Abstract
Purpose
To better understand the mechanistic parameters that govern drug release from polymer micelles with acid-labile linkers.
Methods
A mathematical model was developed to describe drug release from block copolymer micelles composed of a poly(ethylene glycol) shell and a poly(aspartate) core, modified with drug binding linkers for pH-controlled release [hydrazide (HYD), aminobenzoate-hydrazide (ABZ), or glycine-hydrazide (GLY)]. Doxorubicin (Dox) was conjugated to the block copolymers through acid-labile hydrazone bonds. The polymer drug conjugates were used to prepare three polymer micelles (HYD-M, ABZ-M, and GLY-M). Drug release studies were performed to identify the factors governing pH-sensitive release of Dox. The effect of prolonged storage of copolymer material on release kinetics was also observed.
Results
Biphasic drug release kinetics were observed for all three micelle formulations. The developed model was able to quantify observed release kinetics upon the inclusion of terms for unconjugated Dox and two populations of conjugated Dox. Micelle/water partitioning of Dox was also incorporated into the model and found significant in all micelles under neutral conditions but reduced under acidic conditions. The drug binding linker played a major role in drug release as the extent of Dox release at specific time intervals was greater at pH 5.0 than at pH 7.4 (HYD-M > ABZ-M > GLY-M). Mathematical modeling was also able correlate changes in release kinetics with the instability of the hydrazone conjugation of DOX during prolonged storage.
Conclusion
These results illustrate the potential utility of mechanistic modeling to better assess release characteristics intrinsic to a particular drug/nanoparticle system.
Keywords: Nanoparticles, mechanistic modeling, controlled drug release, hydrazone, doxorubicin
INTRODUCTION
Nanoparticles such as dendrimers, liposomes, and polymeric micelles are widely used as drug carriers for the treatment of various human diseases with applications in cancer being particularly prevalent (1–3). Nanoparticle drug carriers that modulate the entrapment and release of drug payloads can favorably alter drug pharmacokinetics by reducing clearance, suppressing non-specific drug accumulation, and enhancing tumor accumulation (2). While many types of nanoformulations have been developed to take advantage of these properties, few have had clinical success, which may be attributable to non-optimal in vivo release rates.
Current methods used to characterize drug release kinetics from nanoparticles are numerous (4–6); however, environmental factors that result in differences between in vitro and in vivo release kinetics must be considered. Mechanistic models that enable separation of environmental effects from intrinsic release parameters can provide a framework for reliable in vitro - in vivo correlations (7). This in turn would reduce the costs incurred during preclinical development due to extensive animal testing and unguided formulation optimization. To this end, considerable efforts have been made to establish mechanistic drug release models for liposomal formulations; however, similar models have yet to be developed for other nanoparticles (7, 8). For other nanoparticles, the unique physicochemical properties of the individual drug and particle in addition to the combined properties of the drug/particle system must be considered.
One particular system which has received considerable attention is polymeric micelles (9, 10). These self-assembled nanoparticles are unique structures that provide a range of options for altering drug release (e.g. drug conjugation, particle size and charge, hydrophobicity). While the numerous options may be advantageous in altering release kinetics, they may also result in complex drug release mechanisms. For example, block copolymer micelles are generally composed of a hydrophilic shell and a hydrophobic core. Drug payloads are typically thought to be entrapped in the micellar core due to the drug’s affinity for hydrophobic regions of the particles (11–13) but in reality the drug distribution may be heterogeneous. Drug release rates may reflect a combination of kinetic factors (i.e. drug diffusion and/or cleavage of the drug-polymer linkage) and thermodynamic factors (i.e. critical micelle concentration, micelle/water partition coefficient, etc.) intrinsic to the drug/polymer system.
Depending on the block copolymer design, drug release can also respond to external stimuli such as heat, electromagnetic waves, enzymatic activity, or pH (11, 13–23). Chemically conjugating drugs to the block copolymers is another way to alter drug release kinetics and adds yet another dimension to the mechanism of drug release. In several instances, these types of micelles often show a biphasic drug release pattern (i.e. burst drug release followed by a sustained, often extremely slow drug release), but a consensus has yet to be reached on the exact cause of the two phases of release (10, 24).
In this study a mechanistic model was developed to describe drug release from block copolymer micelles with hydrazone-conjugated doxorubicin (Dox). These micelles were prepared from block copolymers composed of 12 kDa poly(ethylene-glycol) (PEG) and 16 hydrophobic repeating units, synthesized as previously reported (25). The hydrophobic portion of the block copolymers was modified with hydrazide-based drug linkers (HYD, ABZ, or GLY) which were subsequently used to conjugate Dox to the micelle. Hydrazone bonds have been shown to be pH-sensitive, being relatively stable in neutral conditions but susceptible to hydrolysis in acidic environments (9, 24, 26–29). Furthermore, spacers prior to the hydrazone moiety have been successfully employed to further tune drug release (25). For this reason, GLY and ABZ spacers were employed in this study to alter drug release. A block copolymer without a spacer was also prepared as a control.
Previously, drug release from micelles based on block copolymers with identical drug linkers was modeled using zero and first order release models, combining micelle-dependent and method-dependent release properties into a single kinetic descriptor (25). This oversimplification led to a limited understanding of the specific factors that govern Dox release (e.g., dialysis membrane transport, hydrolysis kinetics, micelle/water partitioning, etc.). Therefore, studies herein focus on developing a mathematical model based on a postulated mechanistic interpretation of individual properties affecting Dox release. Drug release studies using dynamic dialysis as the primary method to monitor release were performed at pH 5.0 and pH 7.4 to elucidate the pH-effect on Dox release from polymeric micelles. In dynamic dialysis, which is typically used to evaluate drug release from various nanoparticle drug carriers, an additional physical barrier (i.e., the dialysis membrane) and the absence of sink-conditions within the dialysis compartment may influence the release rate (7). The kinetics of drug transport across the dialysis membrane as modulated by micelle/water partitioning of the drug were incorporated into the model to isolate their effects on observed drug release profiles and generate intrinsic rate constants for drug release from the block copolymer micelles.
Preliminary modeling results indicated heterogeneous kinetics of hydrazone bond hydrolysis within the micelles. Long term storage of polymer drug conjugates resulted in hydrolysis of the hydrazone bond during storage [similar to a previous study (32)] was identified by mathematical modeling of altered release kinetics after storage. This possible instability was explored and later confirmed experimentally. Additionally, a non-sink release method previously developed to simultaneously determine release kinetics and drug partitioning in liposomal formulations (30) was employed to determine the extent of free Dox partitioning in the HYD micelle formulations and further validate the mechanistic model. Parameter values generated from simultaneous fitting of experimental data to the model served to assist in identifying the factors governing the pH-sensitive release of Dox from these micelle formulations.
MATERIALS AND METHODS
Materials
The following supplies were purchased from Fisher Scientific (USA): Slide-A-Lyzer® dialysis cassettes (10,000 MWCO), Sephadex LH-20 gels, potassium biphthalate sodium hydroxide buffer solution, potassium phosphate monobasic buffer solution, and 96-well plates. Amicon® Ultra centrifugal filters (10,000 MWCO) were purchased from Millipore (USA). Doxorubicin (Dox) HCl was purchased from LC Laboratories (USA).
Micelle preparation and characterization
PEG-p(Asp-HYD-Dox), PEG-p(Asp-ABZ-Hyd-Dox), and PEG-p(Asp-GLY-Hyd-Dox) block copolymers were synthesized and characterized previously (31). Briefly, linker modified block copolymers were dissolved in deionized water (2.0 mg/mL) to prepare HYD, ABZ, and GLY micelles (HYD-M, ABZ-M, and GLY-M), respectively. At this concentration, micelle particle size and ζ-potential were determined using a Zetasizer Nano-ZS (Malvern, UK) equipped with a He-Ne laser (4 mW, 633 nm). A SpectraMax M5 (Molecular Devices, USA) equipped with variable spectrum filters. SoftMax Pro Software was used to quantify Dox.
Drug release studies
Dox release from micelles was observed in triplicate at 37°C in potassium biphthalate sodium hydroxide buffer solution (pH 5.0, 0.005 M ionic strength) and potassium phosphate monobasic buffer solution (pH 7.4, 0.024 M ionic strength). Temperature and pH were monitored throughout the study to ensure no drift from the initial conditions. Two methods were used for drug release studies.
