Abstract
Intra-tumor heterogeneity, variation between individual tumor cells within a patient’s tumor, is increasingly seen as a critical mechanism underlying treatment resistance and therapeutic failure. Despite this growing awareness, few methods to assess intra-tumor heterogeneity exist outside the research laboratory, especially in the absence of a known marker. Mutant allele tumor heterogeneity (MATH) is a novel, non-biased, quantitative method to assess genetic heterogeneity based on tumor next generation exome sequencing. The quantitative aspect of MATH has allowed it to be verified as an actionable biomarker in a retrospective HNSCC data set with available exome sequencing and clinical data. In addition, it was also capable of stratifying patient outcome after controlling for other high-risk features such as p53 mutation, HPV status, and advanced tumor stage. Future work will explore the predictive power of MATH in larger data sets such as The Cancer Genome Atlas and examine the underlying cellular mechanisms responsible for intra-tumor heterogeneity.
Keywords: Intra-tumor heterogeneity, Next generation sequencing, Head and neck squamous cell carcinoma, Biomarker, Personalized medicine, Targeted therapy
Introduction
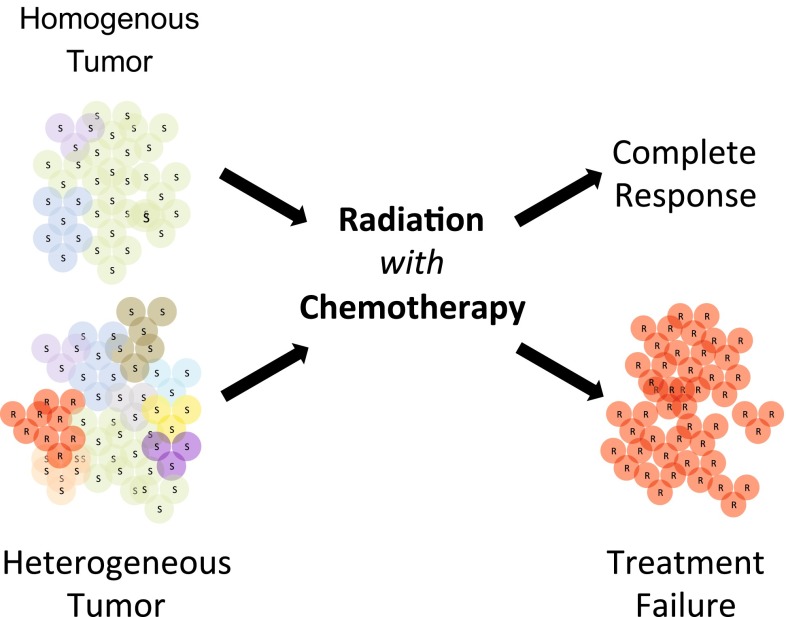
Oncologists and tumor biologists have long been concerned about intra-tumor heterogeneity as a source of variability in tumor behavior resulting in worse clinical outcome (Fig. 1). Consistent with this concern, several recent studies have documented a worrisome role for intra-tumor heterogeneity in tumor development, metastasis, and treatment resistance [1]. Pathologists also share these concerns, although few tools or clinical guidelines exist on how to address intra-tumor heterogeneity as part of routine pathological tumor tissue evaluation. To date, few studies have documented an important role for intra-tumor heterogeneity in treatment resistance and clinical outcomes, and those were often based on the use of known markers as part of traditional FISH and immunohistochemical analysis [2].
Fig. 1.
Intra-tumor heterogeneity as a source of treatment failure. Different tumor subpopulations are represented by colored circles that are either sensitive (S) or resistant (R) to therapy. The homogeneous tumor is depicted as containing few subpopulations, while the heterogeneous tumor is depicted as containing significantly more. Note that the red (R) subpopulation in the heterogeneous tumor is resistant to therapy and enriched after treatment failure. Implied in the figure is the central premise that a heterogeneous tumor with a larger number of subpopulations is more likely to possess a population resistant to therapy
Consequently, there have been no large-scale studies on the prognostic importance of intra-tumor heterogeneity on outcome in any type of cancer. The major reason for this gap in knowledge is that methods typically used for intra-tumor heterogeneity research [3] would be difficult to incorporate widely in clinical research or practice, and are not readily available to the practicing pathologist. For example, pre-identified markers of tumor subpopulations [4, 5] may not generalize well across tumor types and would be difficult to standardize and quantitate. Further, their incorporation would require significant advances in the application of imaging and imaging processing techniques, as well as the creation and validation of guidelines and criteria for scoring and reporting results [6, 7]. Other approaches, involving the separate analyses of multiple portions from single tumor [8] or single-cell analysis [9, 10] would be difficult to scale up for studies of hundreds of tumors, and face many of the same challenges just described for pre-identified markers.
