Abstract
Infrared spectroscopy has played an instrumental role in studying a wide variety of biological questions. However, in many cases it is impossible or difficult to rely on the intrinsic vibrational modes of biological molecules of interest, such as proteins, to reveal structural and/or environmental information in a site-specific manner. To overcome this limitation, many recent efforts have been dedicated to the development and application of various extrinsic vibrational probes that can be incorporated into biological molecules and used to site-specifically interrogate their structural and/or environmental properties. In this Review, we highlight some recent advancements of this rapidly growing research area.
Keywords: Infrared spectroscopy, site-specific probe, vibrational coupling, isotope-editing, unnatural amino acid, Vibrational Stark Effect spectroscopy
1. INTRODUCTION
Molecular vibrations occur on the femtosecond timescale and their frequencies depend on the spatial arrangement and identities of the constituent atoms as well as the multiplicity of the related chemical bonds. Therefore, vibrational spectroscopy has played an indispensable role in the study of structure and dynamics in a wide variety of molecular systems in the gas phase. However, for peptides and proteins in the condensed phase, various inter- and intra-molecular interactions, vibrational couplings, and energy degeneracies often render the intrinsic vibrational modes, such as those arising from polypeptide backbone units, difficult or even impossible to use for providing site-specific information regarding the structure and dynamics of the system in question. For this reason, many past and current efforts have been dedicated to alleviating this limitation and to enhancing the structural and spatial resolution of vibrational spectroscopy in the study of biological molecules, especially proteins (1, 2). In this Review, we will focus on recent advances in the development of site-specific infrared (IR) probes and their applications to a variety of systems and problems.
As pointed out by Getahun et al. (3), for a vibrational mode to be useful as an ideal site-specific probe of local structure, solvation status, electrostatics, and dynamics of proteins, it should meet several criteria. First and foremost, it must exhibit a strong dependence on the physical property of interest, such as the local electric field. In addition, it should be a simple, localized transition that is only sensitive to its immediate environment, since vibrational couplings and spectral overlap can complicate interpretation of the results. Furthermore, it should have a strong transition dipole moment so that low sample concentrations can be used. Also, it should be located in an uncongested region of the IR spectrum of the system of interest where the solvent also has a relatively low absorbance. Additionally, an extrinsic probe should cause minimal structural perturbation to the original system. Finally, for any extrinsic probe to be useful in practical applications, a method must exist to incorporate it into the system of interest. While this is an important and rapidly advancing area, a detailed discussion of how to site-specifically incorporate extrinsic spectroscopic probes into proteins and peptides is beyond the scope of this Review. However, we would like to note that the commonly-used methods to do so include solid-phase peptide synthesis (4), amber codon suppression (5), amino acid replacement (6), native chemical ligation (7), and cysteine alkylation (8); we refer the interested reader to the relevant literature.
Here, we will review recent developments and applications of site-specific IR probes of proteins via the following categories: backbone-based and sidechain-based probes. In addition, we will describe, wherever appropriate, how some of these IR probes could be utilized to increase the structural and spatial resolution of linear and nonlinear IR spectroscopic measurements of proteins.
2. BACKBONE-BASED IR PROBES
Vibrational modes of the protein backbone encode a wealth of structural information. In particular, the amide I mode (1600–1700 cm−1), which arises mainly from the C=O stretching vibration, has been widely used in protein conformational studies owing to its sensitivity to various structural determinants. Since this topic has been extensively reviewed (9–15), here we will describe only a few recent examples, showing the utility of this vibrational mode as a site-specific environmental and/or structural reporter of proteins.
Isotope-editing is currently the method of choice for introducing a site-specific amide I probe into the protein backbone, which replaces a 12C=16O group with either 13C=16O or 13C=18O (16–21). Such an isotopic substitution induces a red-shift of the fundamental frequency of the amide I vibrator of interest, which is often sufficient to break up any couplings with other amide I modes, thus making the isotope-edited C=O group a vibrational reporter of local structures and environments. Since the amide I band of an isotope-edited backbone carbonyl unit could overlap with vibrational bands arising from various sidechains, such as arginine, glutamic acid and aspartic acid, care needs to be taken when applying this method. In addition, due to the natural abundance of 13C, the 13C=16O labeling method may not be applicable to large proteins. Despite these drawbacks, the isotope-edited amide I mode has been employed in numerous studies of protein local environments and conformational changes.
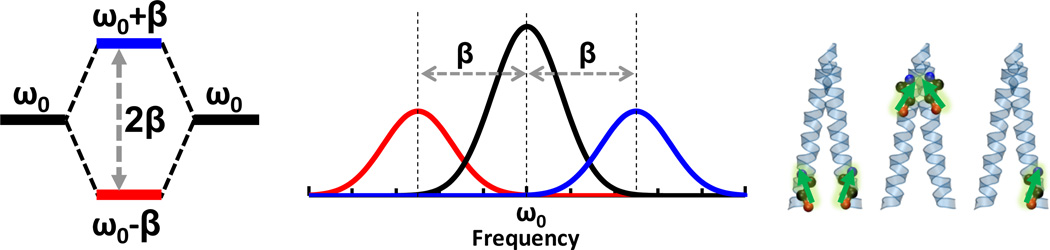
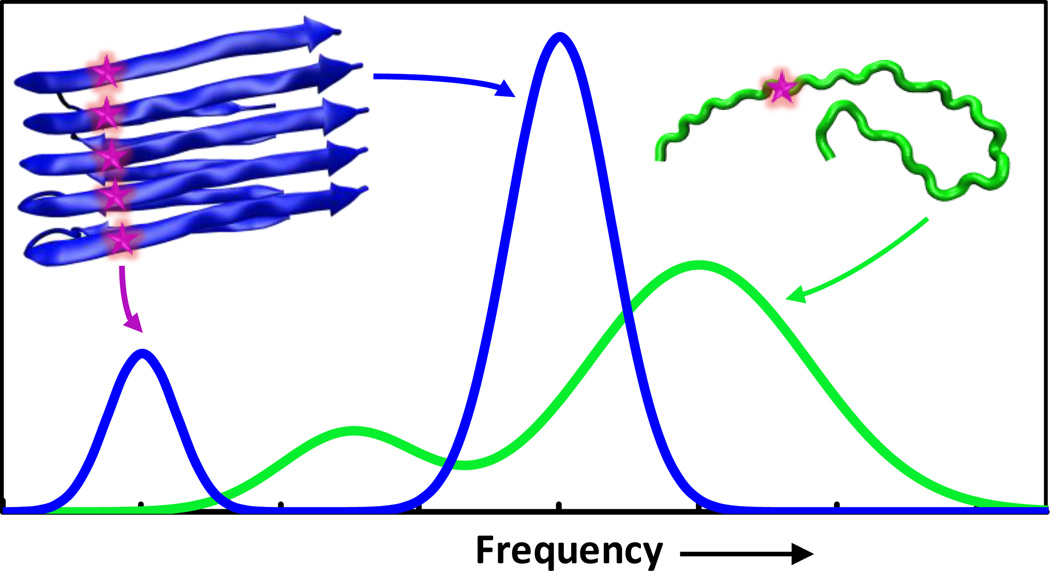
In practice, the IR applications of the aforementioned isotope-editing strategy can be divided into two major classes. In the first class, the labeled C=O group is used as a stand-alone probe of the local structure and environment of proteins (22–26). Recent examples of this include studies of local hydration dynamics (27–29), protein folding thermodynamics and kinetics (30), ligand binding (31–33), peptide-membrane interactions (34, 35), and membrane peptide structure determination (36). In the second class, vibrational coupling between two or more labeled carbonyl groups is used to provide protein structural information at the secondary or tertiary level. An outstanding example demonstrating the power of this coupling strategy is the work of Hochstrasser and coworkers (37, 38), which showed that by assessing the vibrational dipole-dipole couplings of a set of well chosen pair of 13C=18O amide I vibrational modes via two dimensional infrared (2D IR) echo spectroscopy, it is possible to determine the three dimensional structure of a transmembrane (TM) helix dimer. As indicated (Fig. 1), this method is based on the idea that coupling between two degenerate amide I modes, the strength of which depends on distance, orientation and fundamental frequencies, leads to energy splitting and, thus, assessment of this coupling can yield structural information. In other words, by systematically labeling pairs of amide positions through the dimer contact region of the peptide sequence, one can obtain a large number of structural constraints, which, in conjunction with molecular dynamics (MD) simulations, can be used to generate a structural model for the molecule of interest. In principle, this vibrational coupling strategy can be applied to any peptide or protein system to yield single-residue level structural information, especially in structural regions where the dipole-dipole interaction mechanism is solely or predominantly responsible for the resultant coupling between the two isotope-edited vibrators. In a similar application, Zanni and coworkers (39) demonstrated that by strategically placing isotope-edited backbone labels in an aggregation-prone peptide, vibrational coupling of the labeled residues could be used as an indicator of β-sheet formation. As shown (Fig. 2), this method is based on the notion that excitonic coupling among isotope labels on different peptides is a manifestation of the underlying spatial arrangement of these molecules and hence the structure of the aggregates. Using this strategy, Zanni and coworkers studied the aggregation mechanism of human islet amyloid polypeptide (hIAPP) and were able to identify a β-sheet-like intermediate composed of several residues that are disordered in the structure of fully-formed fibril. Similarly, Zanni and coworkers have also applied this strategy to elucidate the aggregation mechanism of other systems (40), including polyglutamine sequences (26), and gamma D-crystallin (41–44).
Figure 1.
Schematic illustration of the coupling strategy (i.e., isotope dilution method) used by Hochstrasser and coworkers (37) for protein structure determination. Energy level splitting due to coupling between two vibrational transitions that are degenerate in energy (left), which is manifested with the appearance of two new peaks that are split in energy (middle). For a helical dimer (right), it is possible to obtain structural constraints based upon the position of the vibrational resonant pair and the experimentally measured coupling parameters.
Figure 2.
Schematic illustration of the coupling strategy used by Zanni and coworkers (39). Upon formation of repeating, ordered parallel β-sheet fibrils, the site-specifically incorporated backbone 13C=18O probe (represented by a star) on one peptide will be brought in close proximity to those bore on other peptides, leading to excitonic coupling among these vibrators. As a result, the IR spectrum of the peptide sample undergoes a redshift, amplification and narrowing; by changing the positions of the isotopic labels and monitoring the appearance of the spectral feature of the exciton band, it is possible to elucidate the structure of the formed fibrils.
Similar to the coupling strategies discussed above, vibrational energy transfer between two site-specific vibrators could also be used to elucidate molecular structures or structural changes. As such, many studies have focused on determining and understanding the physical principles underlying this process (45–50). However, providing an in-depth account of this rapidly expanding field of research is beyond the scope of this Review.
