Abstract
Capuramycin (1) and its analogs are strong translocase I (MurX/MraY) inhibitors. In our SAR studies of capuramycin analogs against M. tuberculosis (Mtb), we observed for the first time that a capuramycin analog, UT-01320 (3) killed non-replicating (dormant) Mtb at low concentrations under low-oxygen conditions, whereas selective MurX inhibitors killed only replicating Mtb under aerobic conditions. Interestingly, 3 did not exhibit MurX enzyme inhibitory activity even at high concentrations, however, 3 inhibited bacterial RNA polymerases with the IC50 values of 100-150 nM range. A new RNA polymerase inhibitor 3 displayed strong synergistic effects with a MurX inhibitor SQ 641 (2), a promising preclinical TB drug.
Keywords: Translocase I, MraY (MurX), RNA polymerase, Non-replicating (dormant) M. tuberculosis, Synergistic effect, TB drug
INTRODUCTION
One-third of the 42 million people living with HIV/AIDS worldwide are co-infected with Mycobacterium tuberculosis (Mtb).1,2 The WHO estimated that 650,000 new cases of multidrug-resistant (MDR)-Mtb emerge each year.3 An outbreak of extensively-drug resistant (XDR)-Mtb threatens TB control and prevention efforts.4 Treatment duration for MDR-Mtb infections is at least 20-28 months. Tuberculosis chemotherapy for XDR-TB takes substantially longer than MDR-TB, and XDR-Mtb strains are responsible for very high mortality rate.5 Therefore, it is very important to discover new drugs that can shorten current TB drug regimens. Mechanisms that enter non-replicating (or dormant) state of Mtb are accounted for a significant factor that requires long-term chemotherapy.6 Wayne et. al. reported that oxygen starvation is linked to TB drug resistance; upon depletion of oxygen in culture, Mtb terminates growth and develops into a characteristic dormant form.7,8 Significantly, the dormant form of Mtb was found to be resistant to most of clinically utilized antimycobacterial agents.8 Thus, new drugs targeting non-replicating Mtb are likely to revolutionize TB chemotherapy.
The cell-wall of Mtb offers many unique targets for drug development.9 However, most of drugs associated with cell-wall biosynthesis have proven difficult to reduce treatment time of TB drug regimens due to the facts that the dormant bacteria are not actively synthesizing cell-walls.10 On the contrary, it was recently reported that a peptidoglycan biosynthesis inhibitor, meropenem (a carbapenem) was effective in killing non-replicating Mtb in combination with clavulanate (a β-lactamase inhibitor).11 Although a mechanism of action of their bactericidal effect against dormant Mtb cells is not known, it is one of few examples that peptidoglycan biosynthesis inhibitors kill dormant form of Mtb. Because several translocase I (MurX/MraY, hereafter referred to as Mur X for Mtb translocase I) inhibitors kill Mtb much faster than other TB drugs under aerobic conditions (Figure 1),12 we commenced SAR studies of capuramycin (1), a known MurX inhibitor antibiotic, to improve efficacy of its antimycobacterial activity in vitro and in vivo (Figure 2).13,14,15 Daiichi-Sankyo and Sequella reported several capuramycin analogs in which in vitro MraY enzyme and antimycobacterial activity could be improved via the modification of the carboxylic group of the capuramycin biosynthetic intermediate, A-500359.16,17,18,19 We have synthesized new capuramycin analogs via our total synthetic scheme,15 in which all analogs are structurally different from the reported molecules and they are difficult to access from A-500359. In screening of new capuramycin analogs against replicating and non-replicating (dormant) Mtb, it was found that a 2′-methylated capuramycin analog, UT-01320 (3) killed both replicating and non-replicating Mtb in microplate alamar blue assay (MABA) and Low-oxygen recovery assay (LORA), respectively.20 To the best of our knowledge, it is the first observation that a capuramycin analog exhibited bactericidal activity against non-replicating Mtb at low concentrations. Herein, we report in vitro biological evaluations of 3, synergistic effect with known MurX inhibitors 1 or 2, and insights into a molecular target of 3 (Figure 2).
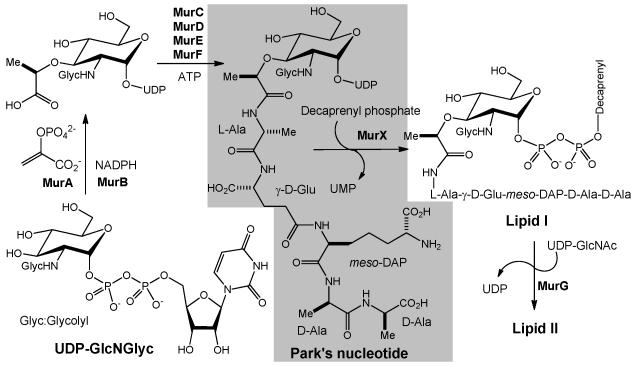
Figure 1.
Biosynthesis of lipid II in M. tuberculosis. Translocase I (MurX) catalyzes the transformation from Park’s nucleotide to lipid I.
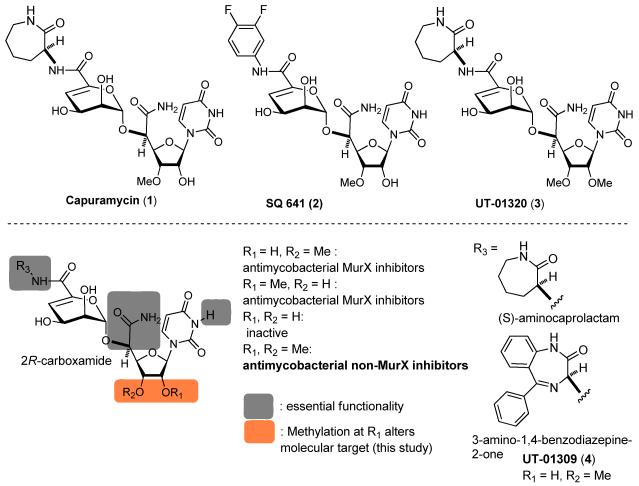
Figure 2.
Structures of capuramycin (1), SQ 641 (2), UT-01320 (3), and UT-01309 (4) and summary of SAR obtained from a 100-membered library.
MATERIALS AND METHODS
General / Chemicals and Reagents
All chemicals were purchased from commercial sources and used without further purification unless otherwise noted. Difco Middlebrook 7H10 agar, Middlebrook 7H9 broth, Tryptic soy agar, Tryptic soy broth, MOPS, tris(hydroxymethyl)aminomethane, 2-mercaptoethanol, sucrose and triton-X 100 were purchased from Sigma-Aldrich. ADC enrichment was purchased from Fisher Scientific. Magnesium chloride and potassium chloride were obtained from VWR. RNA polymerase assay kit (S. aureus and E. coli) was purchased from Profoldin. The kit contained 10× assay buffer, 100× DNA template, 100× NTP mix, 100× S. aureus or E. coli RNA polymerase (RNAP) enzyme and 10× fluorescence dye.
