Abstract
Dysregulation of sleep and metabolism has enormous health consequences. Sleep loss is linked to increased appetite and insulin insensitivity, and epidemiological studies link chronic sleep deprivation to obesity-related disorders including type II diabetes and cardiovascular disease. Interactions between sleep and metabolism involve the integration of signaling from brain regions regulating sleep, feeding, and metabolic function. Investigating the relationship between these processes provides a model to address more general questions of how the brain prioritizes homeostatically regulated behaviors. The availability of powerful genetic tools in the fruit fly, Drosophila melanogaster, allows for precise manipulation of neural function in freely behaving animals. There is a strong conservation of genes and neural circuit principles regulating sleep and metabolic function, and genetic screens in fruit flies have been effective in identifying novel regulators of these processes. Here, we review recent findings in the fruit fly that further our understanding of how the brain modulates sleep in accordance with metabolic state.
Introduction
Sleep represents a nearly universal behavior in the animal kingdom that affects diverse aspects of physiology and behavior. While our understanding of sleep regulation is rapidly improving, much less is known about how sleep interacts with other biological processes including immunity, memory, aging or metabolism. Epidemiological and experimental studies reveal functional interactions between sleep and metabolism. Sleep loss is linked to increased appetite and insulin insensitivity, and short sleeping individuals are more likely to develop obesity, metabolic syndrome, type II diabetes, and cardiovascular disease (Taheri et al. 2004; Chaput et al. 2007; Knutson and Van Cauter 2008). Conversely, metabolic state potently modulates sleep and circadian behavior (Laposky et al. 2008; Green et al. 2008; Froy and Miskin 2010). Flies and mammals suppress sleep when starved, presumably to forage for food, revealing the integration of sleep-wake regulation with metabolic state (Danguir & Nicolaidis, 1979; Keene et al., 2010). Here, we review recent findings in the fruit fly, Drosophila melanogaster, that explore interactions between sleep and metabolism. We suggest that currently, there is a sufficient understanding of sleep and metabolism to investigate more complex questions related to mechanisms of how these processes interact.
Sleep analysis in Drosophila
Sleep can be characterized by physiological changes in brain activity or through behaviors that accompany these changes (Sehgal and Mignot 2011). Flies, like mammals, display distinct electrophysiological patterns that correlate with wake and rest (Nitz et al. 2002; van Alphen et al. 2013). Additionally, flies display all behavioral hallmarks of sleep including extended periods of behavioral quiescence, rebound following deprivation, increased arousal threshold and species-specific changes in posture (Hendricks et al. 2000; Shaw et al. 2000). Multiple systems for behavioural analysis are available for high-throughput detection and measurement of fly activity including infrared monitoring and automated video tracking (Zimmerman et al. 2008; Pfeiffenberger et al. 2010; Donelson et al. 2012; Gilestro 2012). Sleep in Drosophila is typically defined by periods of behavioral quiescence lasting five minutes or longer. This characteristic associates with other behavioral hallmarks of sleep including enhanced arousal threshold and rebound following deprivation (Hendricks et al. 2000; Shaw et al. 2000). Recent findings measuring local field potentials to determine neural activity, and arousal threshold as an indicator for sensory gating, suggest that flies enter a ‘deep sleep’ following approximately 15 minutes of behavioral quiescence, raising the possibility that this is functionally analogous to slow wave sleep in mammals (van Alphen et al. 2013). Similarities are also present at a molecular level, where the sleep suppressing effects of the stimulants caffeine, cocaine, and modafinil are conserved from flies to humans (Shaw et al. 2000; Hendricks et al. 2003; Wu et al. 2009; Lebestky et al. 2009). Therefore, flies present an excellent genetic model for investigating the regulation of sleep in mammalian systems.
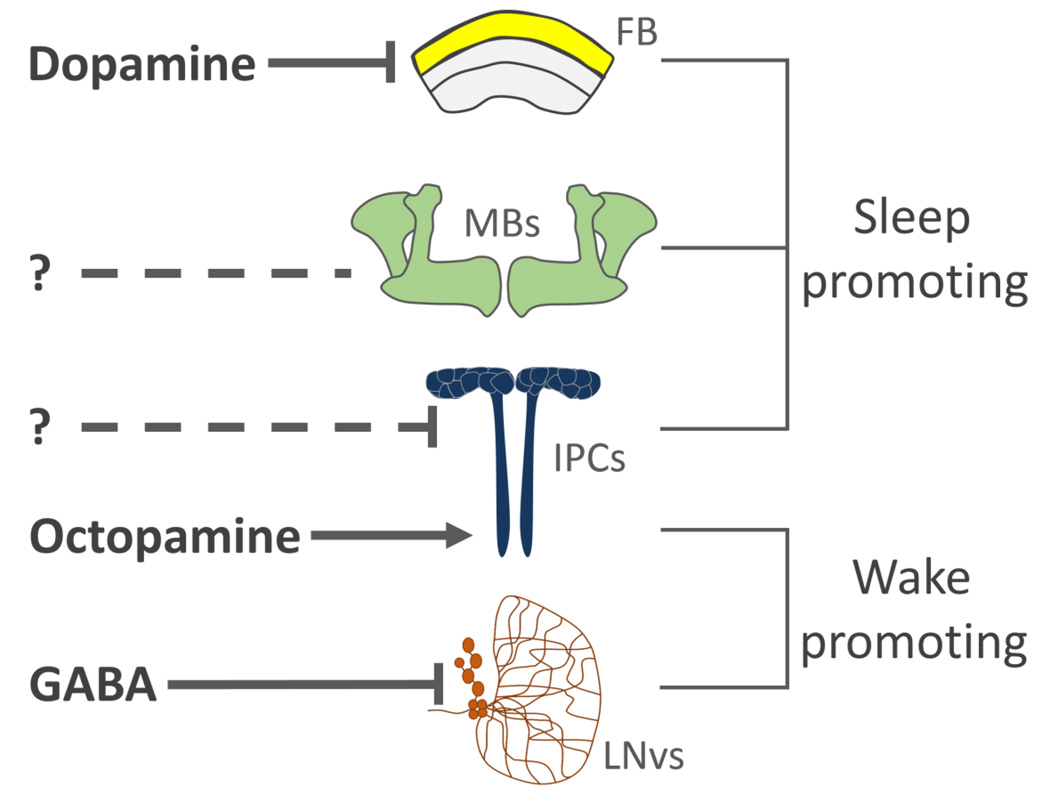
A powerful genetic tool kit in Drosophila allows for the identification of genes and neural circuits that regulate sleep (Griffith 2013). In flies and mammals, sleep-wake regulation involves dynamic interactions between sleep and wake-promoting neural circuits (Griffith 2013). Therefore, it is unlikely that sleep is controlled by a primary ‘sleep center’. Four neural loci involved in sleep-wake regulation appear to be particularly important for sleep-wake regulation: the sleep-promoting mushroom bodies, dopaminergic modulation of dorsal fan-shaped body (dFSB), the hypothalamus-like Insulin Producing Cells (IPCs), as well as a sleep-suppressing role for the circadian pacemaker neurons (reviewed in Griffith et al, 2013; Figure 1). The mushroom bodies are composed of ~5000 neurons that are critical for sensory integration and olfactory memory (Heisenberg 2003; Keene et al. 2004; Davis 2011). Ablation or genetic silencing of the mushroom bodies disrupts sleep, while genetic activation of this structure induces sleep, revealing that sleep is gated by mushroom body activity (Joiner et al. 2006; Pitman et al. 2006). The neurons innervating the mushroom bodies are well characterized and include monoaminergic and peptidergic modulatory neurons that are required for memory formation, and second-order olfactory projection neurons that connect the mushroom bodies to the antennal lobe (Tanaka et al. 2008; Claridge-Chang et al. 2009; Burke et al. 2012). While the sleep-regulating mushroom body-associated neurons are not well understood, the Dorsal Paired Medial (DPM) neurons innervate the mushroom bodies and are required for the formation of both memory and sleep (Yu et al. 2005; Liu et al. 2008; Akalal et al. 2011). Silencing DPM neurons results in fragmented sleep, raising the possibility that DPM-mushroom body connectivity is important for sleep maintenance (Liu et al. 2008).