When combined with a fast initiation or triggering method, the isotope-editing strategies discussed above can be extended to acquire transient structural information of proteins at the single-residue level. For example, Hamm and coworkers (51, 52) used the photoswitchable isomerization of an azobenzene cross-linker to control the folding-unfolding equilibrium of a helical peptide and used isotope-edited amide I labels to assess the heterogeneity of its folding kinetics. Tokmakoff and coworkers (53) have shown that the combination of the laser-induced temperature-jump (T-jump) technique (54–56) with isotope-editing, 2D IR spectroscopy, and MD simulations can allow for a more detailed assessment of protein folding pathways and, in some cases, identification of previously undetected folding intermediates. In a more recent study, Hochstrasser and coworkers (57) demonstrated the feasibility of using transient 2D IR measurements to follow the local structural evolution of a distorted α-helix on the picosecond timescale, initiated by photocleavage of a tetrazine cross-linker. More specifically, using experimentally-determined vibrational coupling information from two isotope-edited amide C=O groups that were located at both ends of the tetrazine moiety, the authors were able to obtain site-specific structural constraints of the isotope-edited amide I pair such as dihedral angle and distance and then used these time-dependent structural information to create molecular snapshots to follow the early conformational events in a “downhill” folding process. Taken together, these examples demonstrate the power of combining isotope-editing with transient 2D IR spectroscopy to elucidate structural and mechanistic details of protein conformational dynamics. With the further development and/or refinement of new and existing conformational triggering and detection methods, we believe that this methodology is capable of yielding ‘structure snapshots’ along the ‘reaction’ coordinate of interest.
The isotope-edited amide I mode will continue to be used to study various biological systems and questions due to its high extinction coefficient, minimal perturbation to protein native structures and relative ease of incorporation. Besides isotope-edited amide carbonyls, other potentially useful site-specific backbone IR probes include thioamides (58), selenoamides (59), and backbone esters (60, 61). While the utility of these moieties as vibrational probes has yet to be confirmed, the fact that they all moderately perturb the native structure of the peptide backbone will certainly limit their applications. On the other hand, we believe that these backbone-based probes are potentially useful in situations where such perturbations are either tolerable or desirable. Therefore, the popularity of this class of probes will continue to grow as future biological questions call for new methods of investigation.
3. SIDECHAIN-BASED IR PROBES
The unique chemical structure and identity of the protein backbone limit the diversity of potential backbone-based IR probes. On the other hand, the chemical diversity of protein sidechains makes it possible to create a large array of sidechain-based IR probes. In addition, since in vitro and in vivo methods for incorporating unnatural amino acids into proteins are becoming more refined and easier to use, the past few years have seen a rapid increase in the employment of various sidechain-based IR probes to study a wide variety of biological questions, ranging from protein folding to enzymatic reactions. For easy comparison, we summarize the basic spectroscopic properties of these probes in Table 1. Through selective examples, below we will showcase some of their most recent applications.
TABLE 1.
Overview of Site-Specific IR Probes
| Name | Frequency Range (cm−1) |
Extinction in H2O (M−1cm−1) |
Ref. | |
|---|---|---|---|---|
| Nitriles | p-cyano-phenylalanine | 2220–2250 | ~220 | (3) |
| Cyano-cysteine | 2150–2180 | ~120a | (147) | |
| 5-cyano-tryptophan | 2210–2240 | ~160b | (63) | |
| Cyanate | 2220–2300 | ~800c | (106) | |
| Azides | Azidohomoalanine | 2100–2140 | 350–400d | (113) |
| p-azido-phenylalanine | 2100–2140 | ~610 | (114) | |
| 3-picolyl azide adenine dinucleotide | 2080–2160 | ~2000 | (148) | |
| Carbonyls | Ketone carbonyl | 1660–1700 | ~1800e | (125) |
| Ester carbonyl | 1690–1770 | ~290 | (126) | |
| Carboxylic acid | 1700–1775 | ~280 | (137) | |
| Carboxylate | 1555–1600 | ~820 | (149) | |
| Metal carbonyls | CpRe(CO)3 | 2010–2030 | ~4100 | (145) |
| CORM-2 | 2040–2100 | NA | (142, 143) | |
| Others | Carbon deuterium | 2100–2400 | 5–10 | (150) |
| Cysteine thiol | 2550–2600 | 5–150 | (138) | |
| Phosphate | 1200–1300 | ~500 | (139) | |
| Fluorocarbon | 1200 | ~700e | (140) |
Solvent conditions other than H2O are indicated:
5050 (v/v) glycerol/water,
60/40 (v/v) water/THF,
THF,
D2O, and
2-MeTHF
3.1 C≡N Stretching Vibration
The frequency of the nitrile (C≡N) stretching vibration is in the range of 2100–2400 cm−1, a transparent region of the protein IR spectrum (62). In addition, various nitrile-containing unnatural amino acids are commercially available. Thus, the C≡N stretching vibration is among the very first sidechain-based extrinsic IR probes to be exploited to study biological processes. The extinction coefficient of the C≡N stretching vibration of alkyl nitriles in water is ~50 cm−1 M−1 (3), whereas that of aromatic nitriles is significantly larger (63–67). For example, the molar extinction coefficient of the C≡N stretching vibration of p-cyano-phenylalanine (PheCN) in water is ~220 cm−1 M−1. As such, PheCN has become one of the most-widely used vibrational probes (68–71). Another common nitrile vibrational probe is thiocyanate (SCN), owing to its ease of incorporation into proteins via chemical modification of solvent-exposed cysteine sidechains (72–76). Below, we will present a few recent works to highlight the use of the C≡N stretching vibration as a site-specific IR probe to study various biological questions (77–79).
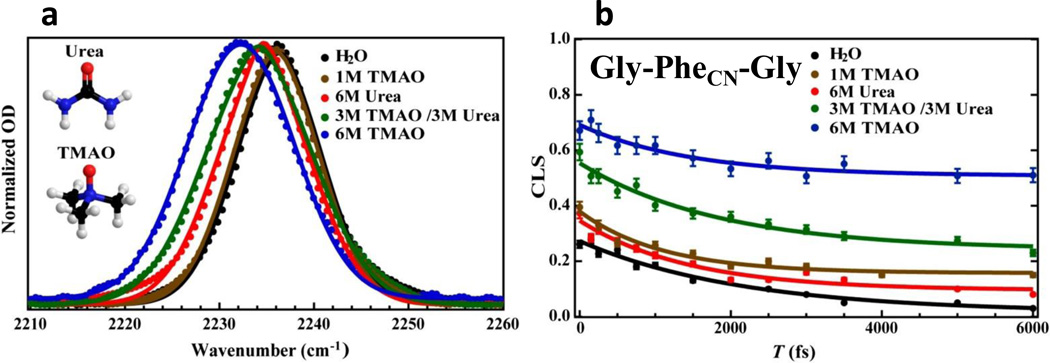
The first example demonstrates the novel use of this vibrational probe in elucidating the microscopic details underlying the protective action of a naturally occurring osmolyte, trimethylamine N-oxide (TMAO) (80). As indicated (Fig. 3), the study of Ma et al. (81) clearly shows that upon addition of TMAO the C≡N stretching frequency of PheCN in both a small peptide and a protein is red-shifted, which implies that the hydrogen bonds (H bonds) formed between the nitrile group and water are weakened. Using 2D IR spectroscopy, the authors further characterized the spectral diffusion dynamics of the C≡N stretching vibration. As shown (Fig. 3), the exponential decay component, which reflects the timescale of H bond fluctuations, becomes faster in the presence of TMAO, consistent with the notion that TMAO weakens the ability of water to form H bonds with the polar groups of a protein. What is more interesting, however, is that the static component, which arises from motions that are too slow to be resolved in these experiments, becomes larger with increasing TMAO concentration. In other words, a high concentration of TMAO makes the peptide become more rigid. A full analysis of the linear IR bands obtained in the presence and absence of TMAO using linear response theory also supports this picture. Based on these findings, the authors concluded that the protein-stabilizing ability of TMAO is realized through a combination of an enthalpy-driven effect, which weakens water-protein H bonds, and an entropy-driven or excluded volume effect, whereby TMAO acts as a nano-crowder (82).
Figure 3.
(a) The C≡N stretching vibrational bands of a tripeptide, Gly-PheCN-Gly, obtained under different solvent conditions, showing the sensitivity of this vibration to environment. (b) Effect of solvent condition on the spectral diffusion dynamics of the C≡N stretching vibration of the tripeptide, showing that TMAO can significantly increase the static component and hence the conformational rigidity of the peptide. These figures are adapted with permission from (81).
Next, we describe examples showing how the C≡N stretching vibration can be used to interrogate the local electric field environment of proteins and, in particular, the role of electrostatics in determining the catalytic activity of enzymes (83–85). In one study, Herschlag and coworkers (86) used Vibrational Stark Effect (VSE) spectroscopy with the SCN probe, in conjunction with NMR measurements, to characterize how modulating the charge of a bound ligand (i.e., phenol) affects the local electrostatic environment in the active site of the bacterial enzyme ketosteroid isomerase. Specifically, by analyzing the lineshape of the C≡N stretching vibration as well as the chemical shift of the nitrile moiety placed in the protein's binding pocket, they were able to develop a model to quantitatively determine the protonation states of the ligand and tyrosine residues in the enzyme active site in a site-specific manner, and account for how the protonation states of these moieties change as a function of the charge density of the ligand. In a similar application, Benkovic and coworkers (87) employed SCN to study the role of electrostatics at each stage of the catalytic cycle of dihydrofolate reductase (DHFR). Through a combination of IR and NMR measurements of the SCN probe and with the help of MD simulations and theoretical calculations, they were able to determine the changes in the structure and local electric field magnitude of the enzyme active site along the protein’s catalytic cycle. They found that variations in the local electric field due to changes in protein conformation play a large role in the hydride transfer ability of DHFR. Both of the examples discussed above highlight the utility of the C≡N stretching vibration in probing the local electrostatic environment of an enzyme active site. However, the described experiments were carried out under equilibrium conditions. Thus, an important extension of the method discussed above would be to perform time-resolved measurements to obtain kinetic insights into the role of electrostatics in enzyme catalysis.