Tetrahydrofuran (THF), methylene chloride (CH2Cl2), dimethyformamide (DMF) were purified via MBRAUN Solvent Purification Systems (MB-SPS) under an Argon atmosphere. Reactions were monitored by thin-layer chromatography (TLC) performed with 0.25 mm coated commercial silica gel plates (EMD, Silica Gel 60F254) using UV light for visualization at 254 nm, or developed with ceric ammonium molybdate or anisaldehyde or copper sulfate or ninhydrin solutions by heating on a hot plate. Reactions were also monitored by using SHIMADZU LCMS-2020 with solvents: A: 0.1% formic acid in water, B: acetonitrile. When necessary, reactions were monitored by SHIMADZU prominence HPLC using Phenomenex Kinetex 1.7 μ XB-C18 100A column (150 × 2.10 mm) and detected at 220, 254 nm. Flash chromatography was performed with Whatman silica gel (Purasil 60 Å, 230-400 Mesh). Proton magnetic resonance (1H-NMR) spectral data were recorded on 400, and 500 MHz instruments. Carbon magnetic resonance (13C-NMR) spectral data were recorded on 100 and 125 MHz instruments. For all NMR spectra, chemical shifts (δ H, δ C) were quoted in parts per million (ppm), and J values were quoted in Hz. 1H and 13C NMR spectra were calibrated with residual undeuterated solvent (CDCl3: δ H =7.26 ppm, δ C =77.16ppm; CD3CN: δ H=1.94ppm, δ C =1.32ppm; CD3OD: δ H =3.31ppm, δ C =49.00 ppm; DMSO-d6: δ H=2.50ppm, δ C =39.5ppm; D2O: δ H=4.79 ppm) as an internal reference. The following abbreviations were used to designate the multiplicities: s=singlet, d=doublet, dd=double doublets, t=triplet, q=quartet, quin=quintet, hept=heptet, m=multiplet, br=broad.
Bacterial strains
The strains used were Mycobacterium smegmatis (ATCC 607) and Mycobacterium tuberculosis H37Rv, H37Rv INHr, H37Rv RFPr, E. coli (ATCC 25019), S. aureus (ATCC 6538D-5), E. faecium (ATCC 349), K. pneumonia (ATCC 8047), and P. aeruginosa (ATCC 27853). These bacteria were obtained from ATCC except for Mycobacterium tuberculosis H37Rv (BEI Resources, NIAID).
MIC assays
Log phase bacterial culture
A single colony of a bacterial strain (M. smegmatis or M. tuberculosis) was grown on a Difco Middlebrook 7H10 nutrient agar (enriched with OADC). Seed cultures and larger cultures were obtained using Middlebrook 7H9 broth enriched with OADC. Single colonies of S. aureus, E. faecium, E. coli, K. pneumonia and P. aeruginosa strains were grown on tryptic soy agar. The flasks were incubated overnight in a shaking incubator at 37 °C with a shaking speed of 200 rpm and cultured to mid-log phase (Optical density - 0.5). The optical density was monitored at 600 nm using a 96 well microplate reader.
Determination of minimum inhibitory concentration (MIC)
The inhibitors were dissolved in DMSO (a final concentration of 1 mg per 100 μL). This concentration was used as the stock solution for all studies. Bacterial cultures at 0.5 optical density, was treated with serial dilutions of inhibitors in aerobic conditions and incubated at 37 °C for 48 hours and 15 days for M. smegmatis and M. tuberculosis respectively. Incubation time was 24 hours for S. aureus, E. faecium, E. coli, K. pneumonia and P. aeruginosa strains. 20 μL of alamar blue was added and incubated on a shaking incubator at 37 °C for 2 h. The lowest concentration at which the color of alamar blue was completely retained as blue was read as the MIC (Pink = Growth, Blue = No growth). The absorbance measurements were also performed using a Biotek Synergy XT, 96 well plate reader at 570 nm and 600 nm.
Luminescence-based low-oxygen-recovery assay (LORA) assay
These assays were performed according to the reported procedures.20 In brief, M. tuberculosis H37Rv cells were transformed by mixing at least 1 μg of purified the plasmid, pFCA-luxAB and incubating at room temperature for 30 min, followed by electroporation.21 M. tuberculosis pFCA-luxAB strain cultured was diluted in Middlebrook 7H12 broth, and sonicated for 15s. The cultures were diluted to obtain an A570 of 0.03 to 0.05 and 3,000 to 7,000 RLUs per 100 μl. Twofold serial dilutions of antimicrobial agents were prepared in black 96-well microtiter plates (100 μl), and 100 μl of the cell suspension was added. The microplate was placed under anaerobic conditions (oxygen concentration, less than 0.16%) by using an Anoxomat model WS-8080 (MART Microbiology) and three cycles of evacuation and filling with a mixture of 10% H2, 5% CO2, and 85% N2. Incubation was continued for 10 days, and transferred to an ambient gaseous condition (5% CO2-enriched air) incubator for a 28h “recovery.” 100 μl culture was transferred to white 96-well microtiter plates for determination of luminescence.
MIC on nutrient starved M. tuberculosis
M. tuberculosis H37Rv was grown in conical flasks with air filter, containing Middlebrook 7H9 medium supplemented with OADC at 37 °C. Exponentially growing bacilli at an OD600 of 0.2 were transferred to PBS supplemented with 0.025% Tween 80 and diluted to a final OD600 of 0.1. 50 ml of this suspension was transferred into a conical flask and starved for 14 days at 37 °C. 100 uL of the culture was transferred to a 96 well plate and MIC experiment was performed according to the procedure described above.22
Kill-curve graph: Determination of colony forming units per milliliter
After the treatment of M. tuberculosis culture with the inhibitors, 10, 100, 1000 and 10,000 fold serial dilutions from each culture tube was sampled every day and 20μL from each dilution was plated on 7H10 agar plates supplemented with OADC enrichment. Plates were incubated for 15 days in a static incubator at 37 °C and colonies were counted.
MurX assay
MurX assay substrates, Park’s nucleotide-Nε-dansylthiourea, neryl phosphate, were chemically synthesized according to the reported procedures.23,24,25,26 M. tuberculosis cells were harvested by centrifugation (4,700 RPM) at 4 °C followed by washing with 0.9% saline solution (thrice) and ~5g of pellet (wet weight) was collected. The cell pellets were suspended in homogenization buffer (containing 50 mM MOPS of pH = 8, 0.25 M sucrose, 10 mM MgCl2 and 5 mM 2-mercaptoethanol) and disrupted by probe sonication on ice (10 cycles of 60s on and 90s off). The resulting suspension was centrifuged at 1,000 ×g for 10 min at 4 °C to remove unbroken cells. The supernatant was centrifuged at 25,000 ×g for 40 min at 4 °C (3 to 4 times). All pellets in each tube were pooled and a second sonication was performed (10 cycles of 60s on and 90s off). The lysate was centrifuged once at 25,000 ×g for 1h and the supernatant was subjected to ultracentrifugation at 60,000 ×g for 1h at 4 °C. The supernatant was discarded and the membrane fraction containing MurX enzyme (P-60) was suspended in the TRIS-HCl buffer (pH 7.5, containing 2-mercaptoethanol).27 Total protein concentrations are about 8~10 mg/mL.28 Aliquots were stored in Eppendorf tubes at −80°C. Similarly, the membrane fractions containing MraY enzyme (P-60) were prepared from M. smegmatis, S aureus, and E. coli, respectively.
