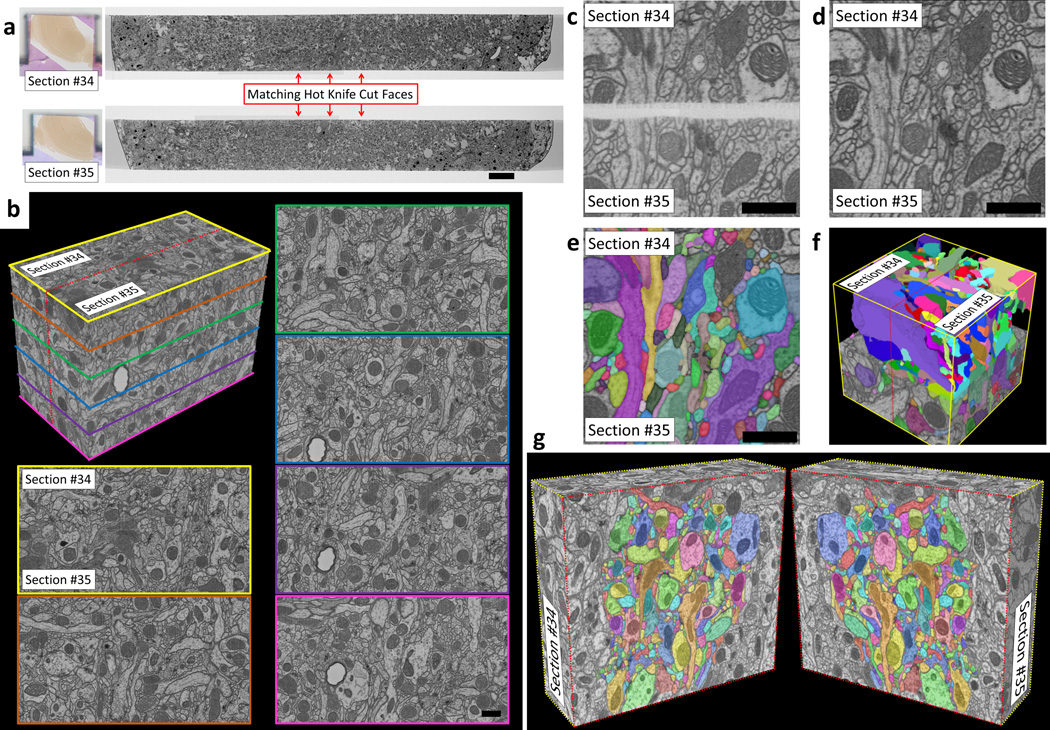
In another test, two sequential 20 µm ultrathick sections cut from a C-PLT (Chemical/ Progressive Lowering of Temperature) prepared Drosophila brain were FIB-SEM imaged and volume stitched (Fig. 3a). The C-PLT process provides high-contrast synaptic features (Supplementary Fig. 9). FIB-SEM datasets were acquired (8×8×8 nm voxels) for matching edge regions of the two sections. A volume stitch over an 8×12 µm region was performed (Fig. 3b-f). Matching neuronal processes are easily identified across the stitch. Processes in Drosophila reach smaller diameters than in mammalian cortex. Given this we expected tracing across the hot knife stitch would be more difficult. We attempted to trace 406 densely-packed processes across the stitch (Fig. 3g). The results of two independent tracers were compared. A total of 8 processes were deemed to be sufficiently ambiguous to be considered “untraceable” by one or both tracers, implying a 98% traceability rate (Supplementary Videos 10–12).
Figure 3.
Volume stitching results on C-PLT prepared fly brain. (a) FIB-SEM images of two sequential 20 µm ultrathick sections (#34 and #35) showing quality of cut surfaces (scale bar 10 µm). (b) Volume stitching results over an 8×12 µm cropped region. The dashed red line on the 3D volume shows the stitch plane. Images with colored borders correspond to cut planes through this 3D stitched volume (Supplementary Video 10) (scale bar 1 µm). (c-d) Side-by-side comparison of aligned hot knife cut edges before (c) and after (d) algorithmic flattening and stitching (scale bars 1 µm). These are slices taken out of a 4×4×4 µm volume which was also traced (e-f). Supplementary Video 12 contains side-by-side movies of all slices of this same volume both before and after stitching as well as after volume tracing. (g) Graphical depiction of a 9×10 µm cropped region which has been split open along the stitch plane. All processes in the densely packed central region were traced across the gap (Supplementary Video 11).