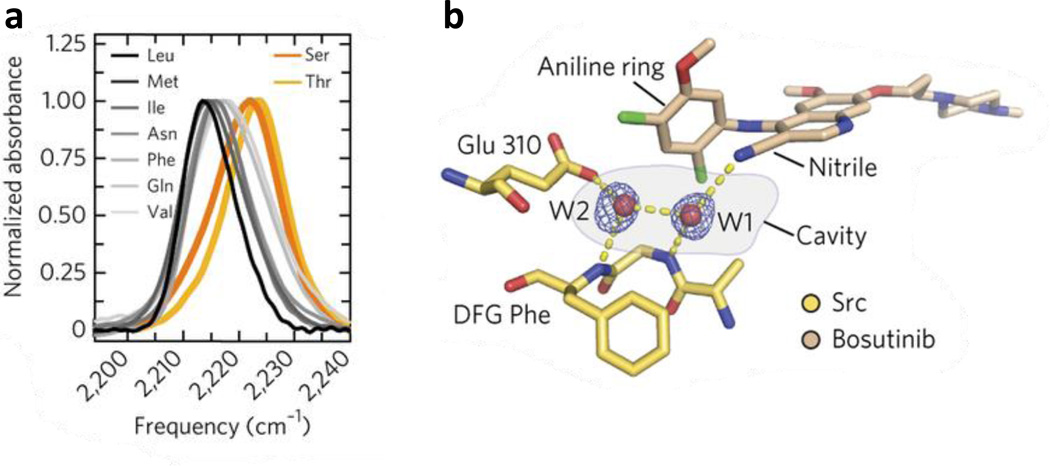
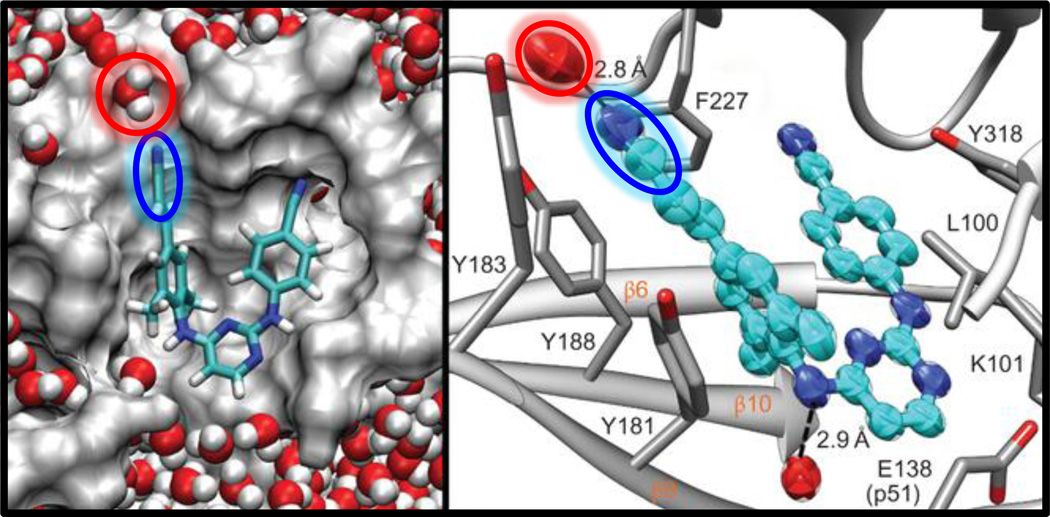
The C≡N stretching vibration of nitriles has also found unique use in investigating the mechanism of ligand-protein interactions, especially in the context of drug binding and recognition. For example, Boxer and coworkers (88) were able to take advantage of the single C≡N group of bosutinib, a Src kinase inhibitor, and used it as an IR reporter to probe the molecular determinants for recognition and binding of this drug to Src kinase (Fig. 4). Based on extensive mutagenesis, IR measurements and the crystal structures of several variants of Src kinase, the authors were able to provide new insights into the role of a key residue, known as the gatekeeper residue, in determining and selecting for inhibitor binding. Also, they identified an additional part of the binding pocket that contains two structured water molecules, which form H bonds with the drug as well as with the binding pocket and thus connect it to a larger H-bonding network that collectively facilitates the underlying protein-drug binding interactions. The important role of structured water in drug binding was also studied by Hochstrasser and coworkers (89, 90), who examined the spectral diffusion dynamics of two nitrile vibrators in an anti-HIV drug, rilpivirine, when it is complexed with HIV-1 reverse transcriptase (RT). Their 2D IR results indicate that the C≡N group of the cinnamonitrile arm of rilpivirine is H-bonded to a structured water molecule close to the binding pocket (Fig. 5). It was proposed that this H bond plays an important role in stabilizing the bound drug because this water molecule also forms H bonds with residues in the protein binding site, thereby linking the drug to a larger H-bonding network. Furthermore, they speculated that the tolerance of rilpivirine to mutations in RT is due to the stabilization effect of this H-bonding network. Since many drug molecules contain the C≡N group and water molecules are frequently found in protein binding pockets, we believe that the methods discussed above can be extended to study the mechanism of ligand or drug binding of other biological systems, such as the Influenza A M2 proton channel (28, 91).
Figure 4.
(a) The C≡N stretching vibrational bands of bosutinib bound to WT Src (yellow) and eight gatekeeper mutants (black, gray and orange). (b) View of the water-mediated hydrogen-bond network in the binding site, with hydrogen bonds shown as dashed yellow lines. Figure adapted with permission from Levinson et al. (88).
Figure 5.
Cartoon representation of the rilpivirine-RT complex, showing the interaction between water (red) and a site-specific nitrile probe (blue) on the drug, determined from MD simulation (left) and X-ray crystallography (right). Figure adapted with permission from Kuroda et al. (90).
Beside the examples discussed above, there are many other interesting and important biological applications of the nitrile probe (69, 92–97). In addition, the effect of the local electric field and hydration on the C≡N stretching frequency has also been extensively studied by means of theoretical and computational approaches (98–100). Due to space limitations, we are unable to describe these studies in detail and would like to refer the interested reader to relevant literature and two of the most recent reviews on nitrile probes (78, 98). Moreover, we would like to point out that aromatic nitriles, such as PheCN, also exhibit useful fluorescence properties, and some of them have been used to study protein folding and binding interactions (101–105). Another point worth mentioning is that some nitrile-containing chemical moieties, such as SeCN and OCN, have unusually long vibrational lifetimes (106, 107), making them appealing candidates for studying dynamic events occurring on the timescale of tens and even hundreds of picoseconds.
3.2 Azide Stretching Vibration
Similar to the C≡N stretching vibration, the asymmetric stretching vibration of azides (R-N3) has also been actively used as a local environmental probe (108–116). Compared to that of nitriles, the azide stretching vibration has a larger extinction coefficient, making it more desirable when low sample concentrations are required. In addition, due to its utility in click chemistry, many methods are available to introduce an azide into proteins and several unnatural amino acids containing the azide moiety are commercially available. On the other hand, the azide stretching frequency shows a lesser dependence on solvent than that of C≡N and, in many cases, its vibrational band is accompanied by features arising from Fermi resonances, making it less attractive in certain applications (117). Examples that illustrate the IR applications of azide probes include using 1) azidohomoalanine to probe differences in folding rates of the protein backbone versus sidechains (118), 2) p-azido-l-phenylalanine to investigate the structures of rhodopsin photointermediates (119), and 3) 3-picolyl azide adenine dinucleotide to examine the local environment and dynamics of the formate dehydrogenase enzyme active site (120).
3.3 C=O Stretching Vibration
The carbonyl stretching vibration of either free or metal-bound carbon monoxides has long been used in protein conformational studies, owing to its large extinction coefficient and sensitivity to local electrostatic and hydration environment. These spectral properties are expected to be conserved for the C=O stretching vibration of carboxylic acid, ketone and ester. For example, the frequency of the C=O stretching mode of acetone is significantly red-shifted in water compared to that in aprotic solvents (121). However, in comparison to the numerous applications of the backbone carbonyl stretching vibration in biological studies, only recently has this vibrational mode gained attention. Below we will discuss several recent studies, highlighting the applicability of this IR probe to addressing various biophysical and biochemical questions and, in particular, to assessing local electric field of proteins.
The sidechain-based C=O probe naturally occurs in several amino acids, including aspartic acid (Asp) and glutamic acid (Glu) residues. It is well known that the C=O stretching frequency of Asp and Glu depends on the protonation status of the respective carboxylate (i.e., 1550–1600 cm−1 for the carboxylate anion and 1700–1775 cm−1 for the carboxylic acid). Thus, this vibrational frequency has been widely used as a convenient marker of the protonation status of these sidechains. In an early study, Xie and coworkers (122) explored the possibility of using the C=O stretching frequency of Asp and Glu to determine their H-bonding status. By examining 31 examples of buried and protonated Asp or Glu residues in proteins such as bacteriorhodopsin and cytochrome c oxidase, they concluded that most buried COOH groups only form one H bond with other protein groups and that the C=O stretching frequency of a given carboxylic acid sidechain is a robust indicator of the number of H bonds thus formed. More interestingly, their results suggest that the C=O stretching frequency of the COOH group in proteins, which is in the range of 1700–1775 cm−1, is linearly dependent on the H-bonding energy. Another application of this vibrational mode was recently demonstrated by Hochstrasser and coworkers (123), who showed, using 2D IR spectroscopy, that the C=O stretching vibration of a specific carboxylate sidechain in a given protein can selectively couple to the amide I mode of specific residues in close proximity and, thus, the resultant coupling patterns could be used to provide structural information. Additionally, Culik et al. (124) recently showed that the carboxylate ion IR mode can be used as a convenient spectroscopic marker of ionic interactions (i.e., salt bridge formation) and utilized it to probe the kinetics of a key sidechain-sidechain interaction in the folding process of a mini-protein, Trp-Cage.
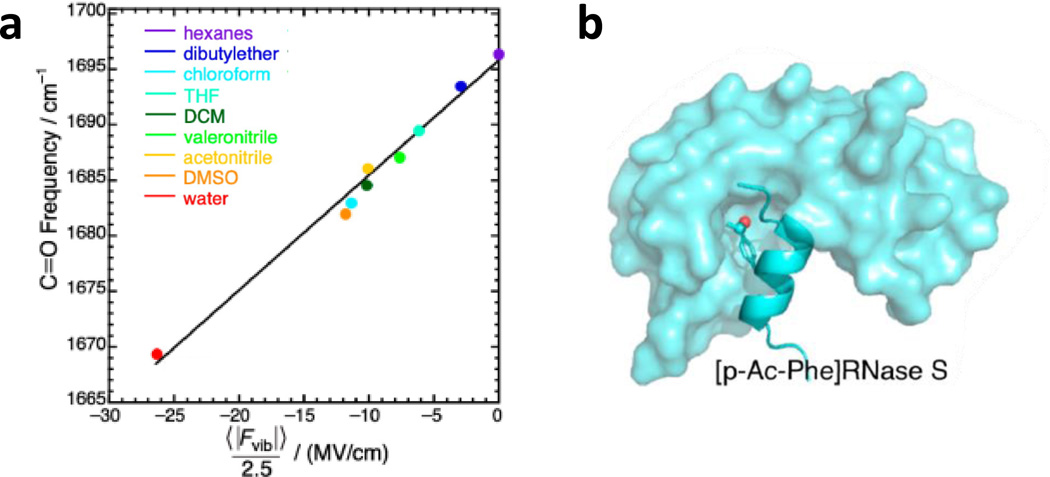
Despite the convenience of their natural occurrence, the ionizability of their sidechains limits the spectroscopic applications of Glu and Asp residues to well chosen systems. As a result, there has been a significant effort in testing and characterizing the IR utility of various unnatural amino acids that consist of a keto or ester functional group. The first example was demonstrated by Boxer and coworkers (125), who demonstrated that the C=O stretching frequency of the unnatural amino acid p-acetyl-l-phenylalanine (p-Ac-Phe) is a convenient and reliable reporter of its local electrostatic field. Specifically, they measured the C=O stretching bands of acetophenone, a model compound of the sidechain of p-Ac-Phe, in a series of protic and aprotic solvents and revealed that the frequency of this vibration shows a sensitive dependence on the solvent. To further quantify this dependence, they carried out MD simulations of this molecule in nine solvents and, for each case, characterized the distribution of the total electric field exerted by the solvent molecules on the C=O bond. As shown (Fig.6), their results indicate a linear frequency-field dependence for this vibration. Using this relationship and the unnatural amino acid p-Ac-Phe, the authors were able to assess the changes in the local electric field strength at the 8th position of the S-peptide upon its binding to the S-protein of Ribonuclease S (Fig. 6).