Park’s nucleotide-Nε-dansylthiourea (2 mM stock solution; 3.75 μL (75 μM)), MgCl2 (0.5 M; 10 μL (50 mM)), KCl (2 M, 10 μL (200 mM)), triton X100 (0.5%; 11.25 μL), tris-buffer (pH = 8; 50mM, 2.5 μL), neryl phosphate (10 mM, 45 μL), and inhibitor (0-100 μM, in DMSO (2.5 μL)) were placed in a 500 μL Eppendorf tube. To a stirred reaction mixture, P-60 (15 μl) was added (the total volume of the reaction mixture: 100 μL). The reaction mixture was incubated for 1h at room temperature (26 °C), and quenched with CHCl3 (200 μL). Two phases were mixed via vortex and centrifuged at 25,000 ×g for 10 min. The upper aqueous phase was assayed via reverse-phase HPLC. The water phase (10 μl) was injected into HPLC (solvent: CH3CN: 0.05 M aq. NH4HCO3 = 25: 75, UV: 350 nm, flow rate: 0.5 mL/min, Column: Kinetex 5u C8 100Å, 150 × 4.60mm), and the area of the peak for lipid I-neryl derivative was quantified to obtain the IC50 value. The IC50 values were calculated from plots of the percent product inhibition versus the inhibitor concentration.26
RNA polymerase assays
M. smegmatis RNAP was prepared according to the procedures described previously by Burgess and Jendrisak.29 M. smegmatis ATCC 607 was cultured to log phase, harvested by centrifugation and washed thrice with 0.9% saline solution. Temperature was maintained at 4 °C for the rest of the procedure unless mentioned otherwise. Cells were suspended in a lysis buffer (0.05 M Tris, 5% v/v glycerol, 2 mM EDTA, 0.1 mM dithiothreitol, 1 mM 2-mercaptoethanol, 0.233 M NaCl, 130 μg/mL lysozyme, 23 μg/mL PhCH2SO2F and 4% w/v sodium deoxycholate) and disrupted by probe sonication on ice (10 cycles of 60s on and 90s off). The resulting suspension was then centrifuged at 8,000 rpm for 45 min at 4 °C. The supernatant was subjected to polyethylenenamine (PEI) fractionation. The supernatant was treated slowly with 10% v/v solution of PEI (pH 7.9) to a final concentration of 0.35%, stirred for 5 min and centrifuged for 15 min at 6000 rpm. Supernatant was discarded. Pellet was suspended in TGED buffer (0.01 M Tris, pH 7.9, 5% v/v glycerol, 0.1 mM EDTA, 0.1 mM dithiothreitol) and 0.5 M NaCl and centrifuged for 15 min at 6000 rpm. Supernatant was discarded and pellet was resuspended in TGED and 1 M NaCl. Centrifuged for 30 min at 6000 rpm. Supernatant was treated with solid ammonium sulfate to 50% saturation, stirred for 20 min and centrifuged for 45 min at 8000 rpm. Pellet was suspended in TGED and was subjected to DNA cellulose chromatography as described in Burgess and Jendrisak. Fractions were pooled and aliquots were stored at −80 °C. The 100× DNA, 100× NTP mix and 10× fluorescence dye was diluted 10 fold with water and the RNAP enzyme was diluted 10 fold with 1× assay buffer. The assay mixture was then prepared in a black-bottom 96 well plate by the addition of 15 μL water, 3 μL of 10× buffer, 3 μL of 10× DNA template, 3 μL of 10× NTP mix, 3 μL of 10× RNA polymerase enzyme (S. aureus or E. coli) or RNAP from M. smegmatis (obtained as described above) and 3 μL of inhibitor (final concentration, 0-10 μM). The total volume of the reaction mixture was kept 30 μL and was incubated for 1h at room temperature. 30 μL of the 1× fluorescent dye was added and incubated for 5 min. The fluorescence intensity was measured at an excitation wavelength of 485 nm and emission wavelength of 528 nm using a Biotek Synergy XT, 96 well plate reader.
Cytotoxicity assays
Cytotoxicity assays will be performed using Vero monkey kidney (ATCC CCL-81) and HepG2 human hepatoblastoma cell (ATCC HB-8065) lines. Vero or HepG2 cells are cultured in 96 well cell culture plates using ATCC-formulated Eagle’s Minimum Essential Medium. Serially diluted aliquots of each test compound at concentrations ranging from 1-25 X of the MIC were added to the cells. Control of compounds with known toxicity such as rifampicin or INH was included on each plate. The plates were incubated and cytopathic effects were determined via the MTT assay.
6-{Carbamoyl-[5-(2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl)-3,4-dimethoxy-tetrahydro-furan-2-yl]-methoxy}-4,5-dihydroxy-5,6-dihydro-4H-pyran-2-carboxylic acid (2-oxo-azepan-3-yl)-amide (UT-01320, 3)
UT-01320 (3) was synthesized according to the scheme reported previously.14,15 [α]20D = + 90 (c 0.1 in H2O); 1H NMR (400 MHz, CD3OD) δ 7.79 (d, J = 8.4 Hz, 1H), 5.90 (d, J = 4.0 Hz, 1H), 5.88 (d, J = 6.4 Hz, 1H), 5.70 (d, J = 8.0 Hz, 1H), 5.12 (d, J = 6.0 Hz, 1H), 4.46 (d, J = 11.2 Hz, 1H), 4.41 (d, J = 3.2 Hz, 1H), 4.32 (t, J = 4.0 Hz, 1H), 4.25 (t, J = 3.2 Hz, 1H), 4.08 (m, 2H), 3.78 (m, 1H), 3.35 (s, 3H), 3.31 (s, 3H), 1.90-1.57 (m, 6H), 1.32 (m, 2H); 13C NMR (101 MHz, CD3OD) δ 176.5, 173.7, 166.0, 162.1, 152.3, 144.3, 142.7, 109.0, 103.6, 103.1, 82.1, 81.0, 79.5, 78.2, 68.8, 63.9, 58.6, 58.1, 55.4, 53.5, 42.5, 29.9, 27.5; HRMS (ESI) calcd for C24H33N5NaO12 ([M + Na]+): 606.2023, found: 606.2031.