Figure 1. Neural regulation of sleep and arousal in Drosophila.
The fan-shaped body (FB), mushroom bodies (MBs) and Insulin Producing Cells (IPC) are sleep-promoting centers within the fly brain. A) Dopaminergic innervation to the dorsal FB inhibits activity and the sleep-promoting effects of this region. B) The sleep-promoting neurons that innervate the MBs are not well understood. C) The IPCs promote sleep through a mechanism that is dependent on the EGFR ligand rhomboid. They also The IPCs also receive excitatory input from octopamine neurons that suppresses sleep. D) The ventral Lateral Neurons (LNvs) regulate circadian function and arousal. These neurons receive inhibitory GABAergic input that functions to promote sleep.
The Drosophila central complex is a neural center regulating locomotion and visual processing (Liu et al. 2006; Poeck et al. 2008; Seelig and Jayaraman 2013). Activation of neurons forming the dorsal fan-shaped body (dFSB), a substructure within the central complex, robustly induces sleep (Donlea et al. 2011). The dFSB receives input from the protocerebral posteriolateral cluster 1 (PPL1) and protocerebral posteriomedial 3 (PPM3) cluster of dopamine neurons that likely inhibit dFSB activity to suppress sleep (Ueno et al. 2012; Liu et al. 2012; Kayser et al. 2014). The excitability of dFSB neurons changes in accordance with sleep debt, supporting the notion that this is a central regulator of sleep drive (Donlea et al. 2014). The sleep-promoting effects of the dFSB are also critical for neuronal, behavioral and structural plasticity. Wake-promoting PPL1 dopamine neurons that innervate the dFSB are less active during early-life, resulting in enhanced sleep which is required for proper brain development (Kayser et al. 2014). Further supporting a role of the dFSB in sleep-dependent plasticity, the detrimental effects of early-life sleep deprivation on memory are rescued by targeted blockade of dopamine signaling, and thermogenetic activation of dFSB neurons facilitate the formation of long-term memory (Seugnet et al. 2011; Donlea et al. 2011). Therefore, in addition to homeostatic regulation of sleep, the dFSB appears to be critical for the integration of sleep with developmental and experience-dependent modification of behavior.
There is also considerable overlap between the neural circuitry regulating sleep and circadian rhythms. The primary pacemaker neurons, the ventral lateral neurons (LNvs), express the neuropeptide pigment dispersing factor (PDF) that promotes arousal and is essential for 24hr locomotor rhythms when animals are placed in constant darkness (Renn et al. 1999; Parisky et al. 2008). Sleep is enhanced in PDF signaling mutants indicating a wake-promoting role the LNvs (Parisky et al. 2008). The LNvs receive inhibitory input through GABA-A receptor activity that enhances sleep duration (Parisky et al. 2008; Agosto et al. 2009). Numerous cellular regulators of metabolism, including insulin signaling pathway components, function in LNvs pacemaker cells to regulate 24hr rhythms indicating that these neurons are involved in integrating metabolic cues with behavior (Zheng et al. 2007). Understanding how the identified sleep-regulating neurons function within a dynamic network that includes the neural circuitry regulating circadian rhythms will be critical for understanding how sleep is modulated within the brain.