Even when intra-tumor heterogeneity is recognized, it is unclear how to stratify patient’s tumor specimens into clinically significant subgroups [11], and consequently intra-tumor heterogeneity is often ignored and rarely reported. Therefore, it has not yet been possible to incorporate information about intra-tumor heterogeneity into clinical trial design and decision-making. The magnitude of this problem is further compounded by the great interest and need to identify patients who are potential candidates for targeted therapy, where high intra-tumor heterogeneity may directly confound tumor cell treatment response [12].
The proliferation of next generation sequencing (NGS) strategies and their application to different tumor types, including head and neck squamous cell carcinoma (HNSCC) [13] has created a previously unexplored opportunity to first develop [14] and then assess [15] a novel marker of genetic intra-tumor heterogeneity, Mutant allele tumor heterogeneity or MATH.
Mutant Allele Tumor Heterogeneity (MATH)
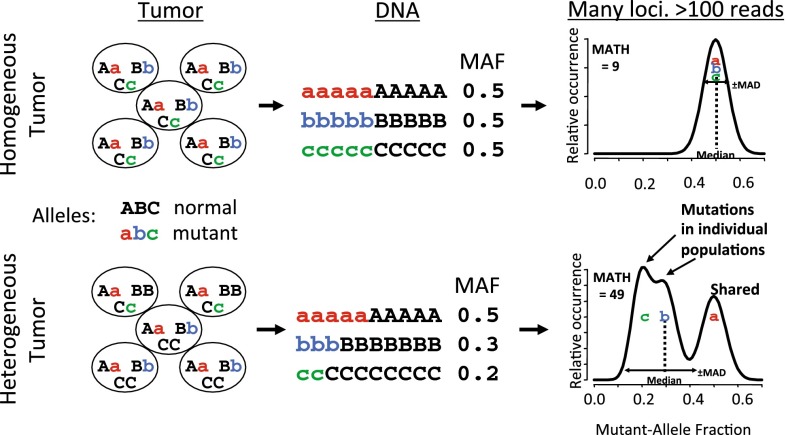
MATH is a quantitative assessment based on NGS of tumor and matched normal DNA as described [14]. NGS provides the percentage of sequencing reads showing a tumor-specific mutation at each genomic locus, the mutant-allele fraction. Typically, a genomic locus mutated early in the clonal evolution of a tumor will be shared among later-arising subclones and consequently will have a higher mutant-allele fraction than a locus mutated later in clonal evolution, which will often just be restricted to one or a few subclones. Consequently, heterogeneous tumors containing genetically distinct subclones will have a wide distribution of mutant-allele fractions among genomic loci-bearing somatic mutations. MATH, the normalized width of this distribution, provides a quantitative measure of this consequence of intra-tumor heterogeneity. A schematic outlining the basic tenets of MATH is shown in Fig. 2, where idealized low (top) and high (bottom) heterogeneity tumors are presented. It is important to note that a heterogeneous tumor will have a broader distribution, shorter peak, and lower median mutant-allele fraction when compared to more homogenous tumors, as shown in the idealized distributions. This is readily seen in three representative distributions of mutant-allele fractions of HNSCC specimens from published NGS data reflecting low, medium, and high MATH scores (Fig. 3).
Fig. 2.
Schematic outline of Mutant Allele Tumor Heterogeneity. Homogenous (top) and heterogeneous (bottom) tumors containing normal (ABC) or mutant (abc) alleles, were subject to NGS and their mutant allele fraction (MAF) determined as part of MATH analysis before plotting their idealized MAF distributions (revised and modified schematic of data previously presented in [14])
Fig. 3.