6-{Carbamoyl-[5-(2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl)-4-hydroxy-3-methoxy-tetrahydro-furan-2-yl]-methoxy}-4,5-dihydroxy-5,6-dihydro-4H-pyran-2-carboxylic acid (3,4-difluoro-phenyl)-amide (SQ 641, 2)
[α]20D = + 105 (c 0.1 in H2O); 1H NMR (400 MHz, CD3OD) δ 7.87 (d, J = 8.4 Hz, 1H), 7.68 (m, 1H), 7.34 (m, 1H), 7.19 (m, 1H), 6.07 (d, J = 4.4 Hz, 1H), 5.74 (s, 1H), 5.63 (d, J = 8.0 Hz, 1H), 5.08 (d, J = 7.2 Hz, 1H), 4.65 (s, 1H), 4.42 (dd, J = 8.8 Hz, 1H), 4.30 (t, J = 4.8 Hz 1H), 4.21 (m, 1H), 3.79 (m, 1H), 3.71 (d, J = 1.2, 1H), 3.48 (s, 3H); HRMS (ESI) calcd for C23H24F2N4NaO11 ([M + Na]+): 593.1307, found: 593.1312.
6-{Carbamoyl-[5-(2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl)-4-hydroxy-3-methoxy-tetrahydro-furan-2-yl]-methoxy}-4,5-dihydroxy-5,6-dihydro-4H-pyran-2-carboxylic acid (2-oxo-azepan-3-yl)-amide (Capuramycin, 1)
Capuramycin was resynthesized according the reported procedure.15 [α]20D = + 98 (c 0.1 in H2O); 1H NMR (400 MHz, CD3OD) δ 7.71 (d, J = 8.0 Hz, 1H), 5.97 (s, 1H), 5.82 (d, J = 8.0 Hz, 1H), 5.73 (s, 1H), 5.35 (s, 1H), 4.59 (d, J = 11.2 Hz, 2H), 4.47 (s, 1H), 4.44 (d, J = 4.8 Hz, 1H), 4.34 (s, 1H), 4.15 (s, 1H), 3.71 (t, J = 4.8 Hz, 1H), 3.26 (s, 3H), 1.94-1.57 (m, 6H), 1.32 (m, 2H); 13C NMR (101 MHz, D2O) δ 176.3, 173.0, 166.1, 161.4, 151.2, 141.5, 141.0, 109.4, 101.8, 99.3, 90.1, 81.6, 78.1, 75.5, 71.9, 64.7, 61.7, 57.8, 52.2, 41.4, 30.3, 27.3; HRMS (ESI) calcd for C23H31N5NaO12 ([M + Na]+): 592.1867, found: 592.1864.
Synergistic effect of UT-01320 (3) with SQ641 or capuramycin
The synergistic or antagonistic activities of MurX inhibitors (capuramycin (1) or SQ 641 (2)) and UT-01320 (3) were assessed in vitro via micro dilution broth checkerboard technique.30,31 The FIC (Fractional Inhibitory Concentration) index was calculated to according to the following equation. ΣFIC = FICA + FICB = CA/MICA + CB/MICB where, MICA and MICB: MIC of drugs A and B, CA and CB = concentrations of drugs A and B used in combination. In these interaction studies, ΣFIC of less than 1 represents synergistic activity.
RESULT
Effectiveness of UT-01320 against non-replicating M. tuberculosis
To date, we have synthesized capuramycin (1), SQ641 (2), and over 100 new capuramycin analogs in which their structures were diversified with optically active amines (R3) and the four uridyl templates, 3′O-methyl (natural form), 2′O-methyl, 2′,3′O-dimethyl-, and 3′O-demethylated (unnatural forms) uridine-manno-pyranuronate derivatives (Figure 2). The syntheses of capuramycin analogs with these templates were accomplished via the synthetic scheme reported previously.14,15 As the results of in vitro evaluation of these molecules in MurX enzyme inhibition26 and Mtb (H37Rv) growth inhibitory activities against replicating and non-replicating (dormant) Mtb via the microplate alamar blue assay (MABA) and low oxygen recovery assay (LORA) assays20, it was identified that UT-01309 (4) possessing 3-amino-1,4-benzodiazepine-2-one exhibited increased MurX enzyme and Mtb growth inhibitory activity (MIC: 2.5 μg/mL vs 12.0 μg/mL for 1, IC50 0.095 μM vs 0.127 μM for 1).15 The analogues synthesized from the 3′O-methyluridine-manno-pyranuronate (natural form) did not exhibit bactericidal activity against non-replicating Mtb even at 400 μg/mL concentrations. The isocapuramycin (2′O-methyl) analogs (R1 = Me, R2 = H in Figure 2) showed equal MurX inhibitory and antimycobacterial activities to the corresponding analogs derived from the natural form (R1 = H, R2 = Me in Figure 2). The antimycobacterial MurX inhibitors, isocapuramycin analogs (2′O-methyluridine-manno-pyranuronate derivatives) were not capable of killing non-replicating Mtb in the LORA assay. The 3′O-demethyl capuramycin analogs (R1, R2 = H in Figure 2) did not exhibit activities in MurX enzyme inhibitory and antimycobacterial assays. Remarkably, 2′O-methylated capuramycin, UT-01320 (3) killed both replicating and non-replicating Mtb at low concentrations with the MICLORA/MABA value of 1.72, whereas the MICLORA/MABA value for rifampicin was 7.35 (Table 1). The MICLORA/MABA value close to 1 or below 1 is considered to be ideal TB drug lead molecules in our programs.
Table 1.
MICs (Mtb) and IC50s (MurX enzyme and mammalian cells) for capuramycin (1) and its analogues, 2-4.
| Molecule | IC50 μM MurX | MIC μg/mL (MABA)a | MIC μg/mL (LORA)b |
IC50 μg/mL HepG2 cellsc |
|---|---|---|---|---|
| Capuramycin (1) | 0.127 | 12.5 | >100 | >400 |
| SQ 641 (2) | 0.098 | 1.56 | >100 | >400 |
| UT-01320 (3) | >100 | 1.50 | 2.58 | >400 |
| UT-01309 (4) | 0.095 | 2.50 | >100 | >400 |
| Rifampicin (RFP) | - | 0.20 | 1.47 | >400 |
| Isoniazid (INH) | - | 0.04-0.10 | >100 | >400 |
| Ethambutol (EMB) | - | 0.78 | >100 | >400 |
MABA: microplate alamar blue assay.
LORA: low-oxygen recovery assay.