Figure 6.
(a) The frequency-field map for the C=O stretching vibration of acetophenone, determined via IR measurements in different solvents and MD simulations. (b) Cartoon representation of the binding of the S-peptide to the S-protein of Ribonuclease S, which brings the site-specific C=O probe (red) to an environment with a smaller electrostatic field. Figure adapted with permission from Fried et al. (125).
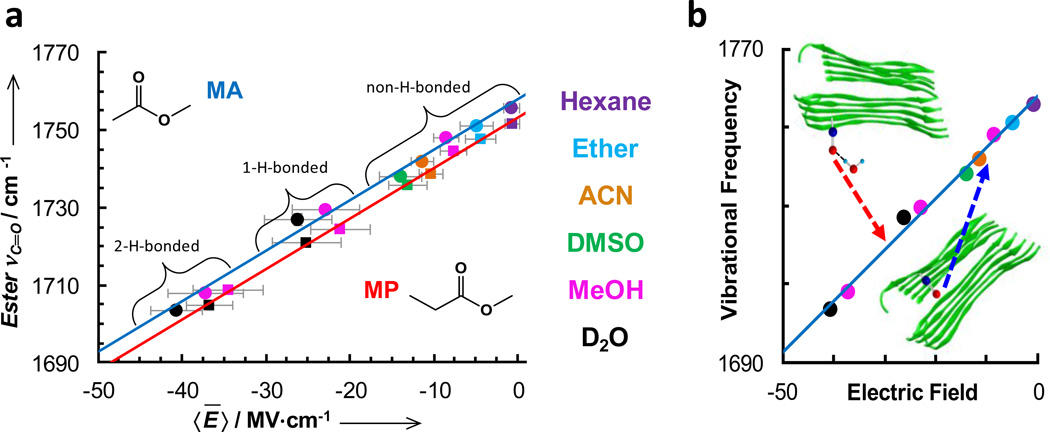
In a second example, Pazos et al. (126) further showed that the C=O stretching frequency of two esters (i.e., methyl propionate and methyl acetate) afford similar spectroscopic properties as that of acetophenone. However, because the C=O stretching vibration of esters is typically in the range of 1710–1750 cm−1, which decreases its spectral overlap with the amide I band of proteins, it offers certain advantages. As shown (Fig. 7), the C=O stretching frequencies of these two esters also display a linear correlation with the averaged local electric field exerted by the solvent molecules and calculated using the method of Boxer and coworkers (125, 127). For both molecules, the slope of the frequency-field correlation is 1.3 MV−1, indicating that this vibration has a very large Stark tuning rate and thus high sensitivity to local electrostatic field variations. The authors further demonstrated that these esters can be incorporated into a peptide or protein by using the unnatural amino acids L-aspartic acid 4-methyl ester (DM) and L-glutamic acid 5-methyl ester (EM). The structures of DM and EM are most similar to, and therefore desirable substitutions for, the natural amino acids glutamine, asparagine, and leucine. Indeed, by replacing the leucine in the Aβ16–22 peptide sequence with DM, the authors found that the resulting mutant exhibits similar aggregation properties as the wild-type peptide; and by using the ester C=O stretching vibration, they were able to probe how the local electrostatic environment of the DM residue changes upon peptide aggregation both in solution and a dry film. Additionally, based on the dependence of the ester C=O stretching frequency on the Onsager field, they were able to determine the dielectric constant (~5.6) of the dry interior of the well-packed fibrils.
Figure 7.
(a) Center frequencies of the carbonyl stretching vibrations of methyl acetate (MA) (circles) and methyl propionate (MP) (squares) versus the calculated local electric field for different solvents. The solid lines are the best fits of these data to a linear equation. (b) Schematic representation of how the ester C=O stretching vibration can be used to probe the local electrostatic environments of peptide fibrils. Figure adapted with permission from Pazos et al. (126).
Due to its high extinction coefficient, sensitivity to the local electric field, and ability to distinguish between different H bond states, it is our belief that the sidechain-based C=O vibrational probes discussed in the last two examples will find more use in future biophysical and biochemical studies.
3.4 Carbon-Deuterium (C-D) Stretching Vibration
Despite its low molar extinction coefficient, the C-H stretching vibration has been widely used as a chromophore in IR and Raman imaging of living cells and organelles. This is because there is a great abundance of C-H groups in proteins and other biological molecules. On the other hand, for this same reason, this vibrational mode is inadequate for providing site-specific information of proteins. Thus, in order to attain site-specificity through the use of this vibration, one needs to covert the C-H group(s) of interest to C-D. Upon deuteration, the vibrational frequency is red-shifted from 2800–3000 cm−1 to 2100–2400 cm−1, making the C-D group a viable local IR probe (128). Indeed, through a series of studies, Romesberg and coworkers have demonstrated the applicability and utility of the C-D stretching vibration as a probe of the local environment of proteins (129–131). They examined the C-D vibrational transitions of a deuterated methionine (i.e. (methyl-d3)methionine) in different protein environments. Using a method previously developed (132), they successfully incorporated this unnatural amino acid into cytochrome c (Cyt c) and showed that the center frequency of the C-D vibrational band is sensitive to the oxidation state and ligand-binding status of Cyt c (133). Using this probe, Romesberg and coworkers also studied the equilibrium folding-unfolding properties of Cyt c and the Src homology 3 (SH3) proteins (134) and showed that the C-D probe is capable of revealing conformational heterogeneity and the existence of folding intermediates. Recently, they further showed that a deuterated glycine (i.e., (d2)Gly) could be incorporated into multiple sites in SH3, and the C-D stretching vibration is sensitive to the position of this amino acid in the protein (130). The biggest advantage of using deuterated amino acids is that their incorporation results in a minimal, if any, perturbation to the native structure and other physical property of interest. In addition, methods for C-D labeling several amino acids are well developed. Thus, we expect more applications of this probe in the future, especially in cases where perturbations arising from the incorporation of bulkier extrinsic/unnatural probes are not tolerable.
One potentially very useful but less explored C-D probe is the Cε-D stretching vibration of histidine (His). Because the imidazole ring of His has a pKa of ~6.5 and can exist in two neutral tautomeric forms, it is frequently found in protein binding sites and enzyme active sites. However, there is currently a lack of convenient vibrational markers that can directly monitor the protonation and tautomerization states of this amino acid, except the possibility of using the amide I mode of isotope-edited His (135). The Cε-D moiety may prove to be a useful candidate in this regard. The recent study of Londergan and coworkers (136) indicated, via Raman spectroscopy, that the Cε-D stretching vibration of His is sensitive to the protonation state of the imidazole. In addition, the deuteration of the epsilon carbon hydrogen of His can easily be achieved via incubation of the amino acid in acidic D2O solution. Currently, we are exploring the IR utility of this Cε-D probe in protein systems.
3.5 Thiol, Phosphate and Fluorocarbon Vibrations
Vibrational modes arising from other chemical moieties, such as thiol (–SH), phosphate (–PO3), and fluorocarbon (–CF), while less utilized, have the potential to be valuable for studies of specific biological systems. For example, the S-H stretching vibration is naturally present in the sidechain of cysteine (Cys) and is located in a transparent window in the IR spectra of proteins (~2550 cm−1) (137). The S-H stretching vibration has a small extinction coefficient when fully solvated; however, as shown by Hamm and coworkers (138), when buried in a hydrophobic environment (i.e., in the core of a protein), its extinction coefficient increases significantly (from 5 to 150 cm−1 M−1). Of particular interest, this transition has a vibrational lifetime of ~6 ps which is significantly longer than the amide I vibrational lifetime (~1 ps), which can be used to study protein dynamics up to tens of picoseconds.
Many biological molecules, such as DNA, lipids and phosphorylated proteins, contain phosphates. The P=O asymmetric stretching vibration of phosphate groups is located between 1200 and 1300 cm−1 and has an extinction coefficient of ~500 M−1 cm−1. Several recent studies have employed this vibrational mode to probe the local environment, for example, the degree of hydration of lipid headgroups in reverse micelles and the water dynamics of hydrated phospholipid surfaces (139). However, in order to use this vibration to achieve site-specificity, one needs to employ the strategy of isotope-editing, which, to the best of our knowledge, has not been implemented thus far.
Fluorine (i.e., 19F) is a popular NMR probe and fluorinated amino acids are frequently used to tune the folding properties of designed peptides and proteins. Thus, these previous applications make it easier to further explore the utility of fluorine-containing unnatural amino acids as site-specific IR probes. The C-F stretching vibration is located at ~1200 cm−1 with an extinction coefficient of ~700 cm−1 M−1 (140). Recently, Cho and coworkers (121) used simulations to examine the solvatochromic and electrochromic properties of fluorine-containing aromatic compounds and showed that the C-F vibration is sensitive to the environment. Furthermore, Boxer and coworkers (140) showed that the C-F stretching vibration has a large Stark tuning rate, suggesting that it will be a useful reporter of the local electric field. For these reasons, we expect that this vibrational mode will become more broadly used in future applications.
3.6 Metal Carbonyl Stretching Vibration
It is well known that upon coordination with metal ions, the stretching vibration strength of carbonyls is significantly enhanced. An environmentally sensitive IR probe with a large extinction coefficient is particularly appealing for biological applications as they typically require low sample concentrations (141, 142). As such, recent years have seen extensive effort in the development and characterization of several metal-carbonyl-based IR probes, as well as methods to incorporate them into proteins. For example, Kubarych and coworkers (143) used synthetic methods to attach a ruthenium dicarbonyl molecule to a His residue (His15) in two lysozymes (hen egg white lysozyme and human lysozyme) and showed that its peak frequency and vibrational lifetime are sensitive to the local hydration dynamics. Interestingly, their results indicated that even for such a small protein there is still spatial heterogeneity in hydration dynamics. In a similar study, Kubarych and coworkers (144) examined the water-membrane interface dynamics using chromium tricarbonyl as a vibrational probe, which can be synthetically incorporated into cholesterol (and hence membranes) through esterification methods. Zanni and coworkers (145) showed that another class of metal carbonyl compound, i.e., tricarbonyl (η5-cyclopentadienyl) rhenium(I) (CpRe(CO)3), has very strong C≡O stretching vibrations and can be used as site-specific IR probes of biological systems. Specifically, they showed that the C≡O vibrational lifetime of CpRe(CO)3 is a useful indicator of the local solvent environment, whereas its frequency is sensitive to the local electric field. Recently, Raleigh and coworkers have also developed multiple methods to incorporate metal carbonyls into proteins, via either a cysteine residue or click chemistry (146).