Dietary regulation of sleep
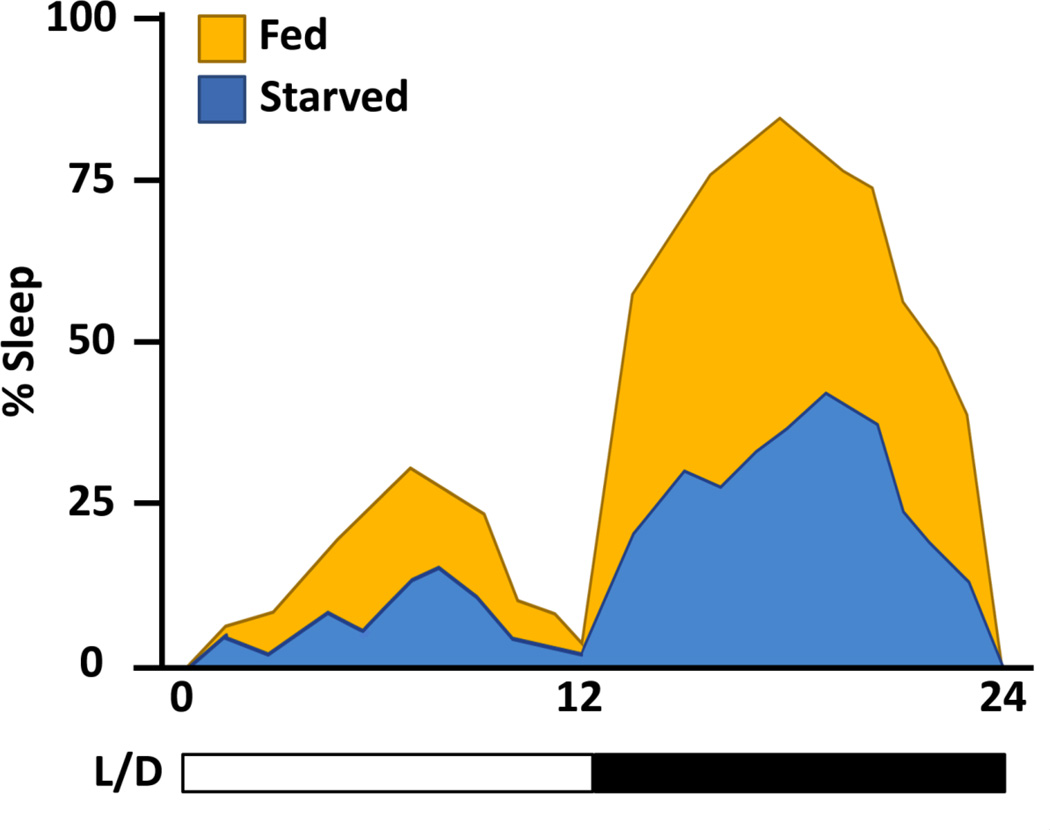
Flies starved on a diet of agar alone become hyperactive and reduce sleep, but the specific dietary components required for normal sleep are not known (Lee and Park 2004; Keene et al. 2010; McDonald and Keene 2010) Figure 2). In the wild, the Drosophila melanogaster diet consists primarily of complex sugars obtained by feeding primarily on rotting fruit (Atkinson and Shorrocks 1977). The laboratory diet of Drosophila is significantly more complex, consisting of carbohydrates, as well as protein and fat from yeast. Altering dietary components robustly affects behavior, physiology, and longevity (Mair et al. 2005; Lee and Micchelli 2013). A proper dietary balance of sugar and yeast is important for the maintenance of homeostasis and fitness. For example, raising dietary sugar concentration increases triglyceride levels, which can be suppressed by simultaneously increasing the yeast concentration (Skorupa et al. 2008). The protein component of yeast is required for proper growth and development in flies and larvae fed a sugar diet are severely undersized (Britton and Edgar 1998; Britton et al. 2002). Additionally, restricting caloric intake has been implicated in increasing lifespan and reducing reproductive output of the animal, revealing diet-related trade-offs between longevity and behavior (Chapman and Partridge 1996; Good and Tatar 2001; Piper et al. 2014).
Figure 2. Starved flies suppress sleep.
The percentage sleep time is depicted over 24hrs of testing. Fed flies (orange) sleep more during than flies starved on agar (blue). The behavior depicted is of flies tested under 12:12 light (L)/Dark (D) conditions.
A number of studies have examined the effects of different diets on fly sleep. Flies fed a diet of 5% sucrose-alone have a similar sleep duration to flies fed normal food suggesting that dietary protein is not required for this behavior (Keene et al. 2010). An alternative study examining the contributions of dietary sugar and yeast to sleep architecture reported no difference in total sleep duration between flies fed a high or low calorie diet of sucrose and yeast (Linford et al. 2012). Interestingly, increasing the dietary sucrose concentration from 5% to 35% does not alter the total sucrose consumed, but suppresses sleep (Catterson et al. 2010). Therefore, these studies seem to suggest that flies sleep normally when fed moderate concentrations of sucrose, but high concentrations of dietary sucrose suppress sleep through a mechanism that is independent of total caloric intake.
Gustatory and olfactory sensory inputs influence many behaviors including locomotor activity and food-searching strategies (Winther et al. 2006; Root et al. 2008). However, the effects of food on sleep duration appear to be independent from these sensory inputs. Flies lacking the sugar-sensing gustatory receptors Gr5a and Gr64a do not respond to sucrose and sleep normally, suggesting that the effects of dietary sugar on sleep are independent from sensory inputs (Dahanukar et al. 2007; Dus et al. 2011). Supporting these findings, flies fed the non-caloric sweetener sucralose suppress sleep similarly to flies starved on agar (Keene et al. 2010). While dietary perception of sugar does not appear to affect sleep duration, it may impact sleep quality. Feeding a sugar-only diet to flies lacking sugar receptors results in fragmented sleep, suggesting that sensory perception of sugar modulates sleep architecture, but not sleep duration (Linford et al. 2012). Therefore, gustatory sensation of sugar may consolidate sleep bouts without affecting total sleep duration.
Yeast represents the primary protein source for flies and its addition to the diet suppresses sleep in male, while enhancing sleep in female flies (Catterson et al. 2010). The nutritional value of yeast is predominantly in the form of amino acids, and it remains to be determined whether specific amino acids within dietary yeast modulate sleep. The amino acid methionine appears to be critical for fecundity and lifespan, raising the possibility that methionine may modulate sleep in aging animals (Grandison et al. 2009). Furthering our understanding of dietary contributions to sleep will require systematic approaches to measure both acute and long-term effects of dietary components on sleep. Taken together, these studies indicate that a diet of sugar alone is sufficient for normal sleep duration and that consolidation of sleep bouts may be dependent on the sensory perception of sugar.
The differences observed among studies examining the effects of diet on sleep may be in part due to different components of ‘standard diets’ and protocols for food preparation (Linford et al. 2012). Recently, a holidic diet has been described to be sufficient for long-term survival in Drosophila (Piper et al. 2014). This diet is composed of purified ingredients that include defined concentrations of amino acids, sucrose, and cholesterol, providing an opportunity to examine contributions of individual nutrients to sleep. Flies fed the holidic diet have similar sleep and activity to those maintained on a standard diet of sugar and yeast, but the effect of individual dietary components has not been tested (Piper et al. 2014). While it will be of great interest to determine the contributions of dietary components to sleep, it will remain difficult to account for changes in food consumption based on diet. Therefore, a confounding factor of these experiments will be the possibility of differences in calories consumed or temporal differences in meal consumption over the course of the sleep assay.