Despite the fact that tracing was possible, it was clear during tracing that some material had been lost between the two matching faces. This is most readily seen when the volume is digitally re-sliced parallel to the stitch. This material loss, combined with residual distortions due to imperfect alignment and ambiguities in the flattening algorithm, adds up to a “jump” that appears visually similar to the loss of one traditional TEM section. By comparing the image correlation “jump” across the hot knife stitch to “virtual” gaps of various amounts we quantified this loss to be ~30nm (Supplementary Figs. 10 and 11). Impacts for the tracing of small neurites are presented (Supplementary Figs. 12 and 13).
Given an optimally prepared sample the ultrathick sectioning process is very reliable. Since sections are so thick, even blocks >>1 mm3 can be reduced to FIB-SEM tabs quickly. We have, however, run into some limitations with particular tissue samples (see Online Methods). In general, tissue must be well fixed and infiltrated, but not overly brittle, for quality ultrathick sectioning.
The volume stitched datasets presented demonstrate that it is possible to subdivide a block of brain tissue into smaller chunks optimally sized for efficient, artifact-free FIB-SEM imaging while retaining the ability to trace neuronal processes. This subdivision is not limited to one dimension. Re-embedding ultrathick sections and cutting them in an orthogonal direction can reduce a large block to square “pillars” or even cubes if desired (Figure 1k and Supplementary Fig. 14). This opens up a range of possible connectomics studies by allowing FIB-SEM’s excellent 3D resolution to be applied to arbitrarily large volumes of brain tissue.
Online Methods
Sample preparation
Drosophila larva (Supplementary Figs. 5a-b, 6 and 7): Brains from wandering 3rd instar larvae of [iso] Canton S G1×w1118 [iso] 5905 flies were obtained by dissection from filleted preps on Sylgard (Dow Corning) plates in PBS and transferred to 2% glutaraldehyde in 0.1 M Na-cacodylate buffer, pH 7.4 fixative for 1 hour at room temp. After rinsing in 0.1 M Na-cacodylate buffer, the samples were post-fixed with 1% OsO4 in 0.1 M Na-cacodylate buffer, rinsed in buffer followed by distilled water, and stained en bloc overnight with 1% aqueous uranyl acetate in a refrigerator. The samples were rinsed in water, dehydrated in an ethanol series followed by propylene oxide, and then infiltrated and embedded in Eponate 12 resin (Ted Pella).
Adult Drosophila brain prepared via HPF-FS fixation (Supplementary Fig. 8): High-pressure freezing/freeze-substitution (HPF-FS) fly brain tissues were vibratome sliced (190 µm) from five-day-old female adult Oregon R wild-type Drosophila heads. After 5 minutes immersion in modified Karnovsky’s fixative15, the slices were cryofixed by Wohlwend Compact 01 HPF machine then processed in Leica EM AFS2 freeze-substitution system. The tissues were warmed from −140 °C to −90 °C at 50 °C/h and freeze-substituted in acetone based cocktail (containing 1% Osmium tetroxide, 0.1% uranyl acetate, 1% methanol and 5% water) at −90 °C for 36 hours. Up to 5% water was added to enhance membrane contrast16. Next, the tissues warmed up to −20 °C at 2 °C/h and keep at −20 °C for 26 h, then brought to 0 °C at 4 °C/h. Following FS, tissues were rinsed in pure acetone, infiltrated and embedded in Poly/Bed 812 (Luft formulations).
Mouse cortex (Fig. 1i-k and Fig. 2): All experimental protocols were conducted according to US National Institutes of Health guidelines for animal research and were approved by the Institutional Animal Care and Use Committee at Janelia Research Campus. A male, adult (3 months old) C57/bl6 mouse was deeply anaesthetised with an intraperitoneal injection of sodium pentobarbitone (30 mg/kg body weight), and immediately perfused with glutaraldehyde (2.5%) and paraformaldehyde (2%) in phosphate buffer (0.1 M, pH 7.4). After two hours, at room temperature, the brain was removed, mounted in 4 % agarose, and coronal slices cut through the cortex with a vibratome (Leica Microsystems VT1200). The slices were washed in cacodylate buffer (0.1M), postfixed with 1.5% potassium ferrocyanide and 1% osmium tetroxide (1 hr), then with 1% osmium tetroxide alone (1 hr), and finally in 1% aqueous uranyl acetate (1 hr). After washing in double distilled water, they were dehydrated with increasing concentrations of ethanol, embedded in Durcupan resin, and hardened at 65 °C for 24 hours, sandwiched between glass microscope slides.