4. CONCLUDING REMARKS
Site-specific backbone and sidechain vibrational probes are incredibly useful tools to overcome the intrinsic structural and spatial resolution limitations of infrared spectroscopy of molecules in the condensed phase. The different moieties and vibrational modes discussed above all have unique strengths and weaknesses, so the choice of a particular probe for a given experiment often depends upon the research question, the system of interest, and the experimental method. While efforts to further develop and identify functional groups that fulfill the requirements of useful vibrational probes will continue, the future of this field is in the application of these probes to more interesting biological questions. One promising direction for site-specific probes is their use in the study of large, complex systems, which may require the use of multiple probes in a single experiment. Additionally, the use of these probes to monitor how a physical property of interest changes during a reaction in real time, such as the variation of local electric field in the active site of a protein during an enzymatic reaction, is pressingly needed. Finally, further expanding the structural determination application of these probes requires studies to better characterize and understand the process of vibrational energy transfer between two well-separated sites in the context of a complex biological molecule.
SUMMARY POINTS.
Experimental assessment of the mechanistic details of how proteins fold and function requires spectroscopic probes that can offer high structural and/or spatial resolution. Many of the infrared probes discussed in this Review meet this requirement.
Isotope-editing either one or multiple atoms in the protein backbone or a sidechain offers a minimal or non-perturbing strategy to introduce a site-specific vibrational probe into proteins.
Using unnatural amino acids that contain an appropriate functional group, such as nitrile, azide, carbonyl, or ester, represents another general strategy to incorporate a site-specific vibrational probe into proteins.
The frequency-field map for the C=O stretching vibration of p-acetyl-l-phenylalanine, l-aspartic acid 4-methyl ester, and l-glutamic acid 5-methyl ester is available, thus making them viable candidates to be used to quantitatively assess local electric field of proteins.
Coupling between two or more site-specifically introduced vibrational probes can provide information on distances and angles between the coupled vibrators. Thus, measurement of vibrational couplings is a powerful strategy to enhance the structural resolution of infrared spectroscopy of proteins.
As already demonstrated in the examples included in this Review, many of the site-specific infrared probes discussed will undoubtedly find more novel applications in addressing important biological questions and processes, such as protein folding, protein-protein interaction, ligand and drug binding, enzymatic reaction, proton and electron transfer, and the role of solvent in mediating protein conformational preferences.
Finally, it is worth to emphasize that all of the probes discussed above have advantages and disadvantages; and the choice of a specific probe depends on the nature of experiment and system of interest.
ACKNOWLEDGMENTS
The authors gratefully acknowledge financial support from the National Institutes of Health (GM-065978 and P41-GM104605). R.M.C. is an NIH Ruth Kirschstein Predoctoral Fellow (GM-008275).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
REFERENCES
- 1.Cho M. Coherent two-dimensional optical spectroscopy. Chem. Rev. 2008;108:1331–1418. doi: 10.1021/cr078377b. [DOI] [PubMed] [Google Scholar]
- 2.Kim H, Cho M. Infrared Probes for Studying the Structure and Dynamics of Biomolecules. Chem. Rev. 2013:5817–5847. doi: 10.1021/cr3005185. [DOI] [PubMed] [Google Scholar]
- 3.Getahun Z, Huang CY, Wang T, De Leon B, DeGrado WF, Gai F. Using nitrile-derivatized amino acids as infrared probes of local environment. J. Am. Chem. Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 4.Merrifield RB. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963;85:2149–2154. [Google Scholar]
- 5.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. A Genetically Encoded Infrared Probe. J. Am. Chem. Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 6.Connor RE, Tirrell DA. Non-canonical amino acids in protein polymer design. Polym. Rev. 2007;47:9–28. [Google Scholar]
- 7.Ayers B, Blaschke UK, Camarero JA, Cotton GJ, Holford M, Muir TW. Introduction of unnatural amino acids into proteins using expressed protein ligation. Pept. Sci. 1999;51:343–354. doi: 10.1002/(SICI)1097-0282(1999)51:5<343::AID-BIP4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 8.Jo H, Culik RM, Korendovych IV, DeGrado WF, Gai F. Selective Incorporation of Nitrile-Based Infrared Probes into Proteins via Cysteine Alkylation. Biochemistry. 2010;49:10354–10356. doi: 10.1021/bi101711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi J-H, Ham S, Cho M. Local Amide I Mode Frequencies and Coupling Constants in Polypeptides. J. Phys. Chem. B. 2003;107:9132–9138. [Google Scholar]
- 10.Ganim Z, Chung HS, Smith AW, Deflores LP, Jones KC, Tokmakoff A. Amide I two-dimensional infrared spectroscopy of proteins. Acc. Chem. Res. 2008;41:432–441. doi: 10.1021/ar700188n. [DOI] [PubMed] [Google Scholar]
- 11.Deflores LP, Ganim Z, Nicodemus Ra, Tokmakoff A. Amide I'–II' 2D IR spectroscopy provides enhanced protein secondary structural sensitivity. J. Am. Chem. Soc. 2009;131:3385–3391. doi: 10.1021/ja8094922. [DOI] [PubMed] [Google Scholar]
- 12.Kim YS, Hochstrasser RM. Applications of 2D IR Spectroscopy to Peptides, Proteins, and Hydrogen-Bond Dynamics. J. Phys. Chem. B. 2009;113:8231–8251. doi: 10.1021/jp8113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maekawa H, Ballano G, Toniolo C, Ge N-H. Linear and two-dimensional infrared spectroscopic study of the amide I and II modes in fully extended peptide chains. J. Phys. Chem. B. 2011;115:5168–5182. doi: 10.1021/jp105527n. [DOI] [PubMed] [Google Scholar]
- 14.Roy S, Lessing J, Meisl G, Ganim Z, Tokmakoff A, et al. Solvent and conformation dependence of amide I vibrations in peptides and proteins containing proline. J. Chem. Phys. 2011;135:234507–234518. doi: 10.1063/1.3665417. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Shi JP, Wang JP. Amide-I Characteristics of Helical beta-Peptides by Linear Infrared Measurement and Computations. J. Phys. Chem. B. 2014;118:94–106. doi: 10.1021/jp4095936. [DOI] [PubMed] [Google Scholar]
- 16.Tadesse L, Nazarbaghi R, Walters L. Isotopically enhanced infrared spectroscopy: a novel method for examining secondary structure at specific sites in conformationally heterogeneous peptides. J. Am. Chem. Soc. 1991;113:7036–7037. [Google Scholar]
- 17.Decatur SM, Antonic J. Isotope-edited infrared spectroscopy of helical peptides. J. Am. Chem. Soc. 1999;121:11914–11915. [Google Scholar]
- 18.Decatur SM. Elucidation of residue-level structure and dynamics of polypeptides via isotopp-edited infrared spectroscopy. Acc. Chem. Res. 2006;39:169–175. doi: 10.1021/ar050135f. [DOI] [PubMed] [Google Scholar]
- 19.Lin YS, Shorb JM, Mukherjee P, Zanni MT, Skinner JL. Empirical Amide I Vibrational Frequency Map: Application to 2D-IR Line Shapes for Isotope-Edited Membrane Peptide Bundles. J. Phys. Chem. B. 2009;113:592–602. doi: 10.1021/jp807528q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Middleton CT, Zanni MT, Skinner JL. Development and Validation of Transferable Amide I Vibrational Frequency Maps for Peptides. J. Phys. Chem. B. 2011;115:3713–3724. doi: 10.1021/jp200745r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang CY, Getahun Z, Zhu YJ, Klemke JW, DeGrado WF, Gai F. Helix formation via conformation diffusion search. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2788–2793. doi: 10.1073/pnas.052700099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bredenbeck J, Hamm P. Peptide structure determination by two-dimensional infrared spectroscopy in the presence of homogeneous and inhomogeneous broadening. Journal of Chemical Physics. 2003;119:1569–1578. [Google Scholar]
- 23.Brewer SH, Song BB, Raleigh DP, Dyer RB. Residue specific resolution of protein folding dynamics using isotope-edited infrared temperature jump spectroscopy. Biochemistry. 2007;46:3279–3285. doi: 10.1021/bi602372y. [DOI] [PubMed] [Google Scholar]
- 24.Backus EHG, Bloem R, Donaldson PM, Ihalainen JA, Pfister R, et al. 2D-IR Study of a Photoswitchable Isotope-Labeled alpha-Helix. J. Phys. Chem. B. 2010;114:3735–3740. doi: 10.1021/jp911849n. [DOI] [PubMed] [Google Scholar]
- 25.Fleming S, Frederix P, Sasselli IR, Hunt NT, Ulijn RV, Tuttle T. Assessing the Utility of Infrared Spectroscopy as a Structural Diagnostic Tool for beta-Sheets in Self-Assembling Aromatic Peptide Amphiphiles. Langmuir. 2013;29:9510–9515. doi: 10.1021/la400994v. [DOI] [PubMed] [Google Scholar]
- 26.Buchanan LE, Carr JK, Fluitt AM, Hoganson AJ, Moran SD, et al. Structural motif of polyglutamine amyloid fibrils discerned with mixed-isotope infrared spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5796–5801. doi: 10.1073/pnas.1401587111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. 2D IR provides evidence for mobile water molecules in β-amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17751–17756. doi: 10.1073/pnas.0909888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh A, Qiu J, DeGrado WF, Hochstrasser RM. Tidal surge in the M2 proton channel, sensed by 2D IR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6115–6120. doi: 10.1073/pnas.1103027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma J, Komatsu H, Kim YS, Liu L, Hochstrasser RM, Axelsen PH. Intrinsic Structural Heterogeneity and Long-Term Maturation of Amyloid β Peptide Fibrils. ACS Chem. Neurosci. 2013;4:1236–1243. doi: 10.1021/cn400092v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang CY, Getahun Z, Wang T, DeGrado WF, Gai F. Time-resolved infrared study of the helix-coil transition using C-13-labeled helical peptides. J. Am. Chem. Soc. 2001;123:12111–12112. doi: 10.1021/ja016631q. [DOI] [PubMed] [Google Scholar]