The first method was dynamic dialysis. All experiments were performed using the same dynamic dialysis method but at varying pH (5.0 or 7.4). Release studies were performed with freshly synthesized material (stored less than one month at − 20 °C) at 1.0 mg copolymer/mL while release studies with 0.1 and 0.5 mg copolymer/mL used material that had been stored for 14 months at − 20 °C. For all of these release studies, block copolymer solutions (3 mL) in 10 mM buffer (pH 5.0 or pH 7.4) were transferred into dialysis cassettes (10,000 MWCO), that were subsequently placed into a 5.0 L reservoir of the same buffer. Aliquots (100 µL) were taken from the dialysis cassettes at the following time points: 0, 0.5, 1, 3, 6, 24, 48, and 72 hours. Drug release experiments performed at 0.1 and 0.5 mg/mL concentrations had these additional time points: 1.5, 2.0, 4.5, and 9.0 hours In the same fashion as the drug loading studies, samples herein were analyzed using UV-Vis spectroscopy. The DOX fingerprint peak at 480 nm was used for detection. Based on a calibration curve (0.98 to 250 µM), the concentration of DOX within each sample was determined. It is important to note that the measured concentration represented the total concentration of conjugated and free DOX. Block copolymer absorbance spectra confirmed that DOX conjugation did not alter its spectrum.
Three additional drug release studies were performed using dynamic dialysis. First, buffer concentration effects were probed by performing drug release studies at low (5 mM) and high (20 mM) buffer concentrations. HYD-M, ABZ-M, and GLY-M solutions (0.5 mg/mL) were transferred into dialysis cassettes which were immediately placed in either 5 or 20 mM buffer solutions (pH 5.0). Samples (100 µL) were removed using a pipetter at predetermined time points (0, 0.5, 1, 1.5, 3, 4.5, 6, 9, 24, 48, and 72 hours).
Secondly, the rate of free Dox diffusion from dialysis cassettes was determined at both pH 5.0 and pH 7.4. Dox was dissolved in 10 mM buffer solution (0.12 mg/mL) and dialyzed under the same conditions used above for Dox-loaded micelle suspensions. In order to monitor Dox binding to the dialysis membrane, a follow-up experiment was performed using the same dialysis cassettes from the Dox diffusion experiments. These previously-used dialysis cassettes were emptied and filled with Dox solutions (0.12 mg/mL). Dox disappearance from dialysis cassettes was then monitored.
Lastly, 0.5 mg/mL suspensions of HYD-M, ABZ-M, and GLY-M which had already undergone release for 72 hours were spiked with free Dox and thoroughly mixed. These suspensions were dialyzed against the same reservoir used in micelle release studies to observe the influence of partitioning of unconjugated, free Dox into the micelle, if any, on Dox transport across the dialysis membrane.
A secondary set of drug release experiments employed a previously developed ultrafiltration method to monitor drug release under non-sink conditions (30). Micelle suspensions (3 mL, 0.5 mg/mL) were transferred to scintillation vials that were placed in a 37°C incubator and gently shaken. At specific intervals, aliquots (250 µL) were removed, diluted to 500 µL with methanol, and ultrafiltered using Amicon® Ultra 0.5 mL centrifugal filters with Ultracel® membranes (10,000 MWCO). Ultrafiltration was achieved by centrifugation of these cartridges at 14,000 rpm for 10 min. Afterwards, the supernatant was collected and diluted to 500 µL with a 50:50 methanol:water solution and the process was repeated twice more. Block copolymers were expected to remain in the concentrate during ultrafiltration due to their high molecular weight (>13,000) while free Dox (544 MW) was removed. Free Dox removal by ultrafiltration was validated by dissolving Dox in a 50% methanol:water mixture and ultrafiltering. After three cycles, spectrometric analysis determined that no Dox was present in the concentrate. An additional confirmation was performed using two identical block copolymer solutions. One sample was spiked with free Dox and vortexed vigorously. Both samples underwent three ultrafiltration cycles. Analysis showed that there was no statistically significant difference between the Dox concentrations in the two concentrates, confirming complete free Dox removal from both.
Determination of percent unconjugated Dox produced during storage
The stability of the Dox-hydrazone bond during storage was a concern due to the time that elapsed between drug release studies (~14 months). Two separate methods were employed to quantify the percent of unconjugated Dox. First, ultrafiltration was used to determine recovery percentages of freshly prepared HYD-M, ABZ-M, and GLY-M. Block copolymers were dissolved in buffer solution and immediately ultrafiltered. Micellar Dox concentrations were determined before and after ultrafiltration. The percent recovered was calculated and reported. Secondly, 14 month-old block copolymers were purified using a Sephadex LH20 column. Briefly, block copolymers were dissolved in methanol and loaded on a Sephadex LH-20 column equilibrated with methanol. Methanol was added drop-wise to the column and flow was controlled by gravity. Due to the higher molecular weight, block copolymers move quicker through the column creating separation between themselves and free DOX. The initial, clear eluted volume was discarded. As the eluted volume turned redthe block copolymer fraction was collected. Methanol was subsequently removed using rotary evaporation to dry and isolate the synthesized copolymer. The amount of DOX was measured before and after purification to determine the extent of drug loading and calculate the percent of conjugated Dox.
Mathematical modeling
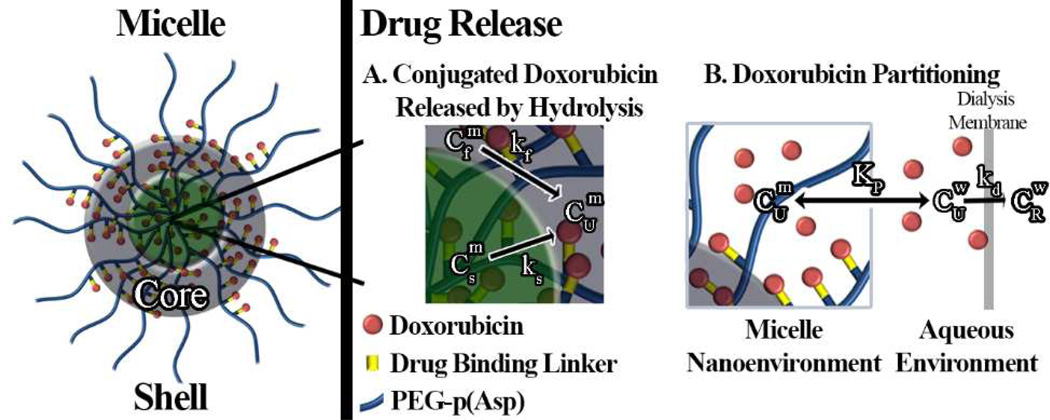
A mathematical model was developed based on Fig. 1 for data fitting and simulations. The model considers three major factors: heterogeneous kinetics of Dox release from the micelle, Dox micelle/water partitioning (Kp), and Dox diffusion through the dialysis membrane (kd). Dox release studies clearly showed biphasic kinetics and therefore two rate constants (kf, ks) were used to represent Dox release from micelles. The remaining variables in the initial model were: conjugated Dox , free Dox partitioned in the micelle , free Dox in the aqueous phase , free Dox in the reservoir (CR), and the percent of Dox initially conjugated to block copolymers (PC).
Figure 1.
Illustration of the mathematical model and parameters used to describe heterogeneous Dox release kinetics from three different micelle formulations (HYD-M, ABZ-M, and GLY-M). Here, the hydrophilic PEG shell surrounds a core consisting of two populations of conjugated Dox corresponding to fast ( ) and slow ( ) hydrolysis governed by rate constants k1 and k2, respectively. After hydrolysis, unconjugated Dox may partition into the micelle ( ) or reside in the aqueous environment ( ), as governed by the partition coefficient Kp. In dynamic dialysis studies, Dox transport though the dialysis membrane is governed by the rate constant kd and unbound Dox concentration,.
The mass balance for total Dox present in dialysis cassettes (MT) includes two main species: unconjugated Dox (MU) and covalently conjugated Dox (MC). Furthermore, unconjugated Dox can either partition into micelles (MUm) or remain in the aqueous phase (MUw) (Eq. 1). MCf and MCs represent two populations of conjugated Dox undergoing fast and slow hydrolysis, respectively (Eq. 2).
| (1) |
| (2) |
The total mass balance of Dox can then be written as:
| (3) |
The total concentration of DOX within the dialysis tube (CT) is measured during dynamic dialysis. Therefore, the mass balance of Eq. 3 is rewritten in terms of the concentrations of the various DOX species using ratios of the aqueous (Vw) and micellar volumes (Vm) to total volume (VT). These ratios, defined as and b , respectively, are employed in Eq. 4:
| (4) |
The density of the micelles was assumed to be similar to that of water, allowing the weight fraction of micelle to be used for volume calculations. Values of a and b depend on the block copolymer concentration during the release studies. The values of a for drug release studies performed at 1.0, 0.5, and 0.1 mg/mL were 0.9990, 0.9995, and 0.9999, respectively.