Adult Drosophila brain prepared via C-PLT fixation (Fig. 3): Chemical/ Progressive Lowering of Temperature (C-PLT) dehydration/ fixation is a modified conventional chemical fixation method. Good tissue preservation and enhanced contrast are achieved (Supplementary Fig. 9). Five-day-old female adult Canton S wild-type flies of the Drosophila were used in this experiment. Isolated brain tissues were prefixed in 2.5% formaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer at ph 7.4 for 2 h at RT. After wash, the tissues were postfixed in 0.5% osmium tetroxide in ddH2O for 20 minutes at 4 °C. The PLT procedure started from 0 °C when the tissues were transferred into 10% acetone. Temperature progressively decreased to −25 °C by acetone concentration gradually increased to 97%. The tissue fixed in 1% Osmium tetroxide, 0.2% uranyl acetate in acetone for 42 hours at −25 °C. After PLT dehydration/ fixation, temperature rose back and tissues were rinsed in pure acetone, infiltrated and embedded in Poly/Bed 812 (Luft formulations).
Ultrathick sectioning
Sample blocks targeted for ultrathick sectioning were first mounted in a standard ultramicrotome tissue chuck (Leica) and trimmed on an UltraCut E ultramicromtome (Reichert-Jung) using a straight-sided diamond ultratrim knife (Diatome). Blocks are trimmed to have a rectangular face approximately 1.5 mm wide and 3 mm long with the tissue centered in that face (Supplementary Fig. 4b). This is to provide a large enough section with ample blank regions for manipulation with vacuum tweezers and forceps. Three of the sides are trimmed straight into the block the full depth of the tissue (which can be >1 mm). The side of the block which will be the trailing edge during hot knife sectioning is given a slope (~45°). If this is not done, we found that this trailing edge would bend out during hot knife sectioning leading to problems.
Sample blocks containing the larva and the adult HPF-FS Drosophila brain were ultrathick sectioned directly on the same UltraCut E using a 35° wedge angle ultrasonic diamond knife and heating jig (Fig. 1b and Supplementary Fig. 2a). This heating jig is machined of a single block of aluminum and fitted with a 120V cartridge heater (CSH-1011001/120V, Omega) and a surface thermocouple (SA1XL-J-SRTC, Omega). These are wired to a benchtop PID temperature controller (CSC32, Omega). Because of heat loss, the knife temperature is always 10–20°C lower than the set point temperature of the heating jig, therefor we always record the temperature of the knife itself by immersing the temperature probe of a digital multimeter (Fluke 179/EFSP) in the oil covering the knife.
Cutting parameters for the adult Drosophila brain prepared via HPF-FS (Supplementary Fig. 8): The ultrasonic knife was tuned to a side-to-side resonance at 31.8 kHz by monitoring the knife’s side with the laser vibrometer (CLV-2534, Polytec). Velocity measurement of the vibrometer was used to calculate the side-to-side motion of the knife to be ~1 µm peak-to-peak. The knife boat was filled with oil (thread cutting oil, Master Plumber) and drops of oil were applied to the knife periodically to maintain lubrication on front and back surfaces of the knife. Cutting speed of the microtome was set to 0.1mm/s. We always take test cuts from a block and adjust temperature of the knife as needed to produce smoothly flowing sections with minimal curl. The temperature needed can vary dramatically with the embedding media. For this run a knife temperature of 54 °C was used. A total of 30 sections (each 20 µm thick) were cut covering all fly tissue in the block. Each section was retrieved from the bottom of the knife boat using metal forceps (we had not yet introduced the vacuum tweezers retrieval method which we now consider superior) and transferred to individual wells in a plastic well plate.
Following trimming on the UltraCut E, sample blocks containing mouse cortex tissue (Fig.1i-k and Fig. 2) and the Adult Drosophila brain prepared via C-PLT fixation (Fig. 3) were transferred (still mounted in tissue chuck) to the Ultrathick Sectioning Testbed (Supplementary Fig. 4) which is designed to accept the same sample chucks as standard ultramicrotomes. Sectioning on the testbed used a 25° wedge angle ‘cryo’ diamond knife (Diatome) (Supplementary Fig. 4d). A drop of oil was applied to the knife prior to cutting each section to maintain lubrication on front and back surfaces of the knife. Supplementary Video 1 shows top and side video microscope views of sectioning and collection process.
Cutting parameters for mouse cortex tissue (Fig.1i-k and Fig. 2): The ultrasonic knife was tuned to a side-to-side resonance at 29.7 kHz (~0.5 µm peak-to-peak motion). Cutting speed = 0.1 mm/s, knife temperature = 60 °C. Sections were removed from the surface of the knife by vacuum tweezers. A total of 24 sections were cut (each 20 µm thick, 16 of which contained tissue) and collected in a plastic well plate.
Cutting parameters for adult Drosophila brain prepared via C-PLT (Fig. 3): The ultrasonic knife was tuned to a side-to-side resonance at 29.9 kHz (~0.5 µm peak-to-peak motion). Cutting speed = 0.1 mm/s, knife temperature = 59 °C. Sections were retrieved by vacuum tweezers. A total of 39 sections were cut (each 20 µm thick) 26 of which contained tissue spanning the entire fly brain. These were collected in a plastic well plate.