- 31.Waldman ADB, Birdsall B, Roberts GCK, Holbrook JJ. 13C-NMR and transient kinetic studies on lactate dehydrogenase [Cys(13CN)165]. Direct measurement of a rate-limiting rearrangement in protein structure. Biochim. Biophys. Acta. 1986;870:102–111. doi: 10.1016/0167-4838(86)90013-0. [DOI] [PubMed] [Google Scholar]
- 32.Doherty GM, Motherway R, Mayhew SG, Malthouse JPG. 13C NMR of cyanylated flavodoxin from Megasphaera elsdenii and of thiocyanate model compounds. Biochemistry. 1992;31:7922–7930. doi: 10.1021/bi00149a025. [DOI] [PubMed] [Google Scholar]
- 33.Fafarman AT, Sigala PA, Herschlag D, Boxer SG. Decomposition of Vibrational Shifts of Nitriles into Electrostatic and Hydrogen-Bonding Effects. J. Am. Chem. Soc. 2010;132:12811–12813. doi: 10.1021/ja104573b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tucker MJ, Getahun Z, Nanda V, DeGrado WF, Gai F. A new method for determining the local environment and orientation of individual side chains of membrane-binding peptides. J. Am. Chem. Soc. 2004;126:5078–5079. doi: 10.1021/ja032015d. [DOI] [PubMed] [Google Scholar]
- 35.Woys AM, Lin Y-S, Reddy AS, Xiong W, de Pablo JJ, et al. 2D IR Line Shapes Probe Ovispirin Peptide Conformation and Depth in Lipid Bilayers. J. Am. Chem. Soc. 2010;132:2832–2838. doi: 10.1021/ja9101776. [DOI] [PubMed] [Google Scholar]
- 36.Manor J, Arbely E, Beerlink A, Akkawi M, Arkin IT. Use of Isotope-Edited FTIR to Derive a Backbone Structure of a Transmembrane Protein. J. Phys. Chem. Lett. 2014 doi: 10.1021/jz501055d. [DOI] [PubMed] [Google Scholar]
- 37.Remorino A, Korendovych IV, Wu Y, DeGrado WF, Hochstrasser RM. Residue-Specific Vibrational Echoes Yield 3D Structures of a Transmembrane Helix Dimer. Science. 2011;332:1206–1209. doi: 10.1126/science.1202997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remorino A, Hochstrasser RM. Three-Dimensional Structures by Two-Dimensional Vibrational Spectroscopy. Acc. Chem. Res. 2012;45:1896–1905. doi: 10.1021/ar3000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchanan LE, Dunkelberger EB, Tran HQ, Cheng P-N, Chiu C-C, et al. Mechanism of IAPP amyloid fibril formation involves an intermediate with a transient β-sheet. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19285–19290. doi: 10.1073/pnas.1314481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woys AM, Almeida AM, Wang L, Chiu C-C, McGovern M, et al. Parallel beta-Sheet Vibrational Couplings Revealed by 2D IR Spectroscopy of an Isotopically Labeled Macrocycle: Quantitative Benchmark for the Interpretation of Amyloid and Protein Infrared Spectra. J. Am. Chem. Soc. 2012;134:19118–19128. doi: 10.1021/ja3074962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moran SD, Woys AM, Buchanan LE, Bixby E, Decatur SM, Zanni MT. Two-dimensional IR spectroscopy and segmental 13C labeling reveals the domain structure of human γD-crystallin amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 2012;109:3329–3334. doi: 10.1073/pnas.1117704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran SD, Decatur SM, Zanni MT. Structural and Sequence Analysis of the Human gamma D-Crystallin Amyloid Fibril Core Using 2D IR Spectroscopy, Segmental C-13 Labeling, and Mass Spectrometry. J. Am. Chem. Soc. 2012;134:18410–18416. doi: 10.1021/ja307898g. [DOI] [PubMed] [Google Scholar]
- 43.Lam AR, Moran SD, Preketes NK, Zhang TO, Zanni MT, Mukamel S. Study of the gamma D-Crystallin Protein Using Two-Dimensional Infrared (2DIR) Spectroscopy: Experiment and Simulation. J. Phys. Chem. B. 2013;117:15436–15443. doi: 10.1021/jp405159v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moran SD, Zhang TO, Decatur SM, Zanni MT. Amyloid Fiber Formation in Human gamma D-Crystallin Induced by UV-B Photodamage. Biochemistry. 2013;52:6169–6181. doi: 10.1021/bi4008353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurochkin DV, Naraharisetty SRG, Rubtsov IV. A relaxation-assisted 2D IR spectroscopy method. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14209–14214. doi: 10.1073/pnas.0700560104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Backus EHG, Nguyen PH, Botan V, Pfister R, Moretto A, et al. Energy transport in peptide helices: A comparison between high- and low-energy excitations. J. Phys. Chem. B. 2008;112:9091–9099. doi: 10.1021/jp711046e. [DOI] [PubMed] [Google Scholar]
- 47.Backus EHG, Bloem R, Pfister R, Moretto A, Crisma M, et al. Dynamical Transition in a Small Helical Peptide and Its Implication for Vibrational Energy Transport. J. Phys. Chem. B. 2009;113:13405–13409. doi: 10.1021/jp904905d. [DOI] [PubMed] [Google Scholar]
- 48.Bian H, Li J, Zhang Q, Chen H, Zhuang W, et al. Ion Segregation in Aqueous Solutions. J. Phys. Chem. B. 2012;116:14426–14432. doi: 10.1021/jp310153n. [DOI] [PubMed] [Google Scholar]
- 49.Li J, Bian H, Wen X, Chen H, Yuan K, Zheng J. Probing Ion/Molecule Interactions in Aqueous Solutions with Vibrational Energy Transfer. J. Phys. Chem. B. 2012;116:12284–12294. doi: 10.1021/jp306369w. [DOI] [PubMed] [Google Scholar]
- 50.Lin Z, Zhang N, Jayawickramarajah J, Rubtsov IV. Ballistic energy transport along PEG chains: distance dependence of the transport efficiency. Phys. Chem. Chem. Phys. 2012;14:10445–10454. doi: 10.1039/c2cp40187h. [DOI] [PubMed] [Google Scholar]
- 51.Ihalainen JA, Bredenbeck J, Pfister R, Helbing J, Chi L, et al. Folding and unfolding of a photoswitchable peptide from picoseconds to microseconds. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5383–5388. doi: 10.1073/pnas.0607748104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ihalainen JA, Paoli B, Muff S, Backus EHG, Bredenbeck J, et al. alpha-Helix folding in the presence of structural constraints. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9588–9593. doi: 10.1073/pnas.0712099105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones KC, Peng CS, Tokmakoff A. Folding of a heterogeneous beta-hairpin peptide from temperature-jump 2D IR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2828–2833. doi: 10.1073/pnas.1211968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams S, Causgrove TP, Gilmanshin R, Fang KS, Callender RH, et al. Fast events in protein folding: Helix melting and formation in a small peptide. Biochemistry. 1996;35:691–697. doi: 10.1021/bi952217p. [DOI] [PubMed] [Google Scholar]
- 55.Thompson PA, Eaton WA, Hofrichter J. Laser temperature jump study of the helix reversible arrow coil kinetics of an alanine peptide interpreted with a 'kinetic zipper' model. Biochemistry. 1997;36:9200–9210. doi: 10.1021/bi9704764. [DOI] [PubMed] [Google Scholar]
- 56.Munoz V, Thompson PA, Hofrichter J, Eaton WA. Folding dynamics and mechanism of beta-hairpin formation. Nature. 1997;390:196–199. doi: 10.1038/36626. [DOI] [PubMed] [Google Scholar]
- 57.Tucker MJ, Abdo M, Courter JR, Chen J, Brown SP, et al. Nonequilibrium dynamics of helix reorganization observed by transient 2D IR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17314–17319. doi: 10.1073/pnas.1311876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miwa JH, Patel AK, Vivatrat N, Popek SM, Meyer AM. Compatibility of the thioamide functional group with beta-sheet secondary structure: Incorporation of a thioamide linkage into a beta-hairpin peptide. Organic Letters. 2001;3:3373–3375. doi: 10.1021/ol0166092. [DOI] [PubMed] [Google Scholar]
- 59.Cohen VI. A Convenient Alkyl, Cycloalkyl and Aralkyl Disulfides Synthesis from Aliphatic and Aromatic Aldehydes, Aliphatic Ketones and Cycloketones. Journal of Organic Chemistry. 1977;42:2645–2647. [Google Scholar]
- 60.Deechongkit S, Nguyen H, Powers ET, Dawson PE, Gruebele M, Kelly JW. Context-dependent contributions of backbone hydrogen bonding to beta-sheet folding energetics. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 61.Bunagan MR, Gao J, Kelly JW, Gai F. Probing the folding transition state structure of the villin headpiece subdomain via side chain and backbone mutagenesis. J. Am. Chem. Soc. 2009;131:7470–7476. doi: 10.1021/ja901860f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Velarde L, Wang H-f. Capturing inhomogeneous broadening of the -CN stretch vibration in a Langmuir monolayer with high-resolution spectra and ultrafast vibrational dynamics in sum-frequency generation vibrational spectroscopy (SFG-VS) J. Chem. Phys. 2013;139 doi: 10.1063/1.4818996. [DOI] [PubMed] [Google Scholar]
- 63.Waegele MM, Tucker MJ, Gai F. 5-Cyanotryptophan as an infrared probe of local hydration status of proteins. Chem. Phys. Lett. 2009;478:249–253. doi: 10.1016/j.cplett.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xue L, Zou F, Zhao Y, Huang X, Qu Y. Nitrile group as infrared probe for the characterization of the conformation of bovine serum albumin solubilized in reverse micelles. Spectrochim. Acta, Part A. 2012;97:858–863. doi: 10.1016/j.saa.2012.07.076. [DOI] [PubMed] [Google Scholar]
- 65.Zhang S, Shi R, Ma X, Lu L, He Y, et al. Intrinsic Electric Fields in Ionic Liquids Determined by Vibrational Stark Effect Spectroscopy and Molecular Dynamics Simulation. Chem-Eur J. 2012;18:11904–11908. doi: 10.1002/chem.201201257. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Z, Guo Y, Lu Z, Velarde L, Wang H-f. Resolving Two Closely Overlapping -CN Vibrations and Structure in the Langmuir Mono layer of the Long-Chain Nonadecanenitrile by Polarization Sum Frequency Generation Vibrational Spectroscopy. Journal of Physical Chemistry C. 2012;116:2976–2987. [Google Scholar]
- 67.Wang X, He X, Zhang JZH. Predicting Mutation-Induced Stark Shifts in the Active Site of a Protein with a Polarized Force Field. J. Phys. Chem. A. 2013;117:6015–6023. doi: 10.1021/jp312063h. [DOI] [PubMed] [Google Scholar]
- 68.Huang CY, Wang T, Gai F. Temperature dependence of the CN stretching vibration of a nitrile-derivatized phenylalanine in water. Chem. Phys. Lett. 2003;371:731–738. [Google Scholar]
- 69.Liu J, Strzalka J, Tronin A, Johansson JS, Blasie JK. Mechanism of Interaction between the General Anesthetic Halothane and a Model Ion Channel Protein, II: Fluorescence and Vibrational Spectroscopy Using a Cyanophenylalanine Probe. Biophys. J. 2009;96:4176–4187. doi: 10.1016/j.bpj.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu W, Webb LJ. Direct Measurement of the Membrane Dipole Field in Bicelles Using Vibrational Stark Effect Spectroscopy. J. Phys. Chem. Lett. 2011;2:1925–1930. [Google Scholar]