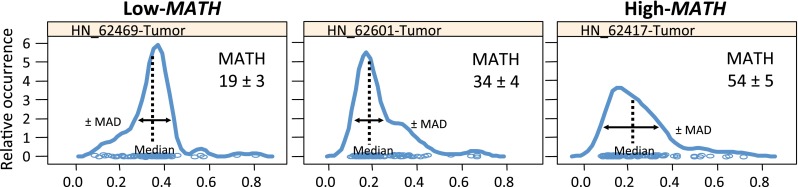
Representative mutant allele distributions from three HNSCCs. Note that a heterogeneous tumor with a high MATH score (right panel) will have a broader distribution, with a shorter peak and lower median mutant allele fraction, when compared to a homogenous, low MATH tumor (shown is revised and modified data that was previously presented in [14])
MATH and Clinical Outcome in HNSCC
The development of a quantitative measure of differences between tumors that reflect intra-tumor heterogeneity based on NGS of bulk tumor DNA allowed us to look at MATH, high-risk markers and clinical outcome among patients with HNSCC [14, 15]. As a first step we looked at the relationship of MATH to several established markers of outcome in HNSCC; cigarette smoking as measured by pack-years, presence of p53 mutation, and HPV status. In all three cases, higher tumor MATH values were associated with the poor outcome classification of these three markers. Cigarette exposure, in addition to being a major cause of HNSCC, is associated with worse outcomes, with each pack-year of cigarette use resulting in a 1 % increase in the hazard ratio for relapse or death. MATH was positively associated with cigarette pack-years, with a 1.1 MATH unit increase per 10 pack-years. The p53 tumor suppressor protein has a well-established tumor suppressor role in HNSCC, and as a result is a common target for inactivation, usually through inactivating “disruptive” mutations that ultimately interfere with DNA binding. High MATH scores were positively associated with p53 mutation, consistent with the relationship of p53 mutation to poor outcome in HNSCC in numerous studies. Increasingly, high-risk HPV infection is directly responsible for a large and growing number of HNSCC cases. These patients are usually younger, lack traditional risk factors and have improved outcomes when compared to their HPV-negative counterparts [16]. In this data set, HPV status was based on aligning sequencing reads to the HPV genome followed by PCR confirmation [13]. Here MATH scores were significantly lower in the HPV-positive tumors when compared to the HPV-negative cases, suggesting these tumors are more homogeneous. This likely reflects the role of the E6 and E7 viral oncogenes targeting the p53 and p16-pRb tumor suppressor pathways for disruption, rather than the need to accumulate inactivating mutations overtime. Importantly, the association of MATH scores and HPV status was independent of the presence of p53 mutations, since HPV-positive tumors are often wild type for p53.
A direct assessment of the impact of MATH on clinical outcome was first performed when the clinical data from Stransky et al. became available for analysis [15]. In this retrospective analysis of 74 HNSCC patients from a single institution, high MATH scores (a MATH value above the median) were found to be significantly associated with shorter overall survival (hazards ratio, 2.5; 95 % confidence interval, 1.3–4.8).