Re-embedding and orthogonal “pillar” cutting test (Fig. 1k): To test the idea of second order subdivision (i.e. pillars and cubes, see Supplementary Fig. 14) we used two of the 20 µm ultrathick sections of Durcupan-embedded mouse cortex tissue cut in the same run as those used in Fig. 2. These sections were then cleaned of oil and re-embedded (separated by a blank Epon spacer) in a hole drilled into the top of a blank Epon block oriented such that subsequent sectioning of these re-embedded 20 µm ultrathick sections would occur in a plane orthogonal to their original sectioning plane. After curing, the block was trimmed and another series of 20 µm ultrathick sections were cut from this new block. These new ultrathick sections contained little tissue “pillars” with square cross sections 20×20 µm wide. These were prepared for FIB-SEM imaging by the method described in Fig. 1c-g (and below) so that a pillar’s four smooth, hot knife cut sides would all show up in FIB-SEM cross section. Fig. 1k shows one image from a FIB-SEM acquired stack of such a tissue pillar.
Known Limitations
Samples prepared with the (R)OTO method17 to enhance osmium concentration tend to crack during sectioning presumably because the increased density of heavy metal makes them brittle. This is a significant limitation since such enhancement is often used in connectomics18. Also, a densely stained block may section fine at 20 µm but crack when cut thicker. We have experimented with post-staining of ultrathick sections using the hot ethanolic technique19 and found that contrast can be significantly increased throughout the full depth (20 µm) of an otherwise lightly-stained section (Supplementary Fig. 15). It is unclear however whether such post-staining can be used to increase contrast in tissue that has already been well prepared with a protocol designed for use with block face imaging like the mouse cortex tissue shown in Fig. 1i-k and Fig 2.
Fly samples prepared using HPF-FS tend to rip at epithelial sheath boundaries under the stress of sectioning, damaging nearby tissue.
The preferred resin for FIB-SEM imaging is Durcupan (since it dramatically reduces streak artifacts), however we have found it difficult to cut relative to Epon so we typically have used Epon embedded tissue for ultrathick sectioning tests. Since these Epon-infiltrated ultrathick sections are eventually flat embedded in Durcupan (Fig. 1f) streaking is minimized but not eliminated. When we must cut Durcupan embedded tissue our solution has been to re-embed the Durcupan-infiltrated tissue in surrounding Epon (tested in Fig. 2) prior to hot knife sectioning –providing improved rigidity during cutting. Since these sections are eventually flat embedded in Durcupan, the resulting FIB-SEM tabs present only Durcupan to the ion beam.
In general, tissue must be well fixed and infiltrated, but not overly brittle, for quality ultrathick sectioning.
Mounting for FIB-SEM imaging
The ultrathick sections collected in plastic well plates are still covered in cutting oil and retain some curl following hot knife sectioning. Under a dissection microscope, individual sections are transferred, using fine tipped forceps, to the surface of a glass slide. Care is taken to ensure that there is still a layer of oil between the section and the glass slide, if not more oil is added. The glass slide is then placed on the top of a hot plate set to 200°C. As the slide heats, the ultrathick section flattens in just a few seconds. We wait 5–10 s after this flattening and then remove the glass slide from the hot plate. Flattened sections from an entire run are arranged in order on the glass slide and then light microscope imaged for later reference.
Removing the cutting oil from sections is done just prior to flat embedding. Uncured Durcupan resin (Durcupan ACM Epoxy Resin Kit (Not water soluble), SPI supplies) is prepared (component formulation: A 11.4 g, B 10.0 g, C 0.3 g, D 0.05–0.10 g) and degassed under vacuum. Five large drops (several millimeters in diameter) of uncured Durcupan are placed on a glass slide under a dissection microscope. A flattened ultrathick section still covered with oil is grasped on a blank corner region with metal forceps and manually dipped repeatedly (~10 times) in the first Durcupan drop. Oil will be seen to disperse into the uncured resin. The top and bottom surfaces of the section are then gently touched to a dry area of the glass slide to help remove excess resin prior to dipping in the next drop. This process is repeated for all five resin drops, then the section (now cleaned of oil) is placed in the groove of a “groove mold” (previously cured out of Durcupan as well, see Fig. 1f and below) in which it will be flat embedded. This process is repeated for all sections of a run, with fresh Durcupan drops being prepared every 3 sections.