- 71.Bazewicz CG, Lipkin JS, Smith EE, Liskov MT, Brewer SH. Expanding the Utility of 4-Cyano-L-Phenylalanine As a Vibrational Reporter of Protein Environments. J. Phys. Chem. B. 2012;116:10824–10831. doi: 10.1021/jp306886s. [DOI] [PubMed] [Google Scholar]
- 72.Bischak CG, Longhi S, Snead DM, Costanzo S, Terrer E, Londergan CH. Probing Structural Transitions in the Intrinsically Disordered C-Terminal Domain of the Measles Virus Nucleoprotein by Vibrational Spectroscopy of Cyanylated Cysteines. Biophys. J. 2010;99:1676–1683. doi: 10.1016/j.bpj.2010.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McMahon HA, Alfieri KN, Clark CAA, Londergan CH. Cyanylated Cysteine: A Covalently Attached Vibrational Probe of Protein-Lipid Contacts. J. Phys. Chem. Lett. 2010;1:850–855. doi: 10.1021/jz1000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stafford AJ, Ensign DL, Webb LJ. Vibrational Stark Effect Spectroscopy at the Interface of Ras and Rap1A Bound to the Ras Binding Domain of RalGDS Reveals an Electrostatic Mechanism for Protein-Protein Interaction. J. Phys. Chem. B. 2010;114:15331–15344. doi: 10.1021/jp106974e. [DOI] [PubMed] [Google Scholar]
- 75.Alfieri KN, Vienneau AR, Londergan CH. Using Infrared Spectroscopy of Cyanylated Cysteine To Map the Membrane Binding Structure and Orientation of the Hybrid Antimicrobial Peptide CM15. Biochemistry. 2011;50:11097–11108. doi: 10.1021/bi200903p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stafford AJ, Walker DM, Webb LJ. Electrostatic Effects of Mutations of Ras Glutamine 61 Measured Using Vibrational Spectroscopy of a Thiocyanate Probe. Biochemistry. 2012;51:2757–2767. doi: 10.1021/bi201225p. [DOI] [PubMed] [Google Scholar]
- 77.Ghosh A, Remorino A, Tucker MJ, Hochstrasser RM. 2D IR photon echo spectroscopy reveals hydrogen bond dynamics of aromatic nitriles. Chemical Physics Letters. 2009;469:325–330. doi: 10.1016/j.cplett.2008.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waegele MM, Culik RM, Gai F. Site-Specific Spectroscopic Reporters of the Local Electric Field, Hydration, Structure, and Dynamics of Biomolecules. J. Phys. Chem. Lett. 2011;2:2598–2609. doi: 10.1021/jz201161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Layfield JP, Hammes-Schiffer S. Calculation of Vibrational Shifts of Nitrile Probes in the Active Site of Ketosteroid Isomerase upon Ligand Binding. J. Am. Chem. Soc. 2013;135:717–725. doi: 10.1021/ja3084384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pazos IM, Gai F. Solute's Perspective on How Trimethylamine Oxide, Urea, and Guanidine Hydrochloride Affect Water's Hydrogen Bonding Ability. J. Phys. Chem. B. 2012;116:12473–12478. doi: 10.1021/jp307414s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma J, Pazos IM, Gai F. Microscopic insights into the protein-stabilizing effect of trimethylamine N-oxide (TMAO) Proc. Natl. Acad. Sci. U.S.A. 2014;111:8476–8481. doi: 10.1073/pnas.1403224111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cho SS, Reddy G, Straub JE, Thirumalai D. Entropic Stabilization of Proteins by TMAO. J. Phys. Chem. B. 2011;115:13401–13407. doi: 10.1021/jp207289b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Levinson NM, Fried SD, Boxer SG. Solvent-Induced Infrared Frequency Shifts in Aromatic Nitriles Are Quantitatively Described by the Vibrational Stark Effect. J. Phys. Chem. B. 2012;116:10470–10476. doi: 10.1021/jp301054e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bagchi S, Fried SD, Boxer SG. A Solvatochromic Model Calibrates Nitriles' Vibrational Frequencies to Electrostatic Fields. J. Am. Chem. Soc. 2012;134:10373–10376. doi: 10.1021/ja303895k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ragain CM, Newberry RW, Ritchie AW, Webb LJ. Role of Electrostatics in Differential Binding of RaIGDS to Rap Mutations E30D and K31E Investigated by Vibrational Spectroscopy of Thiocyanate Probes. J. Phys. Chem. B. 2012;116:9326–9336. doi: 10.1021/jp303272y. [DOI] [PubMed] [Google Scholar]
- 86.Sigala PA, Fafarman AT, Bogard PE, Boxer SG, Herschlag D. Do ligand binding and solvent exclusion alter the electrostatic character within the oxyanion hole of an enzymatic active site? J. Am. Chem. Soc. 2007;129:12104–12105. doi: 10.1021/ja075605a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu CT, Layfield JP, Stewart RJ, French JB, Hanoian P, et al. Probing the Electrostatics of Active Site Microenvironments along the Catalytic Cycle for Escherichia coli Dihydrofolate Reductase. J. Am. Chem. Soc. 2014 doi: 10.1021/ja5038947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Levinson NM, Boxer SG. A conserved water-mediated hydrogen bond network defines bosutinib's kinase selectivity. Nat. Chem. Biol. 2014;10:127–132. doi: 10.1038/nchembio.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fang C, Bauman JD, Das K, Remorino A, Arnold E, Hochstrasser RM. Two-dimensional infrared spectra reveal relaxation of the nonnucleoside inhibitor TMC278 complexed with HIV-1 reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1472–1477. doi: 10.1073/pnas.0709320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuroda DG, Bauman JD, Challa JR, Patel D, Troxler T, et al. Snapshot of the equilibrium dynamics of a drug bound to HIV-1 reverse transcriptase. Nat. Chem. 2013;5:174–181. doi: 10.1038/nchem.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ghosh A, Wang J, Moroz YS, Korendovych IV, Zanni M, et al. 2D IR spectroscopy reveals the role of water in the binding of channel-blocking drugs to the influenza M2 channel. J. Chem. Phys. 2014;140:235105–235113. doi: 10.1063/1.4881188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Urbanek DC, Vorobyev DY, Serrano AL, Gai F, Hochstrasser RM. The Two-Dimensional Vibrational Echo of a Nitrile Probe of the Villin HP35 Protein. J. Phys. Chem. Lett. 2010;1:3311–3315. doi: 10.1021/jz101367d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fafarman AT, Boxer SG. Nitrile Bonds as Infrared Probes of Electrostatics in Ribonuclease S. J. Phys. Chem. B. 2010;114:13536–13544. doi: 10.1021/jp106406p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chung JK, Thielges MC, Fayer MD. Dynamics of the Folded and Unfolded Villin Headpiece (HP35) Measured with Ultrafast 2D IR Vibrational Echo Spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3578–3583. doi: 10.1073/pnas.1100587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jha SK, Ji M, Gaffney KJ, Boxer SG. Direct measurement of the protein response to an electrostatic perturbation that mimics the catalytic cycle in ketosteroid isomerase. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16612–16617. doi: 10.1073/pnas.1113874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bagchi S, Boxer SG, Fayer MD. Ribonuclease S Dynamics Measured Using a Nitrile Label with 2D IR Vibrational Echo Spectroscopy. J. Phys. Chem. B. 2012;116:4034–4042. doi: 10.1021/jp2122856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fafarman AT, Sigala PA, Schwans JP, Fenn TD, Herschlag D, Boxer SG. Quantitative, directional measurement of electric field heterogeneity in the active site of ketosteroid isomerase. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E299–E308. doi: 10.1073/pnas.1111566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choi JH, Oh KI, Lee H, Lee C, Cho M. Nitrile and thiocyanate IR probes: Quantum chemistry calculation studies and multivariate least-square fitting analysis. J. Chem. Phys. 2008;128:8. doi: 10.1063/1.2844787. [DOI] [PubMed] [Google Scholar]
- 99.Lindquist BA, Corcelli SA. Nitrile groups as vibrational probes: Calculations of the C N infrared absorption line shape of acetonitrile in water and tetrahydrofuran. J. Phys. Chem. B. 2008;112:6301–6303. doi: 10.1021/jp802039e. [DOI] [PubMed] [Google Scholar]
- 100.Lindquist BA, Furse KE, Corcelli SA. Nitrile groups as vibrational probes of biomolecular structure and dynamics: an overview. Phys. Chem. Chem. Phys. 2009;11:8119–8132. doi: 10.1039/b908588b. [DOI] [PubMed] [Google Scholar]
- 101.Tucker MJ, Oyola R, Gai F. A novel fluorescent probe for protein binding and folding studies: p-cyano-phenylalanine. Biopolymers. 2006;83:571–576. doi: 10.1002/bip.20587. [DOI] [PubMed] [Google Scholar]
- 102.Tang J, Yin H, Qiu J, Tucker MJ, DeGrado WF, Gai F. Using Two Fluorescent Probes to Dissect the Binding, Insertion, and Dimerization Kinetics of a Model Membrane Peptide. J. Am. Chem. Soc. 2009;131:3816. doi: 10.1021/ja809007f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Serrano AL, Troxler T, Tucker MJ, Gai F. Photophysics of a fluorescent non-natural amino acid: p-Cyanophenylalanine. Chem. Phys. Lett. 2010;487:303–306. doi: 10.1016/j.cplett.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Taskent-Sezgin H, Marek P, Thomas R, Goldberg D, Chung J, et al. Modulation of p-Cyanophenylalanine Fluorescence by Amino Acid Side Chains and Rational Design of Fluorescence Probes of alpha-Helix Formation. Biochemistry. 2010;49:6290–6295. doi: 10.1021/bi100932p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goldberg JM, Batjargal S, Petersson EJ. Thioamides as Fluorescence Quenching Probes: Minimalist Chromophores To Monitor Protein Dynamics. J. Am. Chem. Soc. 2010;132:14718–14720. doi: 10.1021/ja1044924. [DOI] [PubMed] [Google Scholar]
- 106.Tucker MJ, Kim YS, Hochstrasser RM. 2D IR photon echo study of the anharmonic coupling in the OCN region of phenyl cyanate. Chem. Phys. Lett. 2009;470:80–84. doi: 10.1016/j.cplett.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Park KH, Jeon J, Park Y, Lee S, Kwon HJ, et al. Infrared Probes Based on Nitrile-Derivatized Prolines: Thermal Insulation Effect and Enhanced Dynamic Range. J. Phys. Chem. Lett. 2013;4:2105–2110. [Google Scholar]