As described above, micellar Dox concentrations in the dialysis chamber depend on Dox escape from micelles and diffusion through the dialysis membrane (Eqs. 5–8)
| (5) |
| (6) |
| (7) |
| (8) |
where and represent changes in concentrations of covalently attached Dox versus time in the fast and slow phases, respectively; describes the change in free Dox concentration within the dialysis cassette with time; and describes the change in free Dox concentration within the reservoir versus time. The volume ratio of the dialysis cassette to the reservoir was 0.0018. The rate constants for Dox escape from micelles are kf and ks while kd is the rate constant for Dox diffusion through the dialysis membrane. The equilibrium constant for membrane/water partitioning of Dox is . can then be rewritten in terms of total unconjugated Dox:
| (9) |
This term can then be substituted into Eqs. 7–8:
| (10) |
| (11) |
It is important to note that at time zero Dox can be present either in the conjugated or free form (CU). The percent of conjugated Dox (Pc) and the fraction of Dox undergoing fast release, Fkf, are incorporated into the initial conditions (Eqs. 12–15).
| (12) |
| (13) |
| (14) |
| (15) |
Additionally, spike experiments were conducted to monitor the effect of Dox partitioning on the rate of disappearance of unconjugated Dox. For these experiments, block copolymer solutions having undergone 72 hours of release were spiked with free Dox and dialysis was continued. The initial conditions were then altered to account for the remaining conjugated Dox and the initial concentration of unconjugated Dox due to the spike, CTS,0. (Eqs. 16–18).
| (16) |
| (17) |
| (18) |
where Eqs. 16 and 17 account for the monoexponential decline of each population of conjugated Dox during the time that micelles had been dialyzed.
The same model with minor adjustments was used to describe drug release under non-sink conditions. The equation describing total drug concentration was altered due to the analysis method. For non-sink drug release studies, free Dox is separated from conjugated Dox and Dox partitioned within the micelle by ultrafiltration. After free Dox removal, only conjugated and partitioned Dox concentrations in micelles are measured. Then the total measured Dox concentration becomes:
| (19) |
can be expressed in terms of the total concentration of free Dox and the partition coefficient and substituted into Eq. 19 yielding:
| (20) |
| (21) |
The differential equations related to micellar Dox concentrations under non-sink conditions are less complex than those used in dynamic dialysis as there is no reservoir compartment and no Dox transport across a dialysis membrane. For non-sink drug release:
| (22) |
| (23) |
| (24) |
For dynamic dialysis, the model required the term Pc to account for any unconjugated Dox. Initial conditions in non-sink drug release are altered due to removal of free Dox by ultrafiltration.
| (25) |
| (26) |
| (27) |
Drug release profiles for all three types of micelles and both pH conditions studied (i.e. the profiles measure for CT as defined by Eqs. 4 and 19 for dynamic dialysis and ultrafiltration studies, respectively) were fit using Micromath Scientist non-linear regression software using a weighting factor or two.
RESULTS
Micelle preparation and characterization
The HYD-M, ABZ-M, and GLY-M block copolymers having drug loadings of 26 ± 1.6, 17 ± 1.5, and 26 ± 1.1% by weight, respectively, formed micelles in aqueous solution resulting in hydrodynamic diameters of 117 ± 37, 54 ± 12, and 58 ± 11 nm obtained by dynamic light scattering measurements (31). The lower drug loading of ABZ-M has been previously observed and theorized to occur due to the steric hindrance caused by the benzyl moiety (25). The ζ-potentials were 13 ± 0.2, −4.0 ± 0.6, and 0.5 ± 1.5 mV for HYD-M, ABZ-M, and GLY-M, respectively (31). Values reported here are averages ± standard deviation of triplicate measurements.
Validation of drug release experiments
Analyses of release kinetics from the observed release profiles obtained by the two methods employed in this study must account for the effects of the different experimental conditions (7, 8, 30). In the case of dynamic dialysis, significant drug binding or adsorption to the dialysis membrane may alter the kinetics observed during release studies resulting in rate constants and other parameters that are not applicable under other conditions without an appropriate model (7). The disappearance of free Dox from dialysis cassettes at both pH 5.0 and pH 7.4 was monitored to assess this factor. Dox disappearance was first-order indicating that a single rate constant was sufficient to describe Dox transport across the dialysis membrane. Furthermore, disappearance of Dox from a spike experiment conducted to analyze polymer effects on Dox diffusion through the dialysis membrane mimicked that for free Dox. Drug release profiles generated at pH 5.0 and either 5 mM or 20 mM showed no difference, agreeing with previous work in similar systems that ionic strength had no effect on release (29).
Analysis of ultrafiltration studies can be affected by incomplete removal of drug during the centrifugation and washing steps (30). For accurate analysis, ultrafiltration studies herein had to separate aqueous Dox from Dox partitioned into micelles or conjugated to the block copolymers. This was validated first by exposing free Dox to the same ultrafiltration process as the block copolymers. Spectrophotometric analysis of Dox after ultrafiltration indicated complete drug removal. A second confirmation was performed by spike experiments with all micelles (HYD-M, ABZ-M, and GLY-M). Two sets of identical micelle solutions were prepared, one of which was subsequently spiked with free Dox. Both sets of solutions underwent the same ultrafiltration process. Analyses of the concentrates revealed no significant difference between the two solutions, confirming removal of free Dox in the aqueous phase.
Model-predicted hydrazone bond degradation after long-term storage
Unlike the freshly-synthesized 1.0 mg block copolymer/mL studies, initial modeling of the Dox release profiles using copolymer material stored for 14 months at −20 °C (i.e. 0.1 and 0.5 mg block copolymer/mL release studies) required inclusion of a value for Pc representing the percentage of conjugated Dox at the start of a release experiment of less than 100%. At the lower block copolymer concentrations, 35–50% of Dox appeared to be unconjugated at the start of an experiment. As described in the previous methods section, all block copolymers were stored after synthesis as solids at −20°C. Samples were removed and used as needed for experiments. The sole difference between block copolymers used for drug release at lower concentrations and at 1.0 mg/mL was the time at which the experiments were performed. The drug release study at the 1.0 mg/mL concentration was completed approximately 14 months prior to the other experiments.
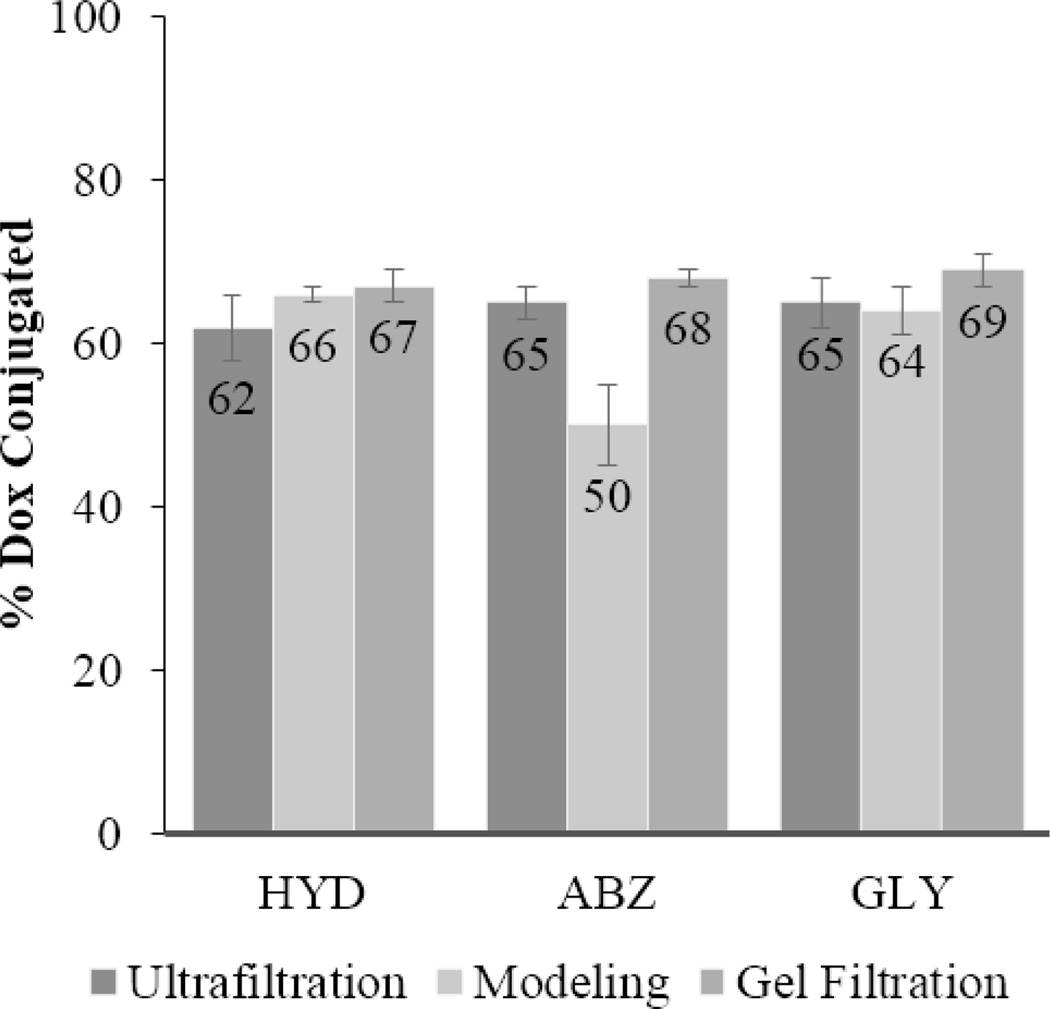
Due to these differences in the apparent percentage of Dox conjugated, partial hydrolysis of the hydrazone linkage connecting Dox to the polymer during storage was suspected. This was verified in two ways: ultrafiltration and Sephadex LH-20 purification (Fig. 2). Both supported the modeling results. Typically greater than 90% recovery is expected after ultrafiltration purification. In the case of the block copolymers stored an additional 14 months, the recovery percentage of Dox was between 62% and 65%. An additional confirmation was performed using Sephadex LH-20 purification. Fourteen month old material was purified to remove any free Dox generated during storage. Drug loading was measured before and after purification. According to this experiment, between 67% and 69% of Dox was retained, depending on the micelle formulation. Thus, the results from the three separate methods were in agreement, all pointing to partial degradation of the hydrazone bond during prolonged storage.