Neurohormonal regulation of metabolism
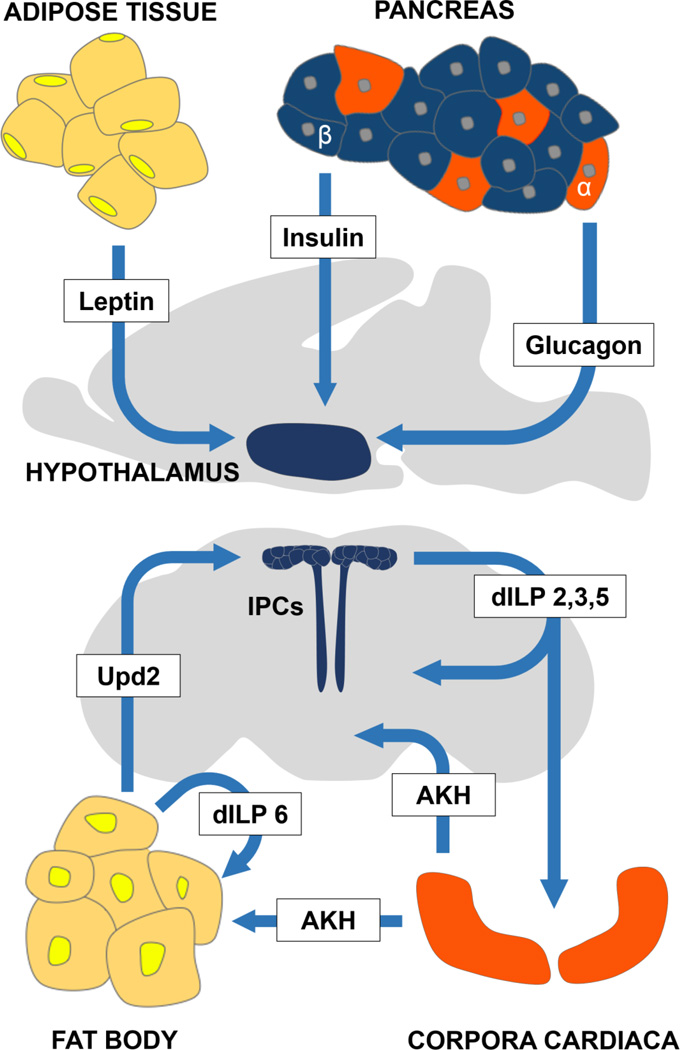
In mammals, the peptide hormones insulin and glucagon are critical for regulation of blood-glucose levels and energy availability (Saltiel and Kahn 2001; Unger and Cherrington 2012). Drosophila possess functional orthologs of insulin and glucagon that appear to have conserved roles in the regulation of metabolic function (Edgar 2006) (Figure 3). The glucagon ortholog adipokinetic hormone (AKH) is expressed in peptidergic secretory cells of the corpora cardiaca (CC) (Kim and Rulifson 2004; Lee and Park 2004; Park et al. 2008). The CC receives input from insulin producing cells (IPCs) and secretes AKH into the hemolymph (Rulifson et al. 2002; Kim and Rulifson 2004). Ablation of the CC results in hypoglycemia, highlighting the importance of these cells in glucose sensing and overall metabolic regulation (Kim and Rulifson 2004). AKH binds to the Adipokinetic Hormone Receptor (AKHR), a G-protein coupled receptor that is expressed in the brain, fat body, and possibly other tissues (Kim and Rulifson 2004; Bharucha et al. 2008). The subsequent breakdown of glycogen and lipids in muscles and fat body are then used for energy (Canavoso et al. 2001; Van der Horst 2003). Manipulations that impair AKH signaling promote glycogen and triglyceride storage, confirming a role for this pathway in controlling carbohydrate and fat metabolism (Kim and Rulifson 2004; Lee and Park 2004; Isabel et al. 2005). AKH function is also implicated in a number of behaviors associated with hunger-induced motivation, including odor-conditioned feeding approach, feeding, and locomotor behavior (Lee and Park 2004; Burt et al. 2014). Ablation of AKH-producing cells reduces the hyperlocomotor and feeding responses to starvation and increases starvation resistance (Lee and Park 2004; Bharucha et al. 2008). Therefore, AKH signaling potently regulates behavior in response to starvation, but it is unclear whether these changes are due to altered energy stores or the acute effects of AKH function.
Figure 3. Functional conservation between organs and cell-types regulating metabolic function.
A) Overview of regions that communicate metabolic regulation to the brain in mammals and Drosophila (B). Regions are color-coded to match their proposed analogous structure. Fat body cells (yellow) secrete unpaired 2 (upd2) the functional ortholog of mammalian leptin. The fat body cells also secrete ILP6 that is proposed to have autoregulatory properties. The Insulin Producing Cells (blue, IPCs) appear to function similarly to mammalian pancreatic beta-cells and the hypothalamus. The Drosophila corpora cardiaca (orange) secretes Adipkinetic Hormone (AKH), which is a functional ortholog of mammalian glucagon that is released from the pancreatic alpha-cells.
The Drosophila genome encodes for 8 distinct insulin-like peptide (ilp) genes that are structurally conserved with mammalian insulin and regulate metabolism, behavior, and growth during development (Brogiolo et al. 2001; Britton et al. 2002; Rulifson et al. 2002; Wu et al. 2005; DiAngelo and Birnbaum 2009). These genes differ in expression, temporal regulation, and function. Ilp2, ilp3 and ilp5 are expressed in 10–14 medial neurosecretory cells that regulate many behaviors including sleep and feeding through the systematic release of ILPs and direct innervation of the AKH-producing cells (Broughton et al. 2005). In larvae, both ilp3 and ilp5 are transcriptionally downregulated in response to starvation, suggesting a role in nutritional state-dependent regulation of behavior and metabolism (Ikeya et al. 2002). The 8 ILPs bind to a single insulin-like receptor (dInR), which is proposed to express ubiquitously (Fernandez et al. 1995). Unlike all other ilps, ilp6 is predominantly expressed in the liver/adipose tissue-like organ, the fat body, raising the possibility that insulin signaling in the fat body is auto-regulated (Okamoto et al. 2009). Activating insulin signaling specifically in the fat body promotes fat storage similar to mammals (Saltiel and Kahn 2001; DiAngelo and Birnbaum 2009). Therefore, ILPs and AKH have diverse functions in regulating metabolism and behavior.