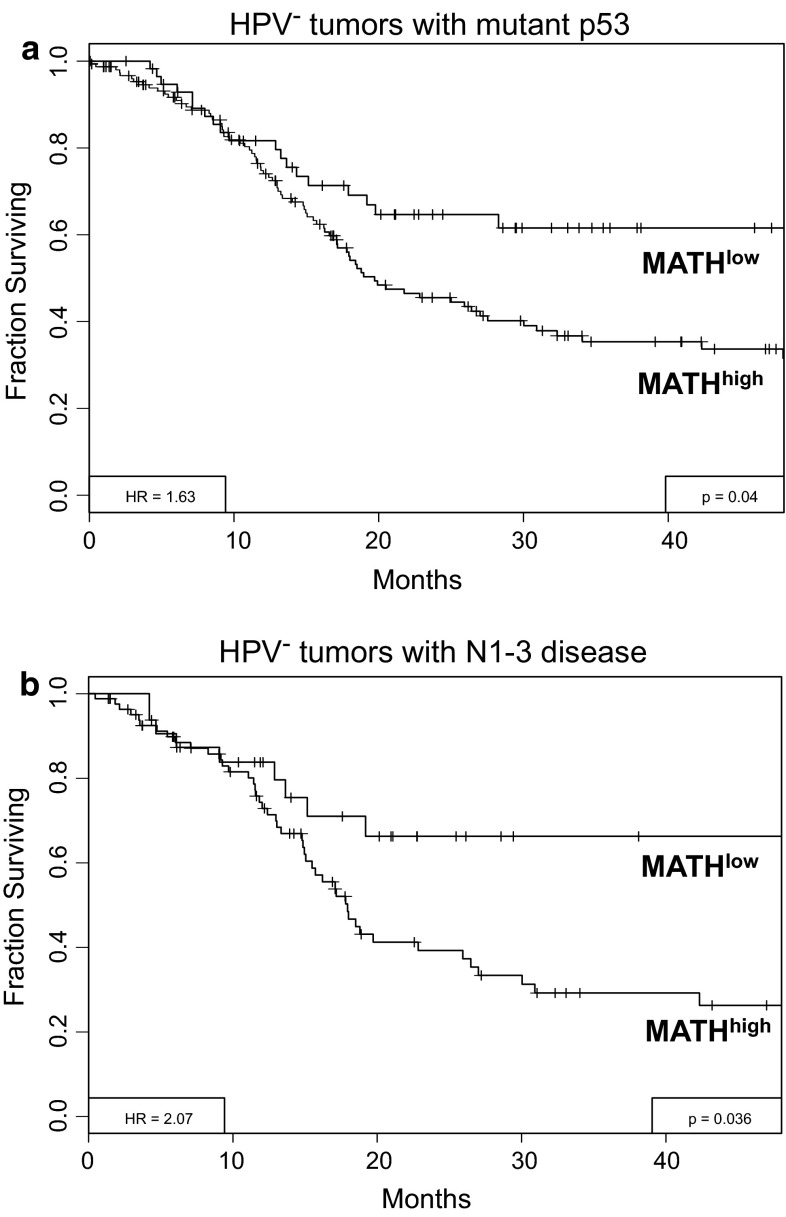
Ongoing analysis based on data from The Cancer Genome Atlas (TCGA) that contains open-access clinical and exome NGS data on HNSCC, has provided an independent validation data set for testing the relationship of MATH to outcome. Our preliminary analysis suggests that similar to our earlier work, high MATH is associated with decreased survival among the 305 HNSCC patients from 14 TCGA institutions. In addition, the increased representation of HPV-positive HNSCC in this larger TCGA data set also allowed us to determine that MATH adds prognostic information beyond that provided by HPV status and other standard prognostic variables. For example, a high MATH score was also found to be associated with poor outcomes in other high-risk patients, such as those with p53 mutation (Fig. 4a) or advanced nodal stage (Fig. 4b), where MATH could further stratify outcome.
Fig. 4.