Figure 2.
The % of Dox conjugated after 14 months of storage determined by different methods. Error bars reflect 95% confidence intervals.
Analyses of drug release profiles
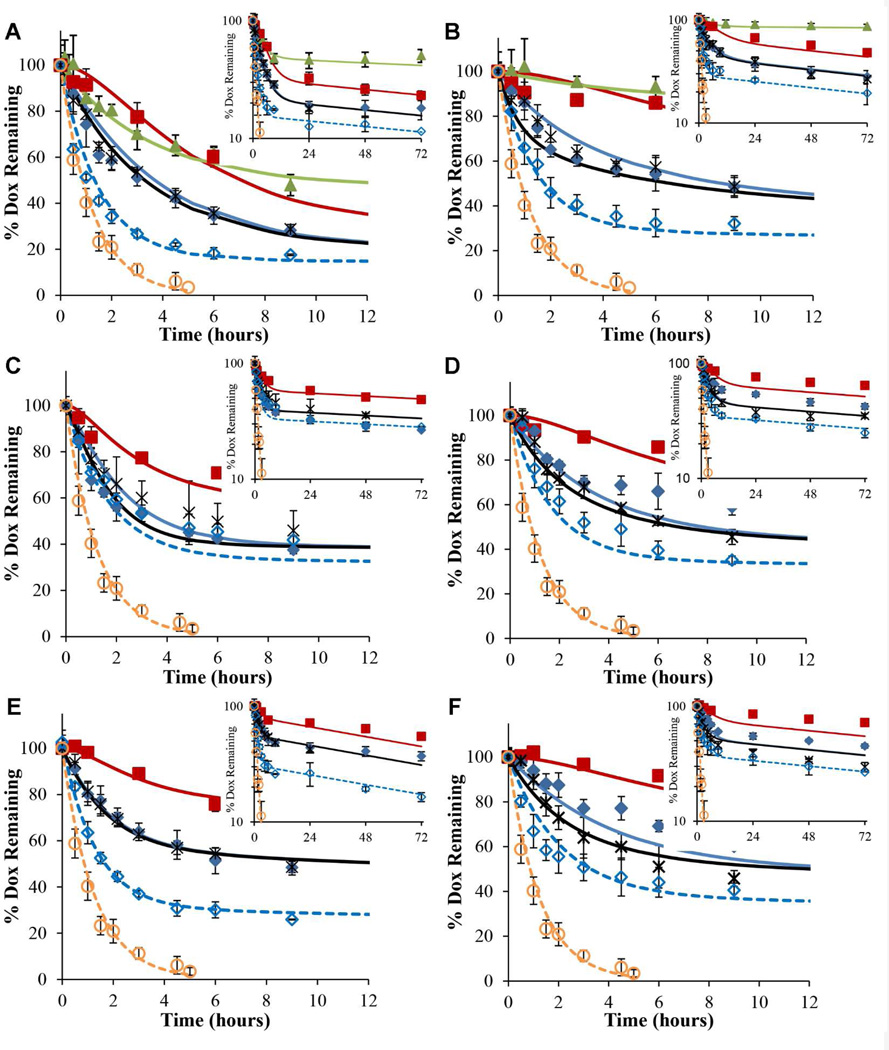
Multiple drug release studies were performed at physiological pH 7.4 and lysosomal pH 5.0 (Fig. 3). First, Dox release profiles from HYD-M, ABZ-M, and GLY-M at 1.0 mg/mL block copolymer concentration were obtained using dynamic dialysis. Irrespective of micelle composition, more Dox was released at pH 5.0 than at pH 7.4. After 72 hours, Dox release from HYD-M was 77% at pH 5.0 and 52% at pH 7.4. At this same time point, ABZ-M released 52% and 35% of Dox at pH 5.0 and pH 7.4, respectively. Similarly, GLY-M exhibited 45% and 28% Dox release at pH 5.0 and pH 7.4, respectively. It was apparent that total drug release followed a pattern of HYD-M releasing the largest percentage of Dox followed by ABZ-M and GLY-M, irrespective of pH. This trend (HYD-M>ABZ-M>GLY-M) was also evident at the 24 and 48 hour time points.
Figure 3.
Dox release profiles at pH 5.0 (A, C, E) and 7.4 (B, D, F) obtained for HYD-M (A and B), ABZ-M (C and D), and GLY-M (E and F). The release profile under non-sink conditions (0.5 mg/mL) is shown ( ) in addition to release profiles obtained by dynamic dialysis at 1.0 (
) in addition to release profiles obtained by dynamic dialysis at 1.0 ( ), 0.5 (
), 0.5 ( ), and 0.1 ( ) mg/mL block copolymer concentrations. Free Dox (0.12 mg/mL) dialysis (
), and 0.1 ( ) mg/mL block copolymer concentrations. Free Dox (0.12 mg/mL) dialysis ( ) and block copolymer (0.5 mg/mL) spiked with free Dox (
) and block copolymer (0.5 mg/mL) spiked with free Dox ( ) profiles are also shown. Lines represent simulated release using the model developed. Error bars indicate the standard deviation of triplicate release studies at each time point.
) profiles are also shown. Lines represent simulated release using the model developed. Error bars indicate the standard deviation of triplicate release studies at each time point.
Additional drug release studies also using dynamic dialysis were performed at different block copolymer concentrations (0.5 and 0.1 mg/mL) (Fig. 3). Decreasing the block copolymer concentrations from 1.0 to 0.5 mg/mL or 0.1 mg/mL resulted in greater Dox release from all micellar systems irrespective of pH but this was largely attributable to the significant fraction of hydrazone bond hydrolysis during storage of the drug copolymer conjugates used in the experiments at lower concentration. Total Dox release from micelles at all concentrations was greater at pH 5.0 than at pH 7.4 and followed the trend: HYD-M >ABZ-M >GLY-M.
Dox release studies from HYD-M (0.5 mg/mL) were also conducted under non-sink conditions at pH 5.0 and 7.4. As opposed to dynamic dialysis release studies, Dox concentrations remaining in the micelles reached a plateau under non-sink conditions (Fig. 3 A & B). Without a reservoir to provide sink conditions, Dox release was presumed to reach equilibrium allowing the pH-dependent micelle/water partitioning effect to be confirmed. More Dox was released at pH 5.0 (52%) than at pH 7.4 (15%) indicative of an increased micelle/water partition coefficient at pH 7.4.
Characterization of pH-sensitive release profiles
Drug release profiles were fitted to the models described in the previous section to determine parameter values for: kf, ks, kd, Kp, and Fkf (Table 1). Pc, the percent of Dox initially conjugated, was not a fitted parameter as it was determined independently by Sephadex purification. At 1.0 mg block copolymer/mL, the percent conjugated was set at 100% for all micelles while the percent conjugated at 0.1 and 0.5 mg block copolymer/mL was set to 67%, 68%, and 69% for HYD-M, ABZ-M, and GLY-M, respectively. Based on the values reported in Table 1, the main parameters contributing to differences in Dox release profiles were the fractions of conjugated Dox undergoing fast release (Fkf) and moderate differences in the rate constant for hydrolysis of the conjugated Dox population remaining after the initial fast release phase (ks). Generally, no significant differences could be discerned in kf values characterizing the rate constant for the conjugated Dox population undergoing rapid release. This was partially due to the significant amount of unconjugated Dox initially present in release studies at the two lower copolymer concentration studied. The partial hydrolysis of conjugated Dox during storage added more drug to the fast phase of release seen in the release studies performed at 0.1 and 0.5 mg copolymer/mL. This made the estimation of a fast release rate constant difficult to distinguish from the rate-limited dialysis of free Dox initially present in the suspensions.
Table 1.