Endocrine integration of sleep and metabolic function
A number of genes have been shown to function within the IPCs to regulate sleep, locomotor activity, and circadian rhythms. Specific subpopulations of IPC neurons express neuropeptide co-transmitters that regulate locomotor behavior (Cavanaugh et al. 2014). Furthermore, the Epidermal Growth Factor Receptor (EGFR) ligand rhomboid-1 (rho) is predominantly expressed in the IPCs, and heat-shock induction of the EGFR ligands, rho and Star, induce sleep, supporting the notion that the IPCs acutely regulate sleep (Foltenyi et al. 2007). Ablation of the IPCs increases sleep sensitivity to a reduced calorie diet, raising the possibility that these cells buffer against diet-induced alterations in sleep (Broughton et al. 2010). Supporting the notion that the IPCs are integrators of diet and activity, mutants for Drosophila Arc1, which is expressed in the IPCs, or flies with disrupted AKH function do not increase locomotor activity in response to starvation (Mattaliano et al. 2007). Therefore, the IPCs appear to be central integrators of sleep/activity and metabolic state and may function similarly to the mammalian hypothalamus to integrate these processes.
The IPCs co-secrete the insulin-like peptides ilp2, Ilp3 and ilp5, along with other neuropeptide co-transmitters. Overexpression of ilp2, as well as ectopic activation of insulin signaling in the fat body or brain does not alter sleep, suggesting that insulin-like signaling is not directly responsible for the IPC-dependent regulation of sleep (Erion et al. 2012). Deletion of all three ilps expressed in the IPCs protects against age-related disruption in sleep, suggesting the ilp release from IPCs regulates age or stress-dependent changes in sleep (Metaxakis et al. 2014). Future work examining the sleep phenotypes of ilp2, ilp3 and ilp5 flies, as well as other neuropeptides that are expressed in the IPCs, may be informative in uncovering the mechanism through which IPCs regulate sleep in response to aging and other environmental factors.
It is possible that IPCs, much like the pancreatic beta-cells, serve as tissue-type autonomous nutrient sensors. There is also strong support for octopamine, the fly analog of norepinephrine, targeting the IPCs to modulate sleep-wake behavior. Feeding flies octopamine or genetically activating octopamine neurons potently promotes wakefulness in Drosophila through the activation of PKA (Crocker and Sehgal 2008). Octopamine is expressed in ~100 cells in the brain that innervate diverse brain regions including the mushroom bodies and IPCs (Sinakevitch and Strausfeld 2006; Busch et al. 2009). Expressing the Na+ bacterial channel NaChBac to hyperactivate distinct classes of octopamine neurons demonstrates the role of the IPC-innervating anterior superior medial protocerebrum (ASM) neurons in suppressing sleep (Sinakevitch and Strausfeld 2006; Crocker et al. 2010). Selectively inhibiting PKA activity in the IPCs through expression of a PKA regulatory subunit blocks the effects of octopamine feeding on sleep, indicating that octopamine targets the IPCs (Crocker et al. 2010). Disruption of PKA function in other sleep promoting centers, including the mushroom bodies and dorsal fan shaped body, does not block the wake-promoting effects of octopamine (Crocker et al. 2010). Physiological imaging with the genetically encoded cAMP sensor EPAC-FRET confirms elevated cAMP levels in IPCs in response to octopamine, supporting the notion that these neurons express a Gαs coupled octopamine receptor (Crocker et al. 2010). In addition to sleep regulation, octopamine also targets the IPCs to regulate metabolism. While the wake-promoting effects are not dependent on ilp2 or ilp3, activating the octopamine-producing neurons enhances triglyceride levels. This increase in triglycerides is partially blocked in ilp2, ilp3 double mutants suggesting that the regulation of fat metabolism by octopamine is partially dependent on ilp2 and ilp3 (Erion et al. 2012). It is likely that the sleep-suppressing octopamine neurons that innervate the IPCs are distinct from those regulating energy stores. These findings reveal that both the wake-promoting and metabolic effects of octopamine are regulated by the IPCs, providing a link between insulin, sleep and metabolism.
A role for the fat body in sleep regulation
Adipose tissue senses overall nutrient levels in the animal and modulates behaviors through metabolic control of energy stores and secreted factors that regulate neural function (Ahima and Lazar 2008; Morton and Schwartz 2011). In Drosophila, the fat body is central to the control of energy homeostasis and represents the primary site of glycogen and triglyceride storage, as well as the main detoxification and immune organ of the fly (Hoshizaki, 2005). The Drosophila fat body has been implicated in regulation of numerous behaviors including courtship, feeding and egg-laying (Lazareva et al. 2007; Xu et al. 2008; Xu et al. 2011; Sassu et al. 2012). Several sleep regulating genes are expressed preferentially in the fat body including Angiotensin-converting enzyme peptidase (ACER). Genetic mutation or pharmalogical blockade of ACER have disrupted nightime sleep, raising the possibility that the fat body functions to promote sleep (Carhan et al. 2011). Fat body function appears to be particularly important for regulating homeostatic sleep changes in response to stressors including starvation and sleep-deprivation. Flies mutant for the adipose triglyceride lipase brummer, a gene highly expressed in the fat body, have elevated triglyceride stores and have an enhanced homeostatic response to sleep deprivation (Grönke et al. 2005; Thimgan et al. 2010). Conversely, flies mutant for lipid storage droplet 2 (lsd2) have reduced triglyceride levels and do not display a homeostatic rebound in response to sleep deprivation, revealing that triglyceride stores in the fat body enhance the homeostatic response to sleep deprivation (Thimgan et al. 2010). Enhanced triglycerides do not appear to acutely regulate sleep because flies mutant for lsd2 have normal sleep architecture (Linford et al. 2012). Therefore, the homeostatic response to sleep deprivation and basal regulation of sleep are likely regulated by distinct mechanisms.
Adipose tissue may modulate sleep through secretion of peptide hormones. In mammals, leptin secreted from adipose tissue binds to hypothalamic receptors to modulate sleep and feeding behavior (Inutsuka and Yamanaka 2013). Sleep deprivation disrupts leptin function, and this likely contributes to the increased feeding and weight gain that accompany chronic sleep deprivation in mammals (Taheri et al. 2004; Barf et al. 2012). Mice that lack the leptin receptor are obese and hypersomnolent, fortifying the notion that leptin inhibits feeding and promotes sleep (Laposky et al. 2006). Recently, the Drosophia cytokine upaired 2 (upd2) was identified as an ortholog of mammalian leptin (Rajan and Perrimon 2012). Secretion of upd2 from the fat body regulates insulin accumulation and release from the IPCs (Laposky et al. 2006). While the role for upd2 in sleep modulation has not been studied, these findings suggest that upd2 may function through the IPCs to regulate sleep in response to metabolic changes.