Cytotoxicity against Vero monkey kidney cells was >400 μg/mL
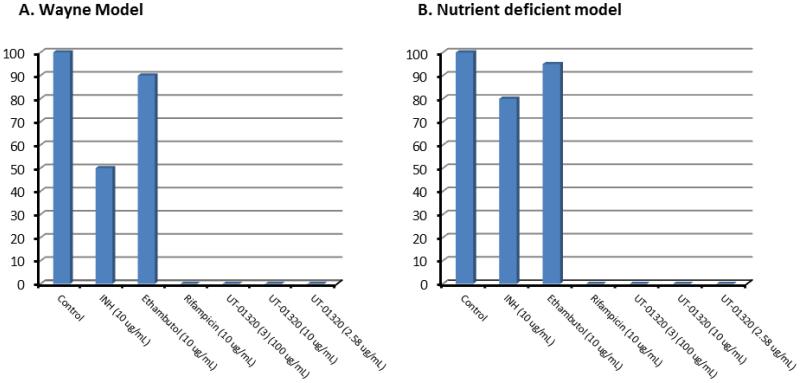
Bactericidal activity of 3 against dormant Mtb was confirmed via the assays using a modified Wayne model under low-oxygen conditions7,32 and a nutrient deficient Mtb cells33; in both assays, no colony-forming unit (CFU) was observed after 15 days’ incubation of the Mtb cultures treated with 3 at 100 μg/mL, 10 μg/mL, and 2.58 μg/mL (MICLORA), respectively (Figure 3).
Figure 3.
Bactericidal activity of UT-01320 against non-replicating Mtb evaluated in Wayne and nutrient deficient models.
M. tuberculosis entered dormant states were cultured with the inhibitors. After 96h under anaerobic (Wayne model) or nutrient deficient conditions, 10-, 100-, 1,000- and 10,000-fold serial dilutions from each culture tube was sampled, and 20μL from each dilution was plated on 7H10 agar plates supplemented with OADC enrichment. Plates were incubated under aerobic conditions for 15 days in a static incubator at 37 °C and colonies were counted. CFU counted for each plate is shown relative to control (100). UT-01320 (3) killed M. tuberculosis at the MICLORA concentrations in both assays. A: Wayne model.; B: a nutrient deficient model.
Rapid antimycobactericidal activity of UT-01320
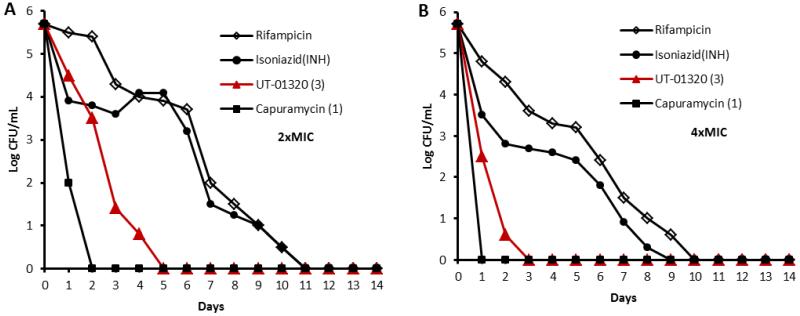
Because of alteration of molecular target, we were interested in whether a non-MurX inhibitor 3 remains rapid bactericidal activity against replicating Mtb under aerobic conditions. The time-kill experiments were performed at two-and four-fold the MIC of 1, 3, and a few 1st-line TB drugs (rifampicin and isoniazid (INH)). Viable cell counting was performed at every 24 h for 14 days. Colony forming units (CFUs) were counted after 15 days of incubation at 37 °C. The rate of killing of 3 against Mtb was compared directly with the reference molecules and the time-kill assessments at 2× and 4× MIC concentrations are shown in Figure 4. The MurX inhibitor, capuramycin (1) yielded log10 CFU/mL reduction of 3.0 or greater for Mtb in 1 day and 1 killed Mtb completely in 2 days at 24.0 μg/mL, albeit, rifampicin and INH required over 7 days to kill 50% of Mtb at 0.4 and 1.0 μg/mL, respectively (Figure 4A).12 UT-01320 (3) killed >50% of Mtb within 3 days and 100% within 5 days at 3.0 μg/mL (2× MIC), and killed 100% of Mtb within 3 days at 6.0 μg/mL (4× MIC) (Figure 4B). Thus, the fast killing profile of 3 was similar to that of capuramycin; 3 killed Mtb faster than rifampicin and INH, but was less effective than capuramycin.
Figure 4.
In vitro time-kill assessment of capuramycin (1), UT-01320 (3), and the first-line TB drugs (rifampicin and INH).
Log-kill reduction in log10 CFU/mL at the concentrations of the molecules at 2× and 4× MIC. A: 2× MIC of rifampicin (0.4 μg/mL), INH (0.2. μg/mL), UT-01320 (3.0 μg/mL), capuramycin (24.0 μg/mL).; B: 4× MIC of rifampicin (0.8 μg/mL), INH (0.4. μg/mL), UT-01320 (6.0 μg/mL), capuramycin (48.0 μg/mL).
Synergistic effect of UT-01320 with MurX inhibitors
The synergistic or antagonistic activities of MurX inhibitors (capuramycin (1) or SQ 641 (2)) and UT-01320 (3) were assessed in vitro via micro dilution broth checkerboard technique.30,34 In the checkerboard analyses of a combination of SQ 641 and UT-01320, the FIC index range of 0.06 to 4 was observed for 47 combinations of two molecules out of 64 different concentrations. Table 2 summarizes the results of FIC index analyses for a combination of 2 plus 3 and 1 plus 3 that showed synergistic combination (ΣFIC < 1). It was demonstrated that UT-01320 (3) exhibited synergy with SQ 641 (2) and capuramycin (1) that killed Mtb at much lower concentrations than the MICA and MICB.
Table 2.
Fractional inhibitory concentration of a combination of two inhibitor molecules.a
| Combination of A and B | MICA (μg/mL) MICB (μg/mL)b |
MIC (μg/mL) CA and CBc |
ΣFICd |
|---|---|---|---|
| A: SQ 641 (2) | 1.56 1.50 |
0.10 0.08 |
0.12 |
| B: UT-01320 (3) | |||
| A: SQ 641 (2) | 1.56 1.50 |
0.10 0.16 |
0.17 |
| B: UT-01320 (3) | |||
| A: SQ 641 (2) | 1.56 1.50 |
0.10 0.32 |
0.28 |
| B: UT-01320 (3) | |||
| A: SQ 641 (2) | 1.56 1.50 |
0.10 0.645 |
0.49 |
| B: UT-01320 (3) | |||
| A: SQ 641 (2) | 1.56 1.50 |
0.10 1.29 |
0.92 |
| B: UT-01320 (3) | |||
| A: Capuramycin (1) | 12.5 1.50 |
1.563 0.08 |
0.18 |
| B: UT-01320 (3) | |||
| A: Capuramycin (1) | 12.5 1.50 |
1.563 0.16 |
0.23 |
| B: UT-01320 (3) | |||
| A: Capuramycin (1) | 12.5 1.50 |
0.78 0.32 |
0.28 |
| B: UT-01320 (3) | |||
| A: Capuramycin (1) | 12.5 1.50 |
0.78 0.645 |
0.49 |
| B: UT-01320 (3) | |||
| A: Capuramycin (1) | 12.5 1.50 |
0.78 1.29 |
0.92 |
| B: UT-01320 (3) |
ΣFIC index for the wells at growth–no growth interface.