Drug release parameters determined through mathematical modelinga
| Micelle | pH | kf (hr−1) | ks × 102(hr−1) | kd (hr−1) | kp × 10−3 | Fkf |
|---|---|---|---|---|---|---|
| HYD-M | 7.4 | 0.24 ± 0.1 | 0.55 ± 0.1 | 0.81 ± 0.03 | 2.5 ± 0.6 | 0.38 ± 0.06 |
| ABZ-M | 7.4 | 0.27 ± 0.2 | 0.36 ± 0.1 | 0.79 ± 0.03 | 1.1 ± 0.8 | 0.34 ± 0.06 |
| GLY-M | 7.4 | 0.30 ± 0.1 | 0.46 ± 0.1 | 0.80 ± 0.03 | 2.2 ± 1 | 0.25 ± 0.04 |
| HYD-M | 5.0 | 0.29 ± 0.05 | 0.45 ± 0.1 | 0.80 ± 0.03 | 0.62 ± 0.2 | 0.68 ± 0.03 |
| ABZ-M | 5.0 | ≥ 1.15b | 0.59 ± 0.2 | 0.78 ± 0.03 | 0 | 0.36 ± 0.05 |
| GLY-M | 5.0 | 0.64 ± 0.5 | 0.82 ± 0.1 | 0.77 ± 0.02 | 0 | 0.20 ± 0.05 |
Data reported as fitted value ± 95% confidence intervals;
Value used for generation of statistics as release of fast phase was rate-limited by Dox transport through the dialysis membrane.
Dox release rates from micelles at pH 7.4 did not differ dramatically with variations in drug-binding linker after accounting for the hydrazone bond degradation during storage (again, the ability to precisely determine kf was diminished due to hydrazone bond degradation during storage). The primary factors accounting for differences between drug release profiles at pH 5.0 and pH 7.4 were the greater fraction of conjugated Dox undergoing fast release (Fkf) which resulted in a greater extent of Dox release from HYD-M at 72 hr and modest but statistically significant increases in the slow release rate constants (ks) at pH 5.0 for ABZ-M and GLY-M micelles. While values of the partition coefficients (Kp) were higher at pH 7.4, the release profiles were only moderately influenced by binding at the micelle concentrations employed (as most readily visualized in the 1 mg/mL release profiles at early time points (Fig. 3B, D, and F)). The secondary phase of Dox release was very slow, irrespective of block copolymer. The primary difference between the model determined drug release parameters was in Fkf (HYD-M>ABZ-M>GLY-M).
DISCUSSION
Previously, apparent release rate constants from Dox-conjugated block copolymer micelles were calculated using zero or first order models, leaving open the question of which factors impacted drug release (25). These apparent release rate constants combined micelle-dependent properties (i.e. hydrazone bond hydrolysis, Dox partitioning) and transport across the dialysis membrane into a single kinetic descriptor. Furthermore, only the pH effect was observed, without considering effects of block copolymer and buffer concentrations. This oversimplified model led to a limited understanding of the release profiles. For this reason, additional studies were performed and a comprehensive mathematical model developed to better elucidate factors (e.g., hydrazone stability, Dox micelle/water partitioning, heterogeneous kinetics, etc.) impacting drug release.
Initial model fitting revealed possible stability issues with the hydrazone bond under the storage times and conditions employed. The degradation mechanism was beyond the scope of this work, but the most likely possibility was hydrolysis during storage at −20°C due to the presence of trace amounts of water. Previously the hydrazone linkage in a Dox conjugate was shown to degrade after lyophilization during storage at 2 – 8°C (32). In that study, a water content of 2% in lyophilized samples resulted in 20% degradation of the hydrazone bond after 12 months. Though polymers were stored at a lower temperature (−20°C) in this work, samples were stored for a longer time. Also, the presence of the hydrophilic PEG may have resulted in higher water retention after freeze drying leading to greater degradation (water contents were not determined in the present study).
Release studies were performed with free Dox to probe dialysis membrane transport as this membrane poses an additional barrier to diffusion that may impact observed release kinetics (33, 34). Irrespective of pH or formulation, the rate constant for Dox diffusion through the dialysis membrane was ~0.80 hr−1. In one additional study, the rate of Dox disappearance was determined a second time with a previously used dialysis cassette. The determined rate constant did not differ significantly from that obtained in the initial study. Another study used a block copolymer solution that had undergone release for 72 hours. The solution was spiked with free Dox and disappearance from the dialysis cassette was monitored. The rate of disappearance was similar to studies with free Dox alone indicating that the presence of micelles under these conditions did not significantly alter release kinetics of free Dox.
The extent of Dox binding to micelles could significantly alter the apparent rate of Dox release in dynamic dialysis when dialysis membrane transport becomes partially rate limiting (35, 36). Release studies were performed at three block copolymer concentrations to probe the effects of drug partitioning. As copolymer concentration decreases, the fraction of micellar bound Dox decreases. When drug release profiles generated at two different block copolymer concentrations overlap, the effect of drug binding to micelles is assumed to be negligible. This was observed at pH 5.0, as drug release profiles from HYD-M, ABZ-M, and GLY-M at 0.1 and 0.5 mg/mL copolymer concentrations overlapped in all three cases. Dox micelle/water partitioning was reduced at pH 5.0 relative to pH 7.4 because Dox (pKa = 8.15) is fully protonated at pH 5.0 but a mixture of protonated and neutral species at pH 7.4 (37).
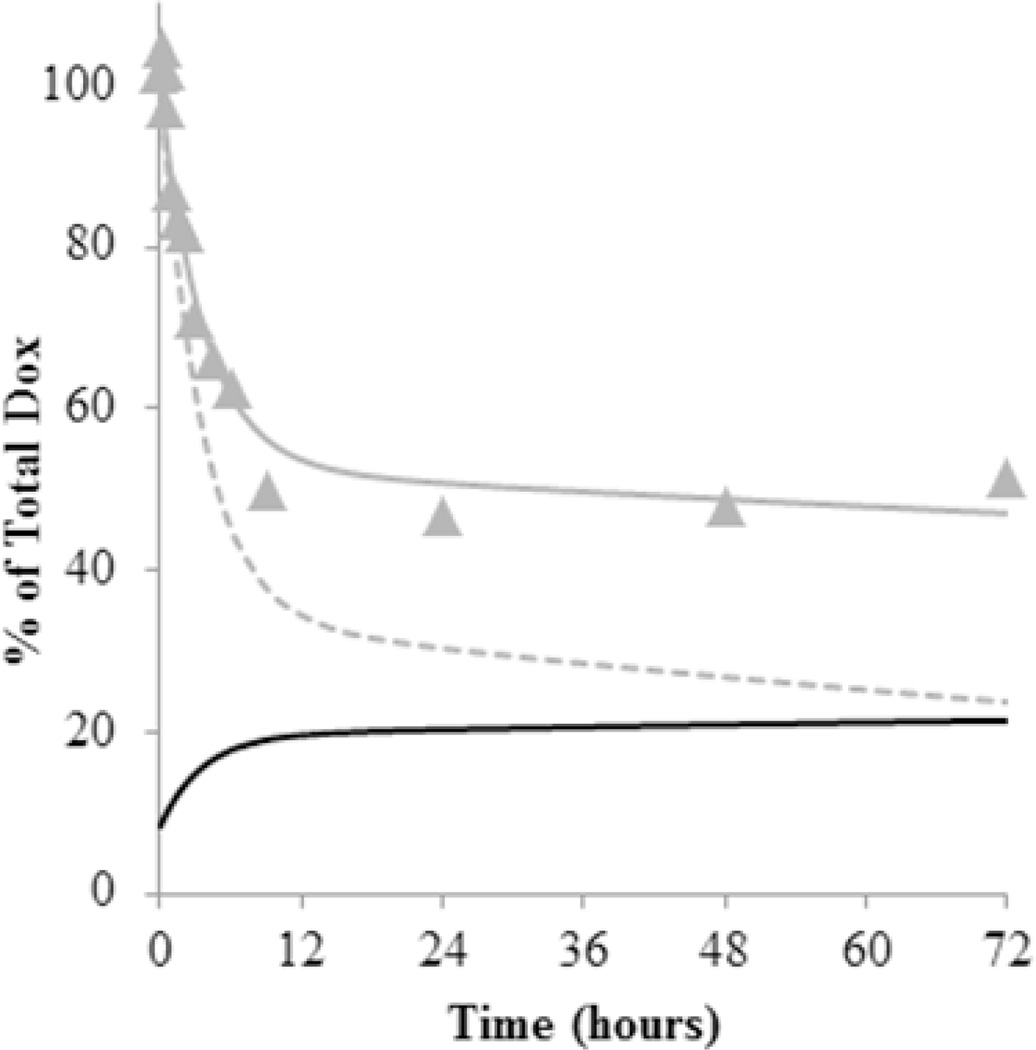
The influence of Dox binding to micelles is illustrated in the simulations shown in Fig. 4. Using the rate constants provided in Table 1, the % of Dox remaining in the ultrafiltered samples analyzed during release are shown along with the fit using the mechanistic model (solid line) and compared to a simulation which assumed binding of unconjugated Dox was negligible (Kp=0). The % of total unconjugated Dox that partitioned into the micelle was also simulated. This clearly shows that partitioning still plays a role in acidic conditions for the HYD-M.