Novel genetic regulators of sleep-metabolism interactions
A number of genes that are required for metabolic regulation of sleep have been identified in Drosophila. Drosophila foraging (for) encodes for a cGMP-dependent Protein Kinase (PKG) that regulates feeding behaviors and context-dependent regulation of sleep. Naturally occurring polymorphisms in for effect a number of behaviors. forrover (forR) flies have higher levels of PKG activity than forSitter (forS) flies and this polymophism results in a number of sleep-related behavioral differences. Flies with the forS polymorphism do not suppress sleep in response to starvation suggesting that modulation of PKG activity is critical for the integration of metabolism and sleep (Keene et al. 2010). In addition to regulating metabolism and sleep, for appears to also regulate trade-offs between resilience to sleep deprivation and starvation. Enhanced PKG activity increases vulnerability to starvation-induced memory loss but protects against mechanically induced sleep deprivation, suggesting that for is involved in a trade-off between sleep and memory loss (Donlea et al. 2012). This effect is localized to the mushroom bodies, a region that is not required for starvation-induced sleep suppression (Keene et al. 2010), raising the possibility that for acts in different regions of the brain to regulate starvation-induced responses and sleep and memory.
Both sleep and metabolism are influenced by the circadian system and disruption of the transcriptional activators Clock or cycle increases sleep loss in response to starvation (Keene et al. 2010). Interestingly, both wild-type and cycle mutant flies lack a compensatory sleep rebound following starvation, suggesting that sleep loss through starvation involves mechanisms that are distinct from mechanical or pharmacological sleep loss (Thimgan et al. 2010). The increased sensitivity of Clock and cycle mutants to starvation-induced sleep suppression is unlikely to be caused by reduced energy stores because adiposity is enhanced in cycle mutants and selectively disrupting Clock in the fat body does not affect sleep suppression during starvation (Thimgan et al. 2010; Keene et al. 2010). The effect of Clk on sleep regulation during starvation appears to localize to the dorsally located populations of circadian neurons that may receive inputs from the arousal promoting LNvs (Keene et al. 2010). These same dorsal clock neurons have previously been suggested to propagate signals through the IPCs, raising the possibility that Clock/cycle modulate insulin release (Rajan and Perrimon 2012). Therefore, a population of circadian neurons regulates interactions between sleep and metabolism, possibly through interactions with IPC neurons.
Conclusions
Powerful genetic tools and behavioral assays are available for investigating the genes and neurons regulating interactions between sleep and metabolism in Drosophila. The metabolic peptide hormones insulin and glucagon-like AKH are key regulators of sleep and locomotor activity. Forward genetic screens in the fly have led to the identification of many novel regulators of sleep and metabolism and it is likely that further study of these genes will advance our understanding of interactions between these processes. In addition, the role of the fat body in Drosophila needs to be further explored in order to define how sleep-regulating neurons are modulated in accordance with metabolic state. Examining interactions between sleep and metabolism provides the opportunity to explore how the brain communicates with peripheral metabolic organs to control physiology, metabolism and behavior.
Bibliography
- Agosto J, Choi JC, Parisky KM, et al. NIH Public Access. 2009;11:354–359. [Google Scholar]
- Ahima RS, Lazar MA. Adipokines and the peripheral and neural control of energy balance. Mol Endocrinol. 2008;22:1023–1031. doi: 10.1210/me.2007-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalal D-BG, Yu D, Davis RL. The long-term memory trace formed in the Drosophila α/β mushroom body neurons is abolished in long-term memory mutants. J Neurosci. 2011;31:5643–5647. doi: 10.1523/JNEUROSCI.3190-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson W, Shorrocks B. Breeding site specificity in the domestic species of Drosophila. Oecologia. 1977;29:223–232. doi: 10.1007/BF00345697. [DOI] [PubMed] [Google Scholar]
- Barf RP, Desprez T, Meerlo P, Scheurink AJW. Increased food intake and changes in metabolic hormones in response to chronic sleep restriction alternated with short periods of sleep allowance. Am J Physiol Regul Integr Comp Physiol. 2012;302:R112–R117. doi: 10.1152/ajpregu.00326.2011. [DOI] [PubMed] [Google Scholar]
- Bharucha KN, Tarr P, Zipursky SL. A glucagon-like endocrine pathway in Drosophila modulates both lipid and carbohydrate homeostasis. J Exp Biol. 2008;211:3103–3110. doi: 10.1242/jeb.016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- Britton JS, Lockwood WK, Li L, et al. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, et al. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MDW, Ikeya T, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton SJ, Slack C, Alic N, et al. DILP-producing median neurosecretory cells in the Drosophila brain mediate the response of lifespan to nutrition. Aging Cell. 2010;9:336–346. doi: 10.1111/j.1474-9726.2010.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke CJ, Huetteroth W, Owald D, et al. Layered reward signalling through octopamine and dopamine in Drosophila. Nature. 2012;492:433–437. doi: 10.1038/nature11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt J, Dube L, Thibault L, Gruber R. Sleep and eating in childhood: a potential behavioral mechanism underlying the relationship between poor sleep and obesity. Sleep Med. 2014;15:71–75. doi: 10.1016/j.sleep.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Busch S, Selcho M, Ito K, Tanimoto H. A map of octopaminergic neurons in the Drosophila brain. J Comp Neurol. 2009;513:643–667. doi: 10.1002/cne.21966. [DOI] [PubMed] [Google Scholar]
- Canavoso LE, Jouni ZE, Karnas KJ, et al. Fat metabolism in insects. Annu Rev Nutr. 2001;21:23–46. doi: 10.1146/annurev.nutr.21.1.23. [DOI] [PubMed] [Google Scholar]
- Carhan A, Tang K, Shirras CA, et al. Loss of Angiotensin-converting enzyme-related (ACER) peptidase disrupts night-time sleep in adult Drosophila melanogaster. J Exp Biol. 2011;214:680–686. doi: 10.1242/jeb.049353. [DOI] [PubMed] [Google Scholar]
- Catterson JH, Knowles-Barley S, James K, et al. Dietary modulation of Drosophila sleep-wake behaviour. PLoS One. 2010;5:e12062. doi: 10.1371/journal.pone.0012062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh DJ, Geratowski JD, Wooltorton JRA, et al. Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell. 2014;157:689–701. doi: 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Partridge L. Female fitness in Drosophila melanogaster: an interaction between the effect of nutrition and of encounter rate with males. Proc Biol Sci. 1996;263:755–759. doi: 10.1098/rspb.1996.0113. [DOI] [PubMed] [Google Scholar]