Sections from an entire run are flat embedded side-by-side in a pre-cured Durcupan ‘groove’ mold against a PET laminate. The objective is to ensure that, in the final FIB-SEM sample tabs, the embedded ultrathick sections are surrounded by a buffer zone of Durcupan on the side of the section facing the ion beam. This buffer zone acts like the front masking material (e.g. platinum) used by other FIB-SEM researchers, helping to prevent milling streaks, and ensures that SEM images will include blank resin zones surrounding the embedded tissue. One way to ensure such a buffer zone is to pre-cure a flat block of blank Durcupan ~100 µm thick, having a ~50 µm deep groove in it (see Fig. 1f).
Making of ‘groove’ mold: We have found that Kapton (polyimide) film provides excellent release properties for molding Durcupan. To make the groove mold we first adhere a 25 mm wide, 25 µm thick strip of Kapton film (CS Hyde) to a glass slide making the base of a mold-making-mold that will be used to make the groove mold. We then cut 2mm wide strips of Kapton adhesive tape (silicone adhesive coated, CS Hyde) and stack these to build up ~100 µm tall walls on the base to serve as the sides of the mold-making-mold. Then a ~50 µm thick strip of adhesive Kapton tape is laid down the center of the base. This mold-making-mold is then filled with Durcupan and flat pressed, in a clamp made from optical cage mounts (Thorlabs), against a top glass slide which is also covered with a strip of Kapton film. The clamp is placed in a 60 °C oven for 24–48 h to cure. One glass slide is removed and Kapton pieces are peeled away to expose the empty groove of the groove mold.
After being cleaned of oil, each section is placed in the bottom of the groove. Since they are covered with uncured Durcupan resin, they are readily arranged and oriented as necessary by manipulation with forceps. Once all sections are placed in the groove it is placed (with no top covering) in a 60 °C oven for ~2 h to partially cure and “tack” the sections lightly in position in the groove. This prevents the sections from shifting and overlapping each other during final flat embedding. A 20 mm wide, 25 µm thick strip of PET film (PP24I, Polymex Clear one side heat sealable one side untreated polyester film, Polyester Converter Ltd.) is then lightly adhered to a glass slide acting as the top of the flat embedding clamp. The PET film is arranged so that its heat-sealable side is facing toward the tissue (resulting in a strong bond). The groove is filled with fresh Durcupan and the arrangement clamped (Fig. 1f) and put in a 60 °C oven for curing for 48 h.
After curing, the flat embedding clamp is removed and Kapton release film peeled away revealing a sturdy slab containing an entire run of ultrathick sections. Individual sections are cut out (using a scalpel) and the resulting tabs adhered to custom metal studs using instant adhesive. A UV laser (LaserMill, New Wave Research) is used to trim each tab to the tissue dimensions. If no part of the tissue extends to the edge of the tab, then we will sometimes drill a small (40×40 µm) laser “via” hole to expose part of the tissue to the surface of the tab. This is done to allow SEM imaging charges to drain away more effectively once the tab’s surface is conductively coated. The sample tab is then mounted on an ultramicrotome and its top surface only is trimmed using a diamond trimming knife (cryotrim20, Diatome). This trimming must be performed with the sample tab oriented so that its long direction is aligned with the cutting direction, otherwise the thin, high aspect ratio tab will bend under the force of trimming. This susceptibility to bending is also why we trim the sides of tabs exclusively with the UV laser. Finally, 20 nm of gold followed by 100 nm of carbon is deposited on the sample while it is tilted and rotated in a vacuum coating machine (Model 682 PECS, Gatan). At this point the tabs are ready for standard FIB-SEM imaging.
FIB-SEM imaging
Sample tabs were FIB-SEM imaged using either a Zeiss Sigma FE-SEM or a Zeiss Merlin FE-SEM. Both were fitted with Magnum FIB columns from FEI. Custom scan generation hardware and control software were used to coordinate the milling and imaging steps.
Imaging parameters for adult Drosophila brain prepared via HPF-FS (optic medula region, Supplementary Fig. 8d-f): Matching regions of the sample tabs containing ultrathick sections #17 and #18 were FIB-SEM imaged in the Merlin FE-SEM using a 7 nA ion beam to mill away ~2 nm surface layers (25 s mill time) between each SEM image. SEM backscatter images were acquired with an 8 nA beam, 1.2 kV landing energy, 2.8 mm working distance, 8×8 nm pixel size, 0.25 µs dwell time. Raw images were aligned (SIFT align, FIJI20) and the data collapsed to 8 nm isotropic voxels prior to volume stitching.