Figure 4.
Simulation of HYD-M (pH 5.0) release profile under non-sink conditions using rate constants in Table 1. The % Dox remaining after ultrafiltration ( ) versus time along with the fit using the mechanistic model (gray line) are compared to a simulation with no partitioning of unconjugated Dox ( =0) into the micelle (gray dotted line). Based on the values in Table 1, the amount of unconjugated Dox that partitioned into the micelle was also simulated as a % of total Dox present in the solution (black line).
) versus time along with the fit using the mechanistic model (gray line) are compared to a simulation with no partitioning of unconjugated Dox ( =0) into the micelle (gray dotted line). Based on the values in Table 1, the amount of unconjugated Dox that partitioned into the micelle was also simulated as a % of total Dox present in the solution (black line).
This partitioning effect in conjunction with the rate of Dox transport through the dialysis membrane has implications on the pH-sensitive nature of the initial phase of release observed from dynamic dialysis. This is illustrated by Fig. 5. Here, the release profile from a 1 mg/mL concentration of GLY-M at pH 7.4 was simulated based on the values reported in Table 1 and compared to the simulated profile of drug remaining in the dialysis tube still conjugated to the block copolymer micelles. It is evident that the lag seen in Dox release is due to the accumulation of free Dox. This is expected based on previous reports of a similar lag in drug release seen with liposomal nanoparticles using dynamic dialysis (30, 38). This effect is exaggerated at pH 7.4 where all formulations have a higher Kp. Substantial partitioning of Dox at pH 7.4 reduces the rate Dox is able to leave the dialysis cassette as a significant portion of unconjugated Dox remains partitioned in the micelles, thus reducing the driving force for Dox diffusion across the dialysis membrane. This is also illustrated by Fig. 5 that compares the release profile from a 1 mg/mL concentration of GLY-M at pH 7.4 with a simulation of this profile if Kp were zero (i.e. no partitioning). In the absence of Dox partitioning, observed release would be faster as the time for the accumulation phase in the dialysis cassette is nearly doubled due to partitioning (Fig. 6).
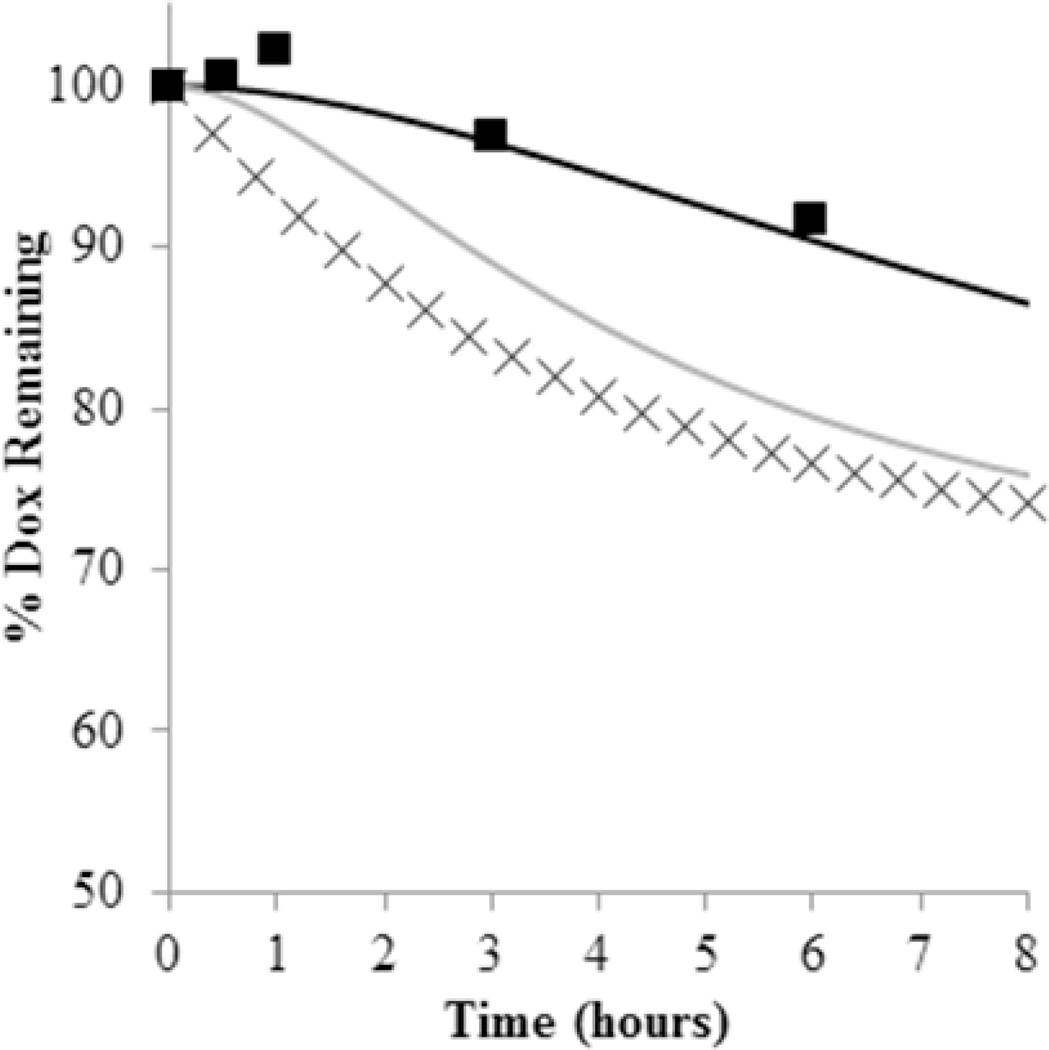
Figure 5.
The effect of micelle/water partitioning of unconjugated Dox in dynamic dialysis release studies. Here, the initial portion of the Dox release profile from a 1.0 mg GLY-M /mL solution at pH 7.4 (■) is shown along with the profile generated by the mathematical model (black line) using the values Table 1. Simulations of the expected release profiles if Kp is assumed to be zero (gray line) and if only conjugated Dox was monitored (×) are also shown to illustrate the lag time in dynamic dialysis release profiles.
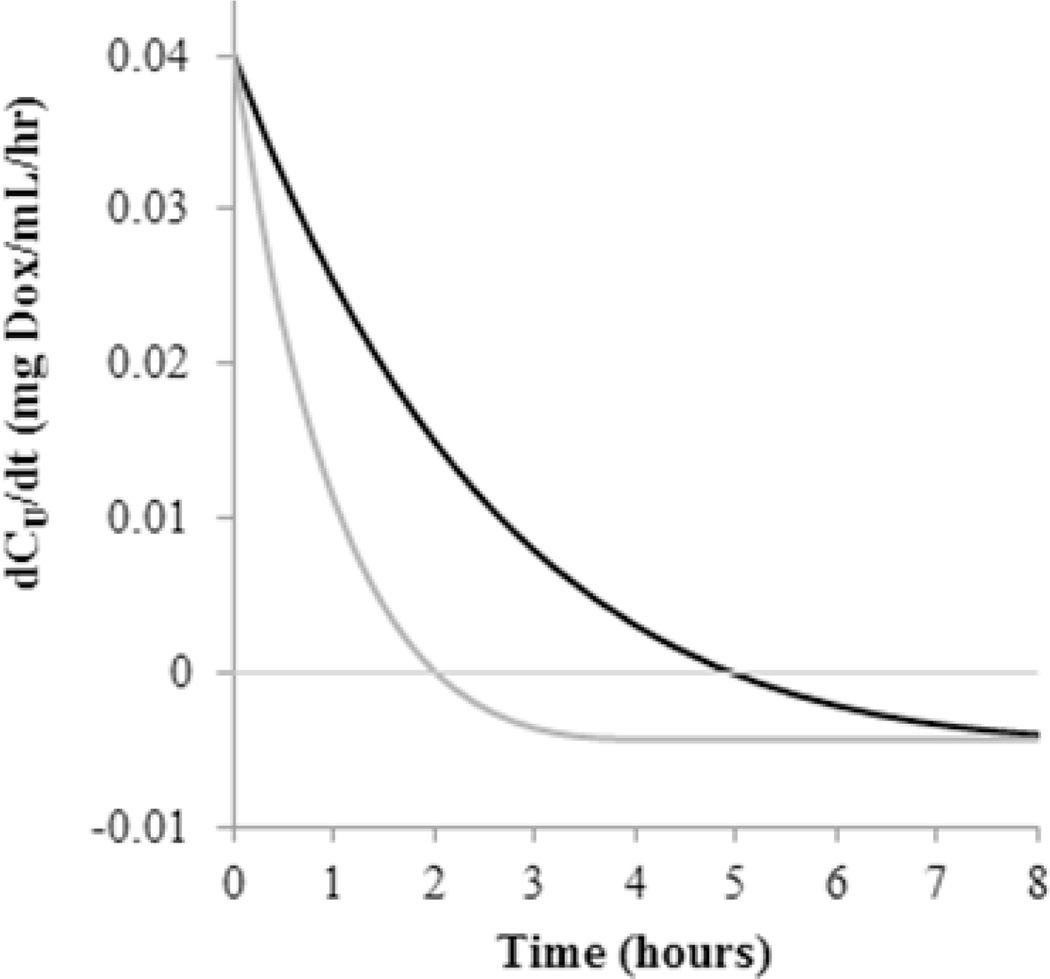
Figure 6.