- 108.Tucker MJ, Gai XS, Fenlon EE, Brewer SH, Hochstrasser RM. 2D IR photon echo of azido-probes for biomolecular dynamics. Phys. Chem. Chem. Phys. 2011;13:2237–2241. doi: 10.1039/c0cp01625j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thielges MC, Axup JY, Wong D, Lee HS, Chung JK, et al. Two-Dimensional IR Spectroscopy of Protein Dynamics Using Two Vibrational Labels: A Site-Specific Genetically Encoded Unnatural Amino Acid and an Active Site Ligand. J. Phys. Chem. B. 2011;115:11294–11304. doi: 10.1021/jp206986v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Choi J-H, Raleigh D, Cho M. Azido Homoalanine is a Useful Infrared Probe for Monitoring Local Electrostatistics and Side-Chain Solvation in Proteins. J. Phys. Chem. Lett. 2011;2:2158–2162. doi: 10.1021/jz200980g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gai XS, Coutifaris BA, Brewer SH, Fenlon EE. A direct comparison of azide and nitrile vibrational probes. Phys. Chem. Chem. Phys. 2011;13:5926–5930. doi: 10.1039/c0cp02774j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wolfshorndl MP, Baskin R, Dhawan I, Londergan CH. Covalently Bound Azido Groups Are Very Specific Water Sensors, Even in Hydrogen-Bonding Environments. J. Phys. Chem. B. 2012;116:1172–1179. doi: 10.1021/jp209899m. [DOI] [PubMed] [Google Scholar]
- 113.Bloem R, Koziol K, Waldauer SA, Buchli B, Walser R, et al. Ligand Binding Studied by 2D IR Spectroscopy Using the Azidohomoalanine Label. J. Phys. Chem. B. 2012;116:13705–13712. doi: 10.1021/jp3095209. [DOI] [PubMed] [Google Scholar]
- 114.Bazewicz CG, Liskov MT, Hines KJ, Brewer SH. Sensitive, Site-Specific, and Stable Vibrational Probe of Local Protein Environments: 4-Azidomethyl-L-Phenylalanine. J. Phys. Chem. B. 2013;117:8987–8993. doi: 10.1021/jp4052598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Garcia-Viloca M, Nam K, Alhambra C, Gao JL. Solvent and protein effects on the vibrational frequency shift and energy relaxation of the azide ligand in carbonic anhydrase. J. Phys. Chem. B. 2004;108:13501–13512. [Google Scholar]
- 116.Ye S, Huber T, Vogel R, Sakmar TP. FTIR analysis of GPCR activation using azido probes. Nat. Chem. Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lipkin JS, Song R, Fenlon EE, Brewer SH. Modulating Accidental Fermi Resonance: What a Difference a Neutron Makes. J. Phys. Chem. Lett. 2011;2:1672–1676. doi: 10.1021/jz2006447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Taskent-Sezgin H, Chung J, Banerjee PS, Nagarajan S, Dyer RB, et al. Azidohomoalanine: A Conformationally Sensitive IR Probe of Protein Folding, Protein Structure, and Electrostatics. Angew. Chem. Int. Ed. 2010;49:7473–7475. doi: 10.1002/anie.201003325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ye S, Zaitseva E, Caltabiano G, Schertler GFX, Sakmar TP, et al. Tracking G-protein-coupled receptor activation using genetically encoded infrared probes. Nature. 2010;464:1386–1389. doi: 10.1038/nature08948. [DOI] [PubMed] [Google Scholar]
- 120.Bandaria JN, Dutta S, Hill SE, Kohen A, Cheatum CM. Fast Enzyme Dynamics at the Active Site of Formate Dehydrogenase. J. Am. Chem. Soc. 2007;130:22–23. doi: 10.1021/ja077599o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Choi J-H, Cho M. Vibrational solvatochromism and electrochromism of infrared probe molecules containing C equivalent to O, C equivalent to N, C=O, or C-F vibrational chromophore. J. Chem. Phys. 2011;134:54513–54525. doi: 10.1063/1.3580776. [DOI] [PubMed] [Google Scholar]
- 122.Nie B, Stutzman J, Xie A. A Vibrational Spectral Maker for Probing the Hydrogen-Bonding Status of Protonated Asp and Glu Residues. Biophys. J. 2005;88:2833–2847. doi: 10.1529/biophysj.104.047639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bagchi S, Falvo C, Mukamel S, Hochstrasser RM. 2D-IR Experiments and Simulations of the Coupling between Amide-I and Ionizable Side Chains in Proteins: Application to the Villin Headpiece. J. Phys. Chem. B. 2009;113:11260–11273. doi: 10.1021/jp900245s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Culik RM, Annavarapu S, Nanda V, Gai F. Using D-amino acids to delineate the mechanism of protein folding: Application to Trp-cage. Chem. Phys. 2013;422:131–134. doi: 10.1016/j.chemphys.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fried SD, Bagchi S, Boxer SG. Measuring Electrostatic Fields in Both Hydrogen-Bonding and Non-Hydrogen-Bonding Environments Using Carbonyl Vibrational Probes. J. Am. Chem. Soc. 2013;135:11181–11192. doi: 10.1021/ja403917z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pazos IM, Ghosh A, Tucker MJ, Gai F. Ester Carbonyl Vibration as a Sensitive Probe of Protein Local Electric Field. Angew. Chem. Int. Ed. 2014;53:6080–6084. doi: 10.1002/anie.201402011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fried SD, Wang L-P, Boxer SG, Ren P, Pande VS. Calculations of the Electric Fields in Liquid Solutions. J. Phys. Chem. B. 2013;117:16236–16248. doi: 10.1021/jp410720y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kumar K, Sinks LE, Wang JP, Kim YS, Hochstrasser RM. Coupling between C-D and C=O motions using dual-frequency 2D IR photon echo spectroscopy. Chem. Phys. Lett. 2006;432:122–127. [Google Scholar]
- 129.Zimmermann J, Gundogdu K, Cremeens ME, Bandaria JN, Hwang GT, et al. Efforts toward Developing Probes of Protein Dynamics: Vibrational Dephasing and Relaxation of Carbon-Deuterium Stretching Modes in Deuterated Leucine. J. Phys. Chem. B. 2009;113:7991–7994. doi: 10.1021/jp900516c. [DOI] [PubMed] [Google Scholar]
- 130.Cremeens ME, Zimmermann J, Yu W, Dawson PE, Romesberg FE. Direct Observation of Structural Heterogeneity in a beta-Sheet. J. Am. Chem. Soc. 2009;131 doi: 10.1021/ja900505e. 5726-+ [DOI] [PubMed] [Google Scholar]
- 131.Naraharisetty SRG, Kasyanenko VM, Zimmermann J, Thielges MC, Romesberg FE, Rubtsov IV. C-D Modes of Deuterated Side Chain of Leucine as Structural Reporters via Dual-frequency Two-dimensional Infrared Spectroscopy. J. Phys. Chem. B. 2009;113:4940–4946. doi: 10.1021/jp8112446. [DOI] [PubMed] [Google Scholar]
- 132.Stillwell WG, Bouwsma OJ, Horning MG. Formation in vivo of Deuterated Methylthio Metabolites of Naphthalene from L-methionine (methyl-d-3) Res. Commun. Chem. Path. 1978;22:329–343. [PubMed] [Google Scholar]
- 133.Chin JK, Jimenez R, Romesberg FE. Direct observation of protein vibrations by selective incorporation of spectroscopically observable carbon-deuterium bonds in cytochrome c. J. Am. Chem. Soc. 2001;123:2426–2427. doi: 10.1021/ja0033741. [DOI] [PubMed] [Google Scholar]
- 134.Yu W, Dawson PE, Zimmermann J, Romesberg FE. Carbon-Deuterium Bonds as Probes of Protein Thermal Unfolding. J. Phys. Chem. B. 2012;116:6397–6403. doi: 10.1021/jp303521t. [DOI] [PubMed] [Google Scholar]
- 135.Ghosh A, Tucker MJ, Gai F. 2D IR Spectroscopy of Histidine: Probing Side-Chain Structure and Dynamics via Backbone Amide Vibrations. J. Phys. Chem. B. 2014 doi: 10.1021/jp411901m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hoffman KW, Romei MG, Londergan CH. A New Raman Spectroscopic Probe of Both the Protonation State and Noncovalent Interactions of Histidine Residues. J. Phys. Chem. A. 2013;117:5987–5996. doi: 10.1021/jp311815k. [DOI] [PubMed] [Google Scholar]
- 137.Barth A. The infrared absorption of amino acid side chains. Prog. Biophys. Mol. Bio. 2000;74:141–173. doi: 10.1016/s0079-6107(00)00021-3. [DOI] [PubMed] [Google Scholar]
- 138.Koziński M, Garrett-Roe S, Hamm P. 2D-IR spectroscopy of the sulfhydryl band of cysteines in the hydrophobic core of proteins. J. Phys. Chem. B. 2008;112:7645–7650. doi: 10.1021/jp8005734. [DOI] [PubMed] [Google Scholar]
- 139.Levinson NM, Bolte EE, Miller CS, Corcelli SA, Boxer SG. Phosphate Vibrations Probe Local Electric Fields and Hydration in Biomolecules. J. Am. Chem. Soc. 2011;133:13236–13239. doi: 10.1021/ja2042589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Suydam IT, Boxer SG. Vibrational Stark effects calibrate the sensitivity of vibrational probes for electric fields in proteins. Biochemistry. 2003;42:12050–12055. doi: 10.1021/bi0352926. [DOI] [PubMed] [Google Scholar]
- 141.Baiz CR, McRobbie PL, Anna JM, Geva E, Kubarych KJ. Two-Dimensional Infrared Spectroscopy of Metal Carbonyls. Acc. Chem. Res. 2009;42:1395–1404. doi: 10.1021/ar9000263. [DOI] [PubMed] [Google Scholar]
- 142.King JT, Kubarych KJ. Site-Specific Coupling of Hydration Water and Protein Flexibility Studied in Solution with Ultrafast 2D-IR Spectroscopy. J. Am. Chem. Soc. 2012;134:18705–18712. doi: 10.1021/ja307401r. [DOI] [PubMed] [Google Scholar]
- 143.King JT, Arthur EJ, Brooks CL, Kubarych KJ. Site-Specific Hydration Dynamics of Globular Proteins and the Role of Constrained Water in Solvent Exchange with Amphiphilic Cosolvents. J. Phys. Chem. B. 2012;116:5604–5611. doi: 10.1021/jp300835k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Osborne DG, Dunbar JA, Lapping JG, White AM, Kubarych KJ. Site-Specific Measurements of Lipid Membrane Interfacial Water Dynamics with Multidimensional Infrared Spectroscopy. J. Phys. Chem. B. 2013;117:15407–15414. doi: 10.1021/jp4049428. [DOI] [PubMed] [Google Scholar]
- 145.Woys AM, Mukherjee SS, Skoff DR, Moran SD, Zanni MT. A Strongly Absorbing Class of Non-Natural Labels for Probing Protein Electrostatics and Solvation with FTIR and 2D IR Spectroscopies. J. Phys. Chem. B. 2013;117:5009–5018. doi: 10.1021/jp402946c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Peran I, Oudenhoven T, Woys AM, Watson MD, Zhang TO, et al. General Strategy for the Bioorthogonal Incorporation of Strongly Absorbing, Solvation-Sensitive Infrared Probes into Proteins. J. Phys. Chem. B. 2014 doi: 10.1021/jp5008279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fafarman AT, Webb LJ, Chuang JI, Boxer SG. Site-specific conversion of cysteine thiols into thiocyanate creates an IR probe for electric fields in proteins. J. Am. Chem. Soc. 2006;128:13356–13357. doi: 10.1021/ja0650403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dutta S, Li Y-L, Rock W, Houtman JCD, Kohen A, Cheatum CM. 3-Picolyl Azide Adenine Dinucleotide as a Probe of Femtosecond to Picosecond Enzyme Dynamics. J. Phys. Chem. B. 2011;116:542–548. doi: 10.1021/jp208677u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Barth A, Zscherp C. What vibrations tell us about proteins. Q. Rev. Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 150.Zimmermann Jr, Thielges MC, Yu W, Dawson PE, Romesberg FE. Carbon–Deuterium Bonds as Site-Specific and Nonperturbative Probes for Time-Resolved Studies of Protein Dynamics and Folding. J. Phys. Chem. Lett. 2011;2:412–416. [Google Scholar]