- Chaput J-P, Després J-P, Bouchard C, Tremblay A. Association of sleep duration with type 2 diabetes and impaired glucose tolerance. Diabetologia. 2007;50:2298–2304. doi: 10.1007/s00125-007-0786-x. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A, Roorda RD, Vrontou E, et al. Writing memories with light-addressable reinforcement circuitry. Cell. 2009;139:405–415. doi: 10.1016/j.cell.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A, Sehgal A. Octopamine regulates sleep in drosophila through protein kinase A-dependent mechanisms. J Neurosci. 2008;28:9377–9385. doi: 10.1523/JNEUROSCI.3072-08a.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron. 2010;65:670–681. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar A, Lei Y-T, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danguir J, Nicolaidis S. Dependence of sleep on nutrients’ availability. Physiol Behav. 1979;22:735–740. doi: 10.1016/0031-9384(79)90240-3. [DOI] [PubMed] [Google Scholar]
- Davis RL. Traces of Drosophila memory. Neuron. 2011;70:8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAngelo JR, Birnbaum MJ. Regulation of fat cell mass by insulin in Drosophila melanogaster. Mol Cell Biol. 2009;29:6341–6352. doi: 10.1128/MCB.00675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson NC, Donelson N, Kim EZ, et al. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS One. 2012;7:e37250. doi: 10.1371/journal.pone.0037250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea J, Leahy A, Thimgan MS, et al. Foraging alters resilience/vulnerability to sleep disruption and starvation in Drosophila. Proc Natl Acad Sci U S A. 2012;109:2613–2618. doi: 10.1073/pnas.1112623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Pimentel D, Miesenböck G. Neuronal machinery of sleep homeostasis in Drosophila. Neuron. 2014;81:860–872. doi: 10.1016/j.neuron.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Thimgan MS, Suzuki Y, et al. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science. 2011;332:1571–1576. doi: 10.1126/science.1202249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dus M, Min S, Keene AC, et al. Taste-independent detection of the caloric content of sugar in Drosophila. 2011:2–7. doi: 10.1073/pnas.1017096108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- Erion R, DiAngelo JR, Crocker A, Sehgal A. Interaction between sleep and metabolism in Drosophila with altered octopamine signaling. J Biol Chem. 2012;287:32406–32414. doi: 10.1074/jbc.M112.360875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez R, Tabarini D, Azpiazu N, et al. The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 1995;14:3373–3384. doi: 10.1002/j.1460-2075.1995.tb07343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- Froy O, Miskin R. Effect of feeding regimens on circadian rhythms: implications for aging and longevity. Aging (Albany NY) 2010;2:7–27. doi: 10.18632/aging.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilestro GF. Video tracking and analysis of sleep in Drosophila melanogaster. Nat Protoc. 2012;7:995–1007. doi: 10.1038/nprot.2012.041. [DOI] [PubMed] [Google Scholar]
- Good TP, Tatar M. Age-specific mortality and reproduction respond to adult dietary restriction in Drosophila melanogaster. J Insect Physiol. 2001;47:1467–1473. doi: 10.1016/s0022-1910(01)00138-x. [DOI] [PubMed] [Google Scholar]
- Grandison RC, Piper MDW, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LC. Neuromodulatory control of sleep in Drosophila melanogaster: integration of competing and complementary behaviors. Curr Opin Neurobiol. 2013;23:819–823. doi: 10.1016/j.conb.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S, Mildner A, Fellert S, et al. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Kirk D, Panckeri K, et al. Modafinil maintains waking in the fruit fly drosophila melanogaster. Sleep. 2003;26:139–146. doi: 10.1093/sleep/26.2.139. [DOI] [PubMed] [Google Scholar]
- Horne J. REM sleep, energy balance and “optimal foraging”. Neurosci Biobehav Rev. 2009;33:466–474. doi: 10.1016/j.neubiorev.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, et al. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- Inutsuka A, Yamanaka A. The physiological role of orexin/hypocretin neurons in the regulation of sleep/wakefulness and neuroendocrine functions. Front Endocrinol (Lausanne) 2013;4:18. doi: 10.3389/fendo.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel G, Martin J-R, Chidami S, et al. AKH-producing neuroendocrine cell ablation decreases trehalose and induces behavioral changes in Drosophila. Am J Physiol Regul Integr Comp Physiol. 2005;288:R531–R538. doi: 10.1152/ajpregu.00158.2004. [DOI] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Kayser MS, Yue Z, Sehgal A. A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science. 2014;344:269–274. doi: 10.1126/science.1250553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, Duboué ER, McDonald DM, et al. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol. 2010;20:1209–1215. doi: 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, Stratmann M, Keller A, et al. Diverse odor-conditioned memories require uniquely timed dorsal paired medial neuron output. Neuron. 2004;44:521–533. doi: 10.1016/j.neuron.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Kim SK, Rulifson EJ. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. 2004;431:316–320. doi: 10.1038/nature02897. [DOI] [PubMed] [Google Scholar]
- Knutson KL, Van Cauter E. Associations between sleep loss and increased risk of obesity and diabetes. Ann N Y Acad Sci. 2008;1129:287–304. doi: 10.1196/annals.1417.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laposky AD, Bass J, Kohsaka A, Turek FW. Sleep and circadian rhythms: key components in the regulation of energy metabolism. FEBS Lett. 2008;582:142–151. doi: 10.1016/j.febslet.2007.06.079. [DOI] [PubMed] [Google Scholar]
- Laposky AD, Shelton J, Bass J, et al. Altered sleep regulation in leptin-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2006;290:R894–R903. doi: 10.1152/ajpregu.00304.2005. [DOI] [PubMed] [Google Scholar]
- Lazareva Aa, Roman G, Mattox W, et al. A role for the adult fat body in Drosophila male courtship behavior. PLoS Genet. 2007;3:e16. doi: 10.1371/journal.pgen.0030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebestky T, Chang J-SC, Dankert H, et al. Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron. 2009;64:522–536. doi: 10.1016/j.neuron.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W-C, Micchelli CA. Development and characterization of a chemically defined food for Drosophila. PLoS One. 2013;8:e67308. doi: 10.1371/journal.pone.0067308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linford NJ, Chan TP, Pletcher SD. Re-patterning sleep architecture in Drosophila through gustatory perception and nutritional quality. PLoS Genet. 2012;8:e1002668. doi: 10.1371/journal.pgen.1002668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Seiler H, Wen A, et al. Distinct memory traces for two visual features in the Drosophila brain. Nature. 2006;439:551–556. doi: 10.1038/nature04381. [DOI] [PubMed] [Google Scholar]