The rate of free Dox transport within the dialysis cassette during release studies performed with GLY micelles at pH 7.4 is simulated based on the mathematical model and values generated in Table 1. The accumulation phase (positive values) of unconjugated DOX in the dialysis cassette when binding is considered (black line) is over double that if there were no binding (gray line).
All drug release studies using polymer micelles in this study resulted in biphasic drug release profiles. Biphasic drug release has been observed in multiple systems, but there is no consensus on the cause (39–43). The exact reason behind the heterogeneous kinetics described herein has not been confirmed, but multiple hypotheses are considered. First, the need to include two populations of conjugated Dox having different release rates may indicate that hydrazone bond hydrolysis rates vary with location in the micelles. Hydrazone linkages closest to the core/shell interface may be hydrolyzed more readily due to higher water and hydronium ion concentrations. Those further within the core of the micelle hydrolyze more slowly due to the hydrophobic nature of the core. Based on this line of thinking, most of the water/hydronium ion likely resides at the interface of the micelle core and the PEG shell. The hydrophilic nature of PEG would reduce the hydrophobic effect at this interface relative to the core of the micelle, increasing the exposure of conjugates near this interface to water/hydronium ions. Due to the high surface-area to volume ratio associated with spherical geometries, the fraction of conjugated Dox susceptible to fast release, Fkf, would only require the core/shell interface to extend to 7–15% of the total radial distance of the micelle’s core for values of Fkf ranging from 0.2 – to 0.38. Even the value of 0.68 obtained for HYD-M at pH 5.0 would only require the interface to include approximately 30% of the core’s total radial distance. Alternatively, the fast rate of release could represent hydrazone bond hydrolysis from single block copolymers while the slow release rate could represent loss of copolymer from the dialysis cassette. This last hypothesis seems unlikely as the copolymer material used in drug release studies was larger than the MWCO of the cassettes. Because the block copolymers should not be able to dialyze through the cassettes, polymer concentrations should remain high within the cassettes and primarily exist as micelles due to the extremely low critical micelle concentrations for these types of polymer micelle systems (44).
While the fitted curves matched the experimental data, the drug release parameters determined from fitting the data had sufficient variability that it was not always possible to arrive at a clear mechanistic explanation for the modest differences in release profiles. The greater extent of release observed for HYD-M, and the pH-sensitive behavior appeared to be heavily dependent on the relative populations of fast and slow releasing Dox conjugates. One difference noted in the nanoparticles was that ABZ-M and GLY-M were roughly 50 nm with a narrow size distribution, while HYD-M were 100 nm with a much wider size distribution. Generally, a pH effect on drug release after 72 hours was modest, with more Dox released in acidic conditions. The introduction of spacers appeared to lower overall drug release over the time frame of these studies.
CONCLUSIONS
Dox release from HYD-M, ABZ-M, and GLY-M was observed, analyzed, and modeled in acidic and neutral conditions. At pH 5.0, total Dox release at specific time intervals varied depending on micellar formulation (HYD-M >ABZ-M >GLY-M). A mechanistic model was developed to estimate parameters governing Dox release. Modeling results identified a stability issue with partial hydrolysis of the hydrazone linkage used to conjugate Dox to the block copolymers during storage and provided a quantitative correlation between druglinker stability and drug release kinetics. The impact of this finding is significant as researchers considering employing hydrazone linkages for drug conjugation should be cognizant of these stability issues and give careful consideration to validation of conjugate stability at the storage conditions selected. In addition to the stability issue, mathematical modeling also revealed pH-sensitive Dox partitioning was present in all three micelle formulations. This partitioning of unconjugated Dox into the micelle in conjunction with its effect on observed Dox transport in dynamic dialysis studies complicated the interpretation of release kinetics for these micelle formulations. Such observed partitioning effects would be negligible in vivo where sink conditions are maintained and not affected by transport through a dialysis membrane. The approach used here to develop a mechanism-based mathematical model to account for method-specific effects on observed drug release profiles should generally be useful for assessing release characteristics intrinsic to a particular drug/nanoparticle system.
ACKNOWLEDGEMENTS
This work is partially supported by the Kentucky Lung Cancer Research Program and the University of Kentucky Cancer Nanotechnology Training Center (UK-CNTC), grant R25CA153954 from the National Cancer Institute. The content herein is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
REFERENCES
- 1.Barenholz Y. Doxil (R) - The first FDA-approved nano-drug: Lessons learned. J Control Release. 2012;160(2):117–134. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 2.Drummond DC, Noble CO, Hayes ME, Park JW, Kirpotin DB. Pharmacokinetics and in vivo drug release rates in liposomal nanocarrier development. J Pharm Sci. 2008;97(11):4696–4740. doi: 10.1002/jps.21358. [DOI] [PubMed] [Google Scholar]
- 3.Ferrari M. Nat Rev Cancer. Nature Publishing Group; 2005. Cancer nanotechnology: opportunities and challenges; pp. 161–171. [DOI] [PubMed] [Google Scholar]
- 4.Domingo C, Saurina J. An Overview of the Analytical Characterization of Nanostructured Drug Delivery Systems: Towards Green and Sustainable Pharmaceuticals: A Review. Anal Chim Acta. 2012;744(0):8–22. doi: 10.1016/j.aca.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Mehnert W, Mäder K. Solid lipid nanoparticles: Production, characterization and applications. Adv Drug Deliver Rev. 2012;64(Supplement)(0):83–101. doi: 10.1016/s0169-409x(01)00105-3. [DOI] [PubMed] [Google Scholar]
- 6.Cho EJ, Holback H, Liu KC, Abouelmagd SA, Park J, Yeo Y. Nanoparticle Characterization: State of the Art, Challenges, and Emerging Technologies. Mol Pharm. 2013;10(6):2093–2110. doi: 10.1021/mp300697h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Modi S, Anderson BD. Determination of Drug Release Kinetics from Nanoparticles: Overcoming Pitfalls of the Dynamic Dialysis Method. Mol Pharm. 2013;10(8):3076–3089. doi: 10.1021/mp400154a. [DOI] [PubMed] [Google Scholar]
- 8.Fugit KD, Anderson BD. The Role of pH and Ring-Opening Hydrolysis Kinetics on Liposomal Release of Topotecan. J Control Release. 2014;174:88–97. doi: 10.1016/j.jconrel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae Y, Fukushima S, Harada A, Kataoka K. Design of environment-sensitive supramolecular assemblies for intracellular drug delivery: Polymeric micelles that are responsive to intracellular pH change. Angew Chem Int Ed. 2003;42(38):4640–4643. doi: 10.1002/anie.200250653. [DOI] [PubMed] [Google Scholar]
- 10.Bae Y, Nishiyama N, Fukushima S, Koyama H, Yasuhiro M, Kataoka K. Preparation and Biological Characterization of Polymeric Micelle Drug Carriers with Intracellular pH-Triggered Drug Release Property: Tumor Permeability, Controlled Subcellular Drug Distribution, and Enhanced in Vivo Antitumor Efficacy. Bioconjugate Chem. 2004;16(1):122–130. doi: 10.1021/bc0498166. [DOI] [PubMed] [Google Scholar]