MICA and MICB are the MIC value of molecule A or B against Mtb (see Table 1).
CA and CB are concentrations of A and B.
ΣFIC is the sum of fractional inhibitory concentration calculated by the equation ΣFIC = FICA + FICB = CA/MICA + CB/MICB.
Spectrum of activity of UT-01320
As summarized in Table 3, UT-01320 (3) has a very narrow spectrum of activity; in growth inhibitory assays against Gram-positive and -negative bacteria including Mycobacterium spp., 3 killed Mtb strains selectively. It is interesting to note that unlike other capuramycin analogs, 3 did not kill a fast-growing M. smegmatis at >200 μg/mL concentrations (e.g. MICs 6.5 and 25 μg/mL for 1 and 4, respectively).
Table 3.
Spectrum of activity of UT-01320 (3).
| Species and strain | MIC (μg/mL)a | ||
|---|---|---|---|
| UT-01320 (3) | RFPd | INHe | |
| M. tuberculosis H37Rv | 1.50 | 0.20 | 0.04-0.10 |
| M. tuberculosis H37Rv INHrb | 2.50 | 0.20-0.10 | >8.0 |
| M. tuberculosis H37Rv RFPrc | 2.45 | >4.0 | 0.05 |
| M. smegmatis ATCC607 | >200 | >200 | >200 |
| S. aureus ATCC6538D-5 | >200 | - | - |
| E. faeciumATCC 349 | >200 | - | - |
| E. coli ATCC 25019 | >200 | - | - |
| K. pneumonia ATCC 8047 | >200 | - | - |
| P. aeruginosa ATCC 27853 | >200 | - | - |
The broth dilution method was used.
INH-resistant M. tuberculosis.
RFP-resistant M. tuberculosis.
RFP: rifampicin.
INH: isoniazid.
A potential molecular target for UT-01320
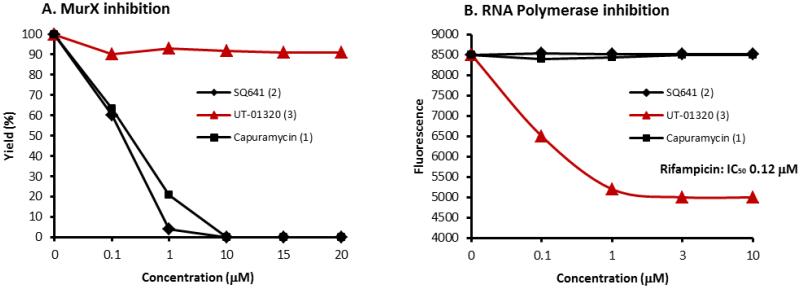
Effectiveness of the collected molecules against non-replicating Mtb has been evaluated in the Wayne model32 as well as the low oxygen recovery assay20. Several electron transport system inhibitors, menaquinone biosynthesis inhibitors,35 a F1F0-ATPase inhibitor, nitroimidazoles, a RNA polymerase inhibitor (e.g. rifampicin), protein biosynthesis inhibitors (e.g. amikacin, capreomycin), and bacterial DNA gyrase inhibitors were effective in killing non-replicating Mtb, however, in most cases their MIC values against non-replicating Mtb cells are significantly higher than those against aerobically growing Mtb cells.35 In preliminary assay screening of UT-01320 (3) against the potential drug targets for non-replicating Mtb, it was observed that 3 inhibited E. coli, S. aureus, and M. smegmatis RNA polymerases with the IC50 values of the 100-150 nM range (vs IC50 120 nM for rifampicin against E. coli RNA polymerase) (Figure 5B), whereas, 3 did not inhibit electron transport systems, menaquinone biosynthesis, Mtb serine/threonine kinases (PKnA and B), and DNA gyrase.36,37,38,39 Capuramycin (1) and SQ 641 (2) did not inhibit bacterial RNA polymerases even at 100 μM concentrations. Figure 5 summaries MurX and RNA polymerase inhibitory activities of Capuramycin (1), SQ 641 (2), and UT-01320 (3).
Figure 5.
MurX and RNA polymerase inhibitory activities of UT-01320, capuramycin and SQ 641.
A. MurX containing P-60 was used.; B. Data summarized here were obtained with E. coli RNA polymerase. UT-01320 (3) inhibited RNA polymerases from M. smegmatis and S. aureus with similar IC50 values.
DISCUSSION
It is important to discover effective TB drugs that kill non-replicating Mtb at low concentrations for shortening the treatment duration of current TB chemotherapies.35 UT-01320 (3) killed non-replicating Mtb at 2.58 μg/mL concentrations (Table 1). Based on the antibacterial activities tested, it was determined that UT-01320 has an antimicrobial spectrum focused against Mtb (Table 3). Selective antimycobacterial agent is preferable for TB chemotherapy due to the fact that TB chemotherapy requires a long regimen, so that broad-spectrum anti-TB agents may cause resistant to other bacteria during TB chemotherapy.35
To date, SAR studies of capuramycin (1) have been performed with the biosynthesis intermediate, A-500359, and the C2’-hydroxy group of capuramycin has been modified with the limited functional groups (i.e. acyl groups).16,19 Capuramycin has not been developed into a pursuable lead by acyl modification at the C’2-hydroxy group. Our total chemical synthetic scheme enables the modification at the C2’- and C3’-hydroxy groups of capuramycin with chemically stable ether groups. As the result of reliable MurX enzyme assays of a small optimized library of capuramycin analogs followed by antimycobacterial growth inhibitory assays,26 several new analogs were identified as improved MurX inhibitors (e.g. UT-01304 (4)) and non-MurX inhibitors having antimycobacterial activity (Figure 2).15,26 Methylation at the C2’-hydroxy group of capuramycin (R1 in Figure 2) alters the molecular target of capuramycin. UT-01320 (3) did not interfere with MurX, but exhibited bactericidal activity against non-replicating (dormant) Mtb with the MICLORA/MICMABA value of 1.72 (MICLORA/MICMABA = 7.35 for rifampicin). Effectiveness of 3 in non-replicating Mtb was confirmed via assays using a modified Wayne model under low-oxygen conditions and a nutrient deficient Mtb cells at the MICLORA concentrations (2.58 μg/mL) (Figure 3). Cytotoxicity (IC50) of 3 was evaluated in vitro using Vero monkey kidney cells and HepG2 human hepatoblastoma cells. As observed for other capuramycin analogs, 3 did not show cytotoxicity even at 400 μg/mL concentrations (Table 1); the selectivity index (IC50 in mammalian cells/MIC against Mtb) of 3 was determined to be >266.