- Liu Q, Liu S, Kodama L, et al. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol. 2012;22:2114–2123. doi: 10.1016/j.cub.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Guo F, Lu B, Guo A. amnesiac regulates sleep onset and maintenance in Drosophila melanogaster. Biochem Biophys Res Commun. 2008;372:798–803. doi: 10.1016/j.bbrc.2008.05.119. [DOI] [PubMed] [Google Scholar]
- Mair W, Piper MDW, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005;3:e223. doi: 10.1371/journal.pbio.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaliano MD, Montana ES, Parisky KM, et al. The Drosophila ARC homolog regulates behavioral responses to starvation. Mol Cell Neurosci. 2007;36:211–221. doi: 10.1016/j.mcn.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald DM, Keene AC. The sleep-feeding conflict: Understanding behavioral integration through genetic analysis in Drosophila. Aging (Albany NY) 2010;2:519–522. doi: 10.18632/aging.100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A, Tain LS, Grönke S, et al. Lowered insulin signalling ameliorates age-related sleep fragmentation in Drosophila. PLoS Biol. 2014;12:e1001824. doi: 10.1371/journal.pbio.1001824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton GJ, Schwartz MW. Leptin and the central nervous system control of glucose metabolism. Physiol Rev. 2011;91:389–411. doi: 10.1152/physrev.00007.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitz DA, van Swinderen B, Tononi G, Greenspan RJ. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr Biol. 2002;12:1934–1940. doi: 10.1016/s0960-9822(02)01300-3. [DOI] [PubMed] [Google Scholar]
- Okamoto N, Yamanaka N, Yagi Y, et al. A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev Cell. 2009;17:885–891. doi: 10.1016/j.devcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Veenstra Ja, Park JH, Taghert PH. Mapping peptidergic cells in Drosophila: where DIMM fits in. PLoS One. 2008;3:e1896. doi: 10.1371/journal.pone.0001896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Locomotor activity level monitoring using the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5518. pdb.prot5518. [DOI] [PubMed] [Google Scholar]
- Piper MDW, Blanc E, Leitão-Gonçalves R, et al. A holidic medium for Drosophila melanogaster. Nat Methods. 2014;11:100–105. doi: 10.1038/nmeth.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Poeck B, Triphan T, Neuser K, Strauss R. Locomotor control by the central complex in Drosophila-An analysis of the tay bridge mutant. Dev Neurobiol. 2008;68:1046–1058. doi: 10.1002/dneu.20643. [DOI] [PubMed] [Google Scholar]
- Rajan A, Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell. 2012;151:123–137. doi: 10.1016/j.cell.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, et al. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Root CM, Masuyama K, Green DS, et al. A presynaptic gain control mechanism fine-tunes olfactory behavior. Neuron. 2008;59:311–321. doi: 10.1016/j.neuron.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Sassu ED, McDermott JE, Keys BJ, et al. Mio/dChREBP coordinately increases fat mass by regulating lipid synthesis and feeding behavior in Drosophila. Biochem Biophys Res Commun. 2012;426:43–48. doi: 10.1016/j.bbrc.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig JD, Jayaraman V. Feature detection and orientation tuning in the Drosophila central complex. Nature. 2013;503:262–266. doi: 10.1038/nature12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Mignot E. Genetics of sleep and sleep disorders. Cell. 2011;146:194–207. doi: 10.1016/j.cell.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L, Suzuki Y, Donlea JM, et al. Sleep deprivation during early-adult development results in long-lasting learning deficits in adult Drosophila. Sleep. 2011;34:137–146. doi: 10.1093/sleep/34.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Sinakevitch I, Strausfeld NJ. Comparison of octopamine-like immunoreactivity in the brains of the fruit fly and blow fly. J Comp Neurol. 2006;494:460–475. doi: 10.1002/cne.20799. [DOI] [PubMed] [Google Scholar]
- Skorupa Da, Dervisefendic A, Zwiener J, Pletcher SD. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell. 2008;7:478–490. doi: 10.1111/j.1474-9726.2008.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri S, Lin L, Austin D, et al. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med. 2004;1:e62. doi: 10.1371/journal.pmed.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka NK, Tanimoto H, Ito K. Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol. 2008;508:711–755. doi: 10.1002/cne.21692. [DOI] [PubMed] [Google Scholar]
- Thimgan MS, Suzuki Y, Seugnet L, et al. The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS Biol. 2010 doi: 10.1371/journal.pbio.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Tomita J, Tanimoto H, et al. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci. 2012;15:1516–1523. doi: 10.1038/nn.3238. [DOI] [PubMed] [Google Scholar]
- Unger RH, Cherrington AD. Glucagonocentric restructuring of diabetes: a pathophysiologic and therapeutic makeover. J Clin Invest. 2012;122:4–12. doi: 10.1172/JCI60016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alphen B, Yap MHW, Kirszenblat L, et al. A dynamic deep sleep stage in Drosophila. J Neurosci. 2013;33:6917–6927. doi: 10.1523/JNEUROSCI.0061-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Horst DJ. Insect adipokinetic hormones: release and integration of flight energy metabolism. Comp Biochem Physiol B Biochem Mol Biol. 2003;136:217–226. doi: 10.1016/s1096-4959(03)00151-9. [DOI] [PubMed] [Google Scholar]
- Winther AME, Acebes A, Ferrús A. Tachykinin-related peptides modulate odor perception and locomotor activity in Drosophila. Mol Cell Neurosci. 2006;31:399–406. doi: 10.1016/j.mcn.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Wu MN, Ho K, Crocker A, et al. The effects of caffeine on sleep in Drosophila require PKA activity, but not the adenosine receptor. J Neurosci. 2009;29:11029–11037. doi: 10.1523/JNEUROSCI.1653-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhao Z, Shen P. Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat Neurosci. 2005;8:1350–1355. doi: 10.1038/nn1540. [DOI] [PubMed] [Google Scholar]
- Xu K, DiAngelo JR, Hughes ME, et al. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 2011;13:639–654. doi: 10.1016/j.cmet.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Keene AC, Srivatsan A, et al. Drosophila DPM neurons form a delayed and branch-specific memory trace after olfactory classical conditioning. Cell. 2005;123:945–957. doi: 10.1016/j.cell.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Zheng X, Yang Z, Yue Z, et al. FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc Natl Acad Sci U S A. 2007;104:15899–15904. doi: 10.1073/pnas.0701599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JE, Raizen DM, Maycock MH, et al. A video method to study Drosophila sleep. Sleep. 2008;31:1587–1598. doi: 10.1093/sleep/31.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]