- 11.Torchilin VP. Structure and design of polymeric surfactant-based drug delivery systems. J Control Release. 2001;73(2–3):137–172. doi: 10.1016/s0168-3659(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 12.Kozlov MY, Melik-Nubarov NS, Batrakova EV, Kabanov AV. Relationship between Pluronic Block Copolymer Structure, Critical Micellization Concentration and Partitioning Coefficients of Low Molecular Mass Solutes. Macromolecules. 2000;33(9):3305–3313. [Google Scholar]
- 13.Batrakova EV, Kabanov AV. Pluronic block copolymers: Evolution of drug delivery concept from inert nanocarriers to biological response modifiers. J Control Release. 2008;130(2):98–106. doi: 10.1016/j.jconrel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frimpong RA, Fraser S, Zach Hilt J. Synthesis and temperature response analysis of magnetic-hydrogel nanocomposites. J Biomed Mater Res A. 2007;80A(1):1–6. doi: 10.1002/jbm.a.30962. [DOI] [PubMed] [Google Scholar]
- 15.Lu D-X, Wen X-T, Liang J, Zhang X-D, Gu Z-W, Fan Y-J. Novel pH-Sensitive Drug Delivery System Based on Natural Polysaccharide for Doxorubicin Release. Chinese J Polym Sci. 2008;26(3):369–374. [Google Scholar]
- 16.Haran G, Cohen R, Bar LK, Barenholz Y. Transmembrane ammonium sulfate gradients in liposomes produce efficient and stable entrapment of amphipathic weak bases. Biochim Biophys Acta. 1993;1151(2):201–215. doi: 10.1016/0005-2736(93)90105-9. [DOI] [PubMed] [Google Scholar]
- 17.Gaber MH, Wu NZ, Hong K, Huang SK, Dewhirst MW, Papahadjopoulos D. Thermosensitive liposomes: Extravasation and release of contents in tumor microvascular networks. Int J Radiat Oncol. 1996;36(5):1177–1187. doi: 10.1016/s0360-3016(96)00389-6. [DOI] [PubMed] [Google Scholar]
- 18.Kong G, Dewhirst MW. Int J Hyperther. Taylor & Francis Ltd; 1999. Review Hyperthermia and liposomes; pp. 345–370. [DOI] [PubMed] [Google Scholar]
- 19.Ono A, Takeuchi K, Sukenari A, Suzuki T, Adachi I, Ueno M. Reconsideration of Drug Release from Temperature-Sensitive Liposomes. Biological Pharm Bull. 2002;25(1):97–101. doi: 10.1248/bpb.25.97. [DOI] [PubMed] [Google Scholar]
- 20.Joguparthi V, Feng S, Anderson BD. Determination of Intraliposomal pH and its Effect on Membrane Partitioning and Passive Loading of a Hydrophobic Camptothecin, DB-67. Int J Pharm. 2008;352(1–2):17–28. doi: 10.1016/j.ijpharm.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai L-A, Tsai P-J, Wang Y-C, Wang Y-J, Lo L-W, Yang C-S. Thermosensitive liposomes entrapping iron oxide nanoparticles for controllable drug release. Nanotechnology. 2009;20(13):9. doi: 10.1088/0957-4484/20/13/135101. [DOI] [PubMed] [Google Scholar]
- 22.Thomas CR, Ferris DP, Lee J-H, Choi E, Cho MH, Kim ES, Stoddart JF, Shin J-S, Cheon J, Zink JI. Noninvasive Remote-Controlled Release of Drug Molecules in Vitro Using Magnetic Actuation of Mechanized Nanoparticles. J Am Chem Soc. 2010;132(31):10623–10625. doi: 10.1021/ja1022267. [DOI] [PubMed] [Google Scholar]
- 23.Amstad E, Kohlbrecher J, Müller E, Schweizer T, Textor M, Reimhult E. Triggered Release from Liposomes through Magnetic Actuation of Iron Oxide Nanoparticle Containing Membranes. Nano Lett. 2011;11(4):1664–1670. doi: 10.1021/nl2001499. [DOI] [PubMed] [Google Scholar]
- 24.Bae Y, Kataoka K. Intelligent polymeric micelles from functional poly(ethylene glycol)-poly(amino acid) block copolymers. Adv Drug Deliver Rev. 2009;61(10):768–784. doi: 10.1016/j.addr.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 25.Ponta A, Bae Y. PEG-poly(amino acid) block copolymer micelles for tunable drug release. Pharm Res. 2010;27(11):2330–2342. doi: 10.1007/s11095-010-0120-z. [DOI] [PubMed] [Google Scholar]
- 26.Lee HJ, Bae Y. Pharmaceutical differences between block copolymer self-assembled and cross-linked nanoassemblies as carriers for tunable drug release. Pharm Res. 2013;30(2):478–488. doi: 10.1007/s11095-012-0893-3. [DOI] [PubMed] [Google Scholar]
- 27.West KR, Otto S. Reversible covalent chemistry in drug delivery. Curr Drug Discov Technol. 2005;2(3):123–160. doi: 10.2174/1570163054866882. [DOI] [PubMed] [Google Scholar]
- 28.Baker MA, Gray BD, Ohlsson-Wilhelm BM, Carpenter DC, Muirhead KA. Zyn-Linked colchicines: controlled-release lipophilic prodrugs with enhanced antitumor efficacy. J Control Release. 1996;40(1–2):89–100. [Google Scholar]
- 29.Etrych T, Chytil P, Jelinkova M, Rihova B, Ulbrich K. Synthesis of HPMA copolymers containing doxorubicin bound via a hydrazone linkage. Effect of spacer on drug release and in vitro cytotoxicity. Macromol Biosci. 2002;2(1):43–52. [Google Scholar]
- 30.Fugit KD, Anderson BD. Dynamic, Non-sink Method for the Simultaneous Determination of Drug Permeability and Binding Coefficients in Liposomes. Mol Pharm. 2014 doi: 10.1021/mp400765n. Article ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponta A, Bae Y. Tumor-preferential sustained drug release enhances antitumor activity of block copolymer micelles. J Drug Target. 0(0):1–10. doi: 10.3109/1061186X.2014.910793. [DOI] [PubMed] [Google Scholar]
- 32.Barbour N, Paborji M, Alexander T, Coppola W, Bogardus J. Stabilization of Chimeric BR96-Doxorubicin Immunoconjugate. Pharm Res. 1995;12(2):215–222. doi: 10.1023/a:1016274825322. [DOI] [PubMed] [Google Scholar]
- 33.Moreno-Bautista G, Tam KC. Evaluation of dialysis membrane process for quantifying the in vitro drug-release from colloidal drug carriers. Colloid Surface A. 2011;389(1–3):299–303. [Google Scholar]
- 34.Gupta PK, Hung CT, Perrier DG. Quantitation of the release of Doxorubicin from colloidal dosage forms using dynamic dialysis. J Pharm Sci. 1987;76(2):141–145. doi: 10.1002/jps.2600760211. [DOI] [PubMed] [Google Scholar]
- 35.Washington C. Evaluation of non-sink dialysis methods for the measurement of drug release from colloids: effects of drug partition. Int J Pharm. 1989;56(1):71–74. [Google Scholar]
- 36.Washington C. Drug release from microdisperse systems: a critical review. Int J Pharm. 1990;58(1):1–12. [Google Scholar]
- 37.Sturgeon RJ, Schulman SG. Electronic absorption spectra and protolytic equilibria of doxorubicin: direct spectrophotometric determination of microconstants. J Pharm Sci. 1977;66(7):958–961. doi: 10.1002/jps.2600660714. [DOI] [PubMed] [Google Scholar]
- 38.Joguparthi V, Xiang T-X, Anderson BD. Liposome Transport of Hydrophobic Drugs: Gel Phase Lipid Bilayer Permeability and Partitioning of the Lactone Form of a Hydrophobic Camptothecin, DB-67. J Pharm Sci. 2008;97(1):400–420. doi: 10.1002/jps.21125. [DOI] [PubMed] [Google Scholar]
- 39.Li F, Danquah M, Mahato RI. Synthesis and characterization of amphiphilic lipopolymers for micellar drug delivery. Biomacromolecules. 2010;11(10):2610–2620. doi: 10.1021/bm100561v. [DOI] [PubMed] [Google Scholar]
- 40.Sutton D, Wang SH, Nasongkla N, Gao JM, Dormidontova EE. Doxorubicin and betalapachone release and interaction with micellar core materials: Experiment and modeling. Exp Biol Med. 2007;232(8):1090–1099. doi: 10.3181/0702-RM-31. [DOI] [PubMed] [Google Scholar]
- 41.Kwon G, Naito M, Yokoyama M, Okano T, Sakurai Y, Kataoka K. Block copolymer micelles for drug delivery: loading and release of doxorubicin. J Control Release. 1997;48(2–3):195–201. [Google Scholar]
- 42.Guo X, Shi C, Wang J, Di S, Zhou S. pH-triggered intracellular release from actively targeting polymer micelles. Biomaterials. 2013;34(18):4544–4554. doi: 10.1016/j.biomaterials.2013.02.071. [DOI] [PubMed] [Google Scholar]
- 43.Lee SJ, Bae Y, Kataoka K, Kim D, Lee DS, Kim SC. In vitro release and in vivo anti-tumor efficacy of doxorubicin from biodegradable temperature-sensitive star-shaped PLGA-PEG block copolymer hydrogel. Polym J. 2008;40(2):171–176. [Google Scholar]
- 44.La SB, Okano T, Kataoka K. Preparation and characterization of the micelle-forming polymeric drug indomethacin-incorporated poly(ethylene oxide)–poly(β-benzyl L-aspartate) block copolymer micelles. J Pharm Sci. 1996;85(1):85–90. doi: 10.1021/js950204r. [DOI] [PubMed] [Google Scholar]