Several TB drugs and drug leads that have the potential to kill non-replicating Mtb in vitro were identified. Bedaquiline is an Mtb ATPase inhibitor that showed effectiveness in killing non-replicating Mtb in vitro. Capreomycin and streptomycin that target protein biosynthesis through binding to the bacterial ribosomes and bacterial topoisomerase II inhibitors (e.g. fluoroquinolones) killed non-replicating Mtb in vitro. It was reported that nitric oxide-mediated unique immune responses by metronidazole and PA-824 (a phase II clinical drug) kill non-replicating Mtb under hypoxic conditions.40 However, in most cases their MIC values against non-replicating Mtb cells are significantly higher than those against Mtb grown under aerobic conditions. Therefore, it is considered to be very important to discover new TB drugs that effectively kill non-replicating Mtb at the same MIC concentrations as those under aerobic conditions. Table 4 summarizes the potential molecular targets whose inhibitors were effective in killing non-replicating Mtb. Recently, Ishizaki et al. reported that caprazamycin (an MraY inhibitor) analogue, CPZEN-45 inhibited mycobacterial WecA, a phosphotransferase responsible for capsular polysaccharide synthesis.41 Although much studies are required to understand WecA in dormant Mtb, CPZEN-45 killed non-replicating Mtb in vitro.42 As such the alternation of molecular target of a uridine-glycosyl peptide MraY inhibitor by chemical modifications has observed. While the exact mechanism of action of UT-01320 (3) is far from completely understood, 3 is a new pharmacophore that inhibits the polymerization of RNA at low concentrations in a dose-response manner (Figure 5).38 UT-01320 (3) killed rifampicin-resistant Mtb at low concentrations. This fact may imply that 3 inhibits RNA-polymerases by interfering with the catalytic sites that are different from the rifampicin binding site(s).43 The structure of UT-01320 (3) is closely related to the MurX inhibitor SQ 641 (2). The PK/PD parameters of 3 may be correlated with those of 2, and thus, a combination of two capuramycin analogs that have distinct drug targets is a great interest in evaluation of their synergistic interactions. The non-MurX inhibitor 3 exhibited strong synergistic effect with a promising preclinical TB drug, SQ 641 (2); a combination of two molecules significantly improved in vitro antimycobacterial activity (Table 2). In addition, UT-01320 (3) retained a fast-bactericidal characteristic observed for capuramycin (Figure 4). These forward-looking biological characteristics for UT-01320 promise that new TB drug leads will be discovered by extensive SAR studies of 2′O-methylated capuramycin analogs. We are currently optimizing UT-01320 (3) to be more effective against both replicating and non-replicating Mtb by modifying the R3 group (Figure 2). Through in vitro biological evaluation of non-MurX inhibitors identified from capuramycin analogs and in vivo evaluation of promising molecules will be reported elsewhere.
Table 4.
Potential targets for non-replicating M. tuberculosis.
|
Supplementary Material
ACKNOWLEGEMENTS
The National Institutes of Health is greatly acknowledged for financial support of this work (AI084411). We also thank University of Tennessee for generous financial support. NMR data were obtained on instruments supported by the NIH Shared Instrumentation Grant. The following reagent was obtained through BEI Resources, NIAID, NIH: Mycobacterium tuberculosis, Strain H37Rv and Gamma-Irradiated Mycobacterium tuberculosis, NR-14819. The authors gratefully acknowledge Drs. William Clemons (California Institute Technology) and Crick (Colorado State University) for useful discussions.
Footnotes
Supplementary Information accompanies the paper on The Journal of Antibiotics website (http://www.nature.com/ja)
REFERENCES
- 1.Nakanjako D, Mayanja-Kizza H, Ouma J, Wanyenze R, Mwesigire D, Namale A, Ssempiira J, Senkusu J, Colebunders R, Kamya MR. Tuberculosis and human immunodeficiency virus co-infections and their predictors at a hospital-based HIV/AIDS clinic in Uganda. Tuberculosis Lung Dis. 2010;14:1621–1628. [PubMed] [Google Scholar]
- 2.Diedrich CR, Flynn JL. HIV-1/Mycobacterium tuberculosis co-infection immunology: How does HIV-1 exacerbate tuberculosis? Infect. Immun. 2011;79:1407–1417. doi: 10.1128/IAI.01126-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chien JY, Lai CC, Tan CK, Chien ST, Yu CJ, Hsueh PR. Decline in rates of acquired multidrug-resistant tuberculosis after implementation of the directly observed therapy, short course (DOTS) and DOTS-Plus programmes in Taiwan. J. Antimicrob. Chemother. 2013;68:1910–1916. doi: 10.1093/jac/dkt103. [DOI] [PubMed] [Google Scholar]
- 4.Ranjbar S, Boshoff HI, Mulder A, Siddiqi N, Rubin EJ, Goldfeld AE. HIV-1 Replication is differentially regulated by distinct clinical strains of Mycobacterium tuberculosis. PLoS One. 2009;4:e6116. doi: 10.1371/journal.pone.0006116. doi: 10.1371/journal.pone.0006116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connolly LE, Edelstein PH, Ramakrishnan L. Why is long-term therapy required to cure tuberculosis? PLoS Med. 2003;4:435–442. doi: 10.1371/journal.pmed.0040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miranda MS, Breiman A, Allain S, Deknuydt F, Altare F. The tuberculous granuloma: an unsuccessful host defense mechanism providing a safety shelter for the bacteria? Clin. Develop. Immun. 2012;2012:1–14. doi: 10.1155/2012/139127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claudia S, Neeraj D, Ruben HC, Zhang M, Ha YH, Schneider P, Cole ST. Simple model for testing drugs against nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2010;54:4150–4158. doi: 10.1128/AAC.00821-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Heijenoort J. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol. Mol. Biol. Rev. 2007;71:620–635. doi: 10.1128/MMBR.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hett EC, Rubin EJ. Bacterial growth and cell division: a Mycobacterial perspective. Micro. Mol. Biol. Rev. 2008;72:126–156. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barry CE, Blanchard JS. The chemical biology of new drugs in development for tuberculosis. Curr. Opin. Chem. Biol. 2010;14:456–466. doi: 10.1016/j.cbpa.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddy VM, Einck L, Nacy CA. In vitro antimycobacterial activity of capuramycin analogs. Antimicro. Agents Chemother. 2008;52:719–721. doi: 10.1128/AAC.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurosu M, Li K. Synthetic studies towards the identification of novel capuramycin analogs with antimycobacterial activity. Heterocycles. 2009;77:217–225. [Google Scholar]