Imaging parameters for adult Drosophila brain prepared via HPF-FS (optic lobula region, Supplementary Fig. 8b,c): Matching regions of the sample tabs containing ultrathick sections #17 and #18 were FIB-SEM imaged in the Merlin FE-SEM using a 7 nA ion beam to mill away ~2 nm surface layers (25 s mill time) between each SEM image. SEM backscatter images were acquired with an 8 nA beam, 1.2 kV landing energy, 2.6 mm working distance, 8×8 nm pixel size, 0.25 µs dwell time. SEM images were taken with ~120×32 µm field of view so that the entire ultrathick section was encompassed (Supplementary Videos 2 and 3). Both sections were FIB-SEM imaged over an ~80 µm depth covering the full depth of the optic lobe’s lobula region in each section. As a test of large-scale volume stitching this dataset was downsampled by taking every 16th image (i.e. spaced ~32 nm distances apart since milling thickness was ~2 nm/image), and these images were themselves scaled by 0.25× in x,y (8×8 nm –> 32×32 nm). The resulting images were then aligned (SIFT alignment, FIJI) into a 32×32×32 nm voxel dataset suitable for testing volume stitching with our current software (Supplementary Videos 4 and 5).
Imaging parameters for mouse cortex (Fig. 2): Matching regions of the sample tabs containing ultrathick sections #21 and #22 were FIB-SEM imaged in the Sigma FE-SEM using a 7 nA ion beam to mill away ~3 nm surface layers (20 s mill time) between each SEM image. SEM backscatter images were acquired with a 2 nA beam, 1.2 kV landing energy, 2.9 mm working distance, 5×5 nm pixel size, 0.8 µs dwell time. Raw images were aligned (SIFT align, FIJI) and the data collapsed to 10 nm isotropic voxels prior to volume stitching.
Imaging parameters for adult Drosophila brain prepared via C-PLT fixation (Fig. 3): Matching regions of the sample tabs containing ultrathick sections #34 and #35 were FIB-SEM imaged in the Sigma FE-SEM using a 7 nA ion beam to mill away ~2 nm surface layers (30 s mill time) between each SEM image. SEM backscatter images were acquired with a 2 nA beam, 1.2 kV landing energy, 2.6 mm working distance, 8×8 nm pixel size, 0.8 µs dwell time. Raw images were aligned (SIFT align, FIJI) and the data collapsed to 8nm isotropic voxels prior to volume stitching.
Volume stitching
Our methodology for volume stitching is diagrammed in Supplementary Fig. 5. This process is broken into two main steps: 1. Flattening each stack separately so that their hot knife cut surfaces reside in a single plane. 2. Transforming one stack so that it aligns with the other stack, allowing the two to be joined together across their flattened cut surfaces into a single 3D volume. We have written custom Matlab programs to perform each of these steps.
Flattening
We have written a custom GUI-based Matlab program in which the user opens up a FIB-SEM stack containing a hot knife cut edge in cross section. The GUI allows the user to interactively test different image filtering and thresholding parameters which are used by the algorithm for edge finding. The GUI also allows the user to graphically designate areas of the stack images which the edge finding algorithm should ignore and, instead, simply extrapolate over based on neighboring regions. This is required to prevent dirt particles (which sometime are found clinging to the cut edge of the section) from causing deviations from the true cut edge. It is also required so that “empty” biological features like the Drosophila trachea air tubes (shown in Supplementary Fig. 5 step #1 images) and the blood vessels in cortex are not treated as true edges by the algorithm. A user opens up a FIB-SEM stack in the program and scrolls through the stack designating with mouse drags and clicks what areas are to be ignored and extrapolated over. Then the user uses a scrollbar to adjust a gray scale threshold used to optimally separate the tissue of the ultrathick section from the blank resin it is embedded in. Once set, the program automatically finds the image coordinates of the hot knife cut edge in all stack images. It does this by first applying an anisotropic low pass spatial filter (approximately aligned parallel to the edge) to the image and then walking along each pixel column from the blank resin region toward the tissue looking for the first row position in which the pixel’s grayscale value is below threshold (i.e. the first pixel containing tissue). Pixels at the physical cut boundary which contain stained membranes are easily found by this algorithm, but pixels at the physical cut boundary which contain lightly-stained cytoplasm are often bypassed, resulting in the found edge containing many erroneous invaginations (see Supplementary Fig. 6). A smoothed curve is fit to this “approximate edge” and all edge positions which significantly deviate from this smoothed curve are deleted. This subset of points is then used to fit another smoothed curve and this process is iterated (3 times) to get a final smoothed curve which best approximates the edge in that image. This process is done for all images in the stack separately, however the previous image’s found edge position is used to set the range over which the next stack’s image is searched.
Once this process is completed, the program uses all of the found edges to fit a smoothed 3D surface approximating the full hot knife cut boundary of this ultrathick section across the FIB-SEM stack. The program’s GUI can be used to display, in different colors overlaid on the currently-viewed FIB-SEM stack image, the initial “approximate edge”, the smoothed edge based on this stack image, and the final smoothed edge based on the 3D smoothed surface, giving the user the ability to adjust parameters if necessary.