- 14.Kurosu M, Li K, Crick DC. A concise synthesis of capuramycin. Org. Lett. 2009;11:2393–2396. doi: 10.1021/ol900458w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Siricilla S, Aleiwi BA, Kurosu M. Improved synthesis of capuramycin and its analogues. Chem. A. Europ. J. 2013;19:13847–13858. doi: 10.1002/chem.201302389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koga T, Fukuoka T, Harasaki T, Inoue H, Hotoda H, Kakuta M, Muramatsu Y, Yamamura N, Hoshi M, Hirota T. Activity of capuramycin analogs against Mycobacterium tuberculosis, Mycobacterium avium and Mycobacterium intracellular in vitro and in vivo. J. Antimicro. Chemother. 2004;54:755–760. doi: 10.1093/jac/dkh417. [DOI] [PubMed] [Google Scholar]
- 17.Nikonenko BV, Reddy VM, Protopopova M, Bogatcheva E, Einck L, Nacy CA. Activity of SQ641, a capuramycin analog, in a murine model of tuberculosis. Antimicrob. Agents Chemother. 2009;53:3138–3139. doi: 10.1128/AAC.00366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubuisson T, Bogatcheva E, Krishnan MY, Collins MT, Einck L, Nacy CA, Reddy VM. In vitro antimicrobial activities of capuramycin analogues against non-tuberculous mycobacteria. J. Antimicrob. Chemother. 2010;65:2590–2597. doi: 10.1093/jac/dkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogatcheva E, Dubuisson T, Protopopova M, Einck L, Nacy CA, Reddy VM. Chemical modification of capuramycins to enhance antibacterial activity. J. Antimicrob. Chemother. 2011;66:578–587. doi: 10.1093/jac/dkq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho SH, Warit S, Wan B, Hwang CH, Pauli GF, Franzblau SG. Low-oxygen-recovery assay for high-throughput screening of compounds against nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2007;51:1380–1358. doi: 10.1128/AAC.00055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snewin VA, Gares MP, Gaora PO, Brown IN, Young DB. Assessment of immunity to mycobacterial infection with luciferase reporter constructs. Infect. Immun. 1999;67:4586–4593. doi: 10.1128/iai.67.9.4586-4593.1999. Z. H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gengenbacher M, Rao SPS, Pethe K, Dick T. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology. 2010;156:81–87. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- 23.Kurosu M, Mahapatra S, Narayanasamy P, Crick DC. Chemoenzymatic synthesis of park’s nucleotide: toward the development of high-throughput screening for MraY inhibitors. Tetrahedron Lett. 2007;48:799–803. [Google Scholar]
- 24.Li K, Kurosu M. Synthetic studies on Mycobacterium tuberculosis specific fluorescent park’s nucleotide probe. Heterocycles. 2008;76:455–469. [Google Scholar]
- 25.Mitachi K, Mohan P, Siricilla S, Kurosu M. One-pot protection-glycosylation reactions for synthesis of lipid II analogues. Chem. A. Europ. J. 2014;20:4554–4558. doi: 10.1002/chem.201400307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siricilla S, Mitachi K, Skorupinska-Tudek K, Swiezewska E, Kurosu M. Biosynthesis of a water-soluble lipid I analogue and a convenient assay for translocase I. Anal. Biochem. 2014;461:36–35. doi: 10.1016/j.ab.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandana R, Marie-Antoinette L, Peter S, Mamadou D. Breaking down the wall: Fractionation of mycobacteria. J. Microbiol. Methods. 2007;68:32–39. doi: 10.1016/j.mimet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 28.Bouhss A, Crouvoisier M, Blanot D, Mengin-Lecreulx D. Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis. J. Biol. Chem. 2004;279:29974–29980. doi: 10.1074/jbc.M314165200. [DOI] [PubMed] [Google Scholar]
- 29.Burgess RR, Jendrisak JJ. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving polymin P precipitation and DNA-cellulose chromatography. Biochem. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh MH, Chen MY, Victor LY, Chow JW. Synergy assessed by checkerboard. A critical analysis. Diagn. Microbiol. Infect. Dis. 1993;16:343–349. doi: 10.1016/0732-8893(93)90087-n. [DOI] [PubMed] [Google Scholar]
- 31.Ohrt C, Willingmyre GD, Lee P, Knirsch C, Milhous W. Assessment of azithromycin in combination with other antimalarial drugs against Plasmodium falciparum in vitro. Antimicrob. Agents Chemother. 2002;46:2518–2524. doi: 10.1128/AAC.46.8.2518-2524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wayne LG, Sramek HA. Antigenic differences between extracts of actively replicating and synchronized resting cells of Mycobacterium tuberculosis. Infect. Immun. 1979;24:363–370. doi: 10.1128/iai.24.2.363-370.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gengenbacher M, Rao SPS, Pethe K, Dick T. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology. 2010;156:81–87. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- 34.Meletiadis J, Pournaras S, Roilides E, Walsh TJ. Defining fractional inhibitory concentration index cutoffs for additive interactions based on self-drug additive combinations, monte carlo simulation analysis, and in vitro-in vivo correlation data for antifungal drug combinations against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2010;54:602–609. doi: 10.1128/AAC.00999-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Debnath J, Siricilla S, Wan B, Crick DC, Lenaerts AJ, Franzblau SG, Kurosu M. Discovery of selective menaquinone biosynthesis inhibitors against Mycobacterium tuberculosis. J. Med. Chem. 2012;55:3739–3755. doi: 10.1021/jm201608g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levin ME, Hatfull GF. Mycobacterium smegmatis RNA polymerase: DNA supercoiling, action of rifampicin and mechanism of rifampicin resistance. Mol. Microbiol. 1993;8:277–285. doi: 10.1111/j.1365-2958.1993.tb01572.x. [DOI] [PubMed] [Google Scholar]
- 37.Siegmund V, Santner T, Micura R, Marx A. Screening mutant libraries of T7 RNA polymerase for candidates with increased acceptance of 2′-modified nucleotides. Chem. Commun. 2012;48:9870–9872. doi: 10.1039/c2cc35028a. [DOI] [PubMed] [Google Scholar]
- 38.Kuhlman P, Duff HL, Galant A. A fluorescence-based assay for multisubunit DNA-dependent RNA polymerases. Analy. Biochem. 2004;324:183–190. doi: 10.1016/j.ab.2003.08.038. [DOI] [PubMed] [Google Scholar]
- 39.Onodera K, Kawasaki T, Kamijo S. Discovery of novel antimicrobial agents targeting the bacterial RNA polymerase by high-throughput virtual screening. Chem-Bio. Informat. J. 2011;11:52–62. [Google Scholar]
- 40.Boshoff HIM, Barry CE. Tuberculosis - metabolism and respiration in the absence of growth. Nature Rev. Microbiol. 2005;3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 41.Ishizaki Y, Hayashi C, Inoue K, Igarashi M, Takahashi Y, Pujari V, Crick DC, Brennan PJ, Nomoto A. Inhibition of the first step in synthesis of the Mycobacterial cell wall core, catalyzed by the GlcNAc-1-phosphate transferase WecA, by the novel caprazamycin derivative CPZEN-45. J. Biol. Chem. 2013;288:30309–30319. doi: 10.1074/jbc.M113.492173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engohang-Ndong J. Antimycobacterial drugs currently in phase II clinical trials and preclinical phase for tuberculosis treatment. Expert Opin. Investig. Drugs. 2012;21:1789–1800. doi: 10.1517/13543784.2012.724397. [DOI] [PubMed] [Google Scholar]
- 43.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.