Once the user is satisfied with the found edge, they press a button to direct the entire stack to be “flattened”. This is done by shifting columns of pixels in each image so that all of their found edge pixels now reside in a single plane in the 3D flattened stack (see Supplementary Fig. 5).
Transforming and stitching
Once both the “top” and “bottom” stacks are flattened, we digitally re-slice each stack (using FIJI) to make their flattened hot knife cut surface parallel to the stack’s image plane. (Note, this cut surface does not actually reside in a single image plane of this digitally re-sliced stack, but is actually spread across about 3 image planes which are nosier and of lower-contrast than other parts of the stack. This is due to loss of staining at the very surface of the cut as well as the approximate nature of the edge detection algorithm.) These digitally re-sliced stacks are then concatenated to each other using FIJI, creating a single stack containing tissue on both sides of the hot knife cut. However, in this combined stack, there are still many images containing blank resin separating the two sides of the tissue, and both sides are not aligned to each other.
This stack is put into our second custom Matlab program which displays the top stack’s surface image and the bottom stack’s surface image side by side in Matlab’s cpselect() alignment tool. This allows the user to choose corresponding landmarks in both images. For the volume stitched stacks presented in this paper the user selected ~100–200 points of correspondence spread across the surfaces. Once selected, these correspondence points are used by our program to define an image transformation which is applied to all of the digitally re-sliced images in the “bottom” stack using the Matlab function cp2tform(). (The type of image transformation used for Supplementary Fig. 8d-f and Fig. 3 was pure affine, whereas cp2tform’s “local weighted mean” transform was used for Supplementary Fig. 8c, and Fig. 2.) This process creates a new combined stack where the tissue on both sides of the cut are aligned with each other, but they are still separated by blank resin frames. The user simply removes these blank resin frames, and any frames containing some surface voxels but which are overall too noisy or low contrast to be gainfully included, from the stack. The result is the final “volume stitched” stack, which is digitally re-sliced again to put it back into the same plane as the original FIB-SEM images (Supplementary Fig. 5).
Note that the stitch line in Fig. 3b,d is harder to see than in Fig. 2c. This is because we digitally re-sliced the Fig. 3 volume parallel to the stitch plane and contrast adjusted all images to have same gray scale mean and standard deviation. This compensated for a loss of stain contrast (described above) at the surface of the hot knife cut (Supplementary Video 10).
Neurite tracing
In order to verify that processes could be traced across the stitch, volume stitched stacks were digitally re-sliced (so that the stitch plane was parallel to the image plane) and were loaded into TrakEM221. We endeavored to trace all processes crossing the stitch plane within a given area. For tracing of the cortex sample shown in Fig. 2b, we decided to trace 200 total processes, densely tracing until we reached that number. For tracing the C-PLT Drosophila sample shown in Fig. 3g, we decided to trace 400 processes. Using TrakEM2’s arealists, we manually colored only the profiles on image planes 2, 3, and 4 pixels from the stitch plane on both sides of the stitch, however context information was used over planes further from the stitch plane as well. Processes where any ambiguity existed were examined by re-slicing the isotropic stack in a different plane. As described in the text, <2% of processes crossing the stitch (in the C-PLT fly tissue) were sufficiently ambiguous to be considered “untraceable” by one or both of the two independent tracers. The TrakEM2 colored stacks are available online as Supplementary Video 9 and Supplementary Video 11.
We also performed volume tracing across the hot knife stitch in the 4×4×4 µm volume shown in Fig. 3c-f and in Supplementary Video 12. This volume tracing was performed by loading the stitched volume into the ilastik 1.1 software22.
Red-green overlays and image mismatch comparisons
To generate the red-green overlay images shown in Supplementary Fig. 10, the volume stitched stack was first digitally re-sliced so that the stitch plane was parallel to the image plane of the stack. Then the stack’s images were collapsed to faux 40nm slices by averaging neighboring stack images making sure not to average across the stitch plane itself. A control plane within this faux TEM stack was chosen at random, and that Nth image was red-green overlaid with the N+1th image to make the 0nm gap overlay, with the N+2th image to make the 40nm gap, and with the N+3th image to make the 80nm gap. The images on either side of the actual stitch plane were red-green overlaid to make the hot knife overlay.
We used image correlation to help quantify our intuitive judgment that the hot knife image jump was approximately equivalent to the 40nm gap’s jump. For this, the images used for the red-green overlays were contrast inverted (to convert them back to signal levels) and filtered with a Gaussian filter of radius 4 (to remove noise that was clearly not relevant to the shift of membranes). The two images’ grayscale values were then pixel-wise subtracted and the absolute values of these subtractions were averaged over all pixels to give a measure of image mismatch. For the cortex tissue of Supplementary Fig. 10a these image mismatches were: 0nm gap = 3.1, 40nm gap = 5.2, 80nm gap = 6.3, hot knife jump = 5.3. For the C-PLT Drosophila tissue of Supplementary Fig. 10b these image mismatches were: 0nm gap = 4.7, 40nm gap = 6.8, 80nm gap = 7.8, hot knife jump = 6.4.
In Supplementary Fig. 11 we tried to quantify this hot knife gap further for the C-PLT fly volume stitch. Four separate control plane within its faux TEM stack were chosen at random and at each of these control planes the 0nm, 40nm, and 80nm gap image mismatches were calculated as above. These image mismatches are plotted on the Y-axis of Supplementary Fig. 11 vs. nanometers removed on the X-axis. Microsoft Excel was used to fit a second-order polynomial to this data and this fit was used to determine what amount of nanometers removed was equivalent to the observed hot knife gap image mismatch (~30nm).
Supplementary Material
Acknowledgments
We would like to thank Shinya Takemura and Stephen Plaza for assistance with tracing, and Richard Schalek for assistance with early EM imaging.
Footnotes
Author Contributions
K.J.H. conceived, designed, and performed the experiments and wrote the manuscript. C.S.X., H.F.H., & K.J.H. constructed the custom FIB-SEM systems used. C.S.X. designed and implemented the custom FIB-SEM control hardware and software used. Z.L. developed the C-PLT technique used and prepared the C-PLT tissue. G.W.K. prepared mouse cortex tissue displayed in manuscript. R.D.F. prepared larva tissue. R.D.F. & Z.L. prepared HPF-FS tissue. J.C.T. prepared mouse cortex tissue displayed in supplementary figures. Early research (Supplementary Figs. 1–3) was performed in J.W.L. laboratory. All FIB-SEM research was performed in H.F.H. laboratory.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Knott G, Marchman H, Wall D, Lich B. J. Neurosci. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu CS, Hess H. Microscopy & Microanalysis. 2011;17:664–665. [Google Scholar]
- 3.Briggman KL, Bock DD. Curr. Opin. Neurobiol. 2011;22:1–8. doi: 10.1016/j.conb.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 4.Harris KM, et al. J. Neurosci. 2006;26:12101–12103. doi: 10.1523/JNEUROSCI.3994-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bock DD, et al. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayworth KJ, et al. Front. Neural Circuits. 2014;8:1–18. doi: 10.3389/fncir.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denk W, Horstmann H. PLoS Biology. 2004;2:1900–1909. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denk W, Briggman KL, Helmstaedter M. Nature Reviews Neurosci. 2012;13:351–358. doi: 10.1038/nrn3169. [DOI] [PubMed] [Google Scholar]
- 9.Morgan JL, Lichtman JW. Nature Methods. 2013;10:494–500. doi: 10.1038/nmeth.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayat MA. Principles and Techniques of Electron Microscopy. Cambridge, UK: Cambridge University Press; 2000. [Google Scholar]
- 11.McGee-Russell SM, Gosztonyi G. Nature. 1967;214:1204–1206. doi: 10.1038/2141204a0. [DOI] [PubMed] [Google Scholar]
- 12.McGee-Russell SM, De Bruijn WC, Gosztonyi G. J. Neurocytology. 1990;19:655–661. doi: 10.1007/BF01188034. [DOI] [PubMed] [Google Scholar]
- 13.West RW. Stain Technology. 1972;47:201–204. doi: 10.3109/10520297209116485. [DOI] [PubMed] [Google Scholar]
- 14.Studer D, Gnaegi H. J Microsc. 2000;197:94–100. doi: 10.1046/j.1365-2818.2000.00638.x. [DOI] [PubMed] [Google Scholar]
References for online methods
- 15.Meinertzhagen IA, O’Neil SD. J Comp. Neurol. 1991;305:232–263. doi: 10.1002/cne.903050206. [DOI] [PubMed] [Google Scholar]
- 16.Walther P, Ziegler A. J. Microsc. 2002;208:3–10. doi: 10.1046/j.1365-2818.2002.01064.x. [DOI] [PubMed] [Google Scholar]
- 17.Seligman AM, Wasserkrug HL, Hanker JS. J Cell Biol. 1966;30:424–432. doi: 10.1083/jcb.30.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tapia JC, et al. Nat. Protocols. 2012;7:193–206. doi: 10.1038/nprot.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locke M, Krishnan N. J. Cell Biology. 1971;50:550–557. doi: 10.1083/jcb.50.2.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schindelin J, et al. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, Douglas RJ. PLoS One. 2012;7:e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sommer C, Strähle C, Köthe U, Hamprecht FA. Eighth IEEE International Symposium on Biomedical Imaging (ISBI) Proceedings. 2011:230–233. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.