MATH can further stratify high-risk HNSCC patients. a Survival of high-risk HPV negative patients with mutant p53 can be further stratified by MATH analysis. b Survival of high-risk HPV negative patients with N1-3 nodal status can be further stratified by MATH analysis. Data based on ongoing analysis of 305 patients from the HNSCC TCGA data set ([14], unpublished data)
Discussion
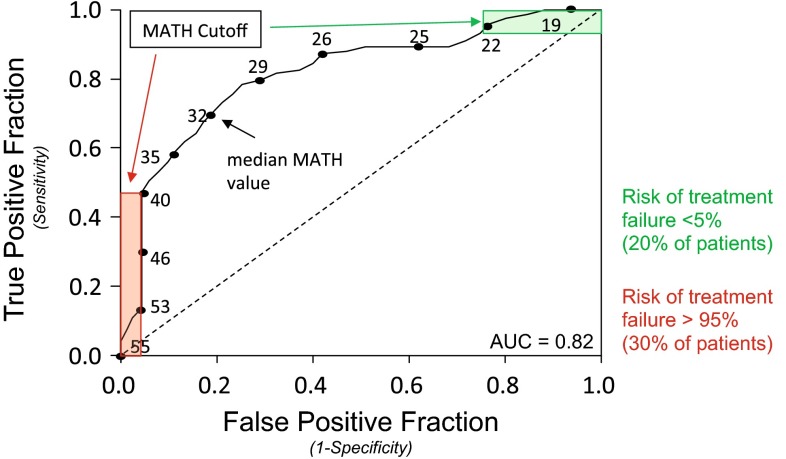
In our analysis, high intra-tumor heterogeneity as assessed by MATH analysis is associated with markers of poor outcome and decreased overall survival among HNSCC patients. This result suggests that the level of heterogeneity itself, as represented by MATH, can serve as an important biomarker. Here tumors with higher MATH scores are more heterogeneous and consequently statistically more likely to contain subclones that are resistant to therapy. It is important to emphasize that a heterogeneous tumor is just more likely to possess a resistant subclone when compared to a more homogenous tumor. It is possible that a highly heterogeneous tumor could be sensitive to therapy and a homogenous tumor resistant, however the likelihood is very low. For example, in our prior analysis [15] the chance that HNSCCs with MATH scores >40 (representing 30 % of tumors) would be sensitive to treatment was <5 % (Fig. 5).
Fig. 5.
Predicting outcome with MATH. Use of the receiver operating characteristic curve and specified MATH cutoff value allows the outcome of a subset of patients to be predicted with high specificity (revised and modified schematic of data initially presented in [15]). Selection of specific MATH cutoffs of >40 (red) or <22 (green) allows treatment outcome to be predicted with >95 % specificity in 50 % of patients
Intra-tumor heterogeneity represents an important challenge to overcome in the clinical care of the cancer patient [17]. The resistant subclones likely found in heterogeneous tumors are capable of tumor repopulation and tumor progression, even if there was an initial response to therapy. Incorporation of MATH into clinical care has the potential to stratify patient tumors into specific response categories, allowing treatment to be tailored to the level of heterogeneity present. Low MATH tumors would be excellent candidates for either treatment de-escalation to minimize toxicity, or for targeted therapy where the homogeneity of the tumor may suggest the presence of an oncogene addiction pathway. High MATH tumors would remain problematic, although the prognostic value of MATH could provide important insights regarding the likely outcome of traditional treatment, and consequently could be used to recruit and stratify high-risk patients into specific clinical trials.
Scientifically, further insight into the true source of intra-tumor heterogeneity needs to be gained and the relative contributions of the two competing models of tumor evolution (cancer stem cell versus clonal evolution) defined [18]. However, specific approaches to targeting intra-tumor heterogeneity are beginning to be discussed. Clinical strategies to target intra-tumor heterogeneity must at their core address and reverse Darwinian forces [19]. In traditional chemotherapy, the fit cells of a tumor are targeted as the proliferative population, with the subsequent selection of the slower growing resistant cells. Two novel approaches to target these slow growing cells include Metronomic therapy [20], where chemotherapy is delivered more frequently at a lower dose, and Adaptive therapy [21], which attempts to turn the cancer into a chronic condition by maintaining a fit population of chemotherapy sensitive cells. Alternate strategies which also demonstrate promise include the identification of novel shared trunk mutations for targeted therapy through the use of phylogenetic analysis within a branched evolution model [22], and immunotherapeutic approaches that take advantage of the tremendous diversity of the immune response to target tumor antigens [23]. Finally, one potential source of intra-tumor heterogeneity, genomic instability, may be targeted directly through the use of checkpoint inhibitors [24].
Conclusion
The role of intra-tumor heterogeneity in treatment resistance is increasingly being recognized as a major barrier to curative cancer therapy. MATH, a quantitative measure of genetic intra-tumor heterogeneity based on NGS of bulk tumor, represents an unbiased way to characterize intra-tumor heterogeneity without assessing specific genes or their protein products. The relationship of MATH to clinically important variables and treatment outcome support the use of MATH as a quantitative biomarker to assess intra-tumor heterogeneity. In our analysis, high intra-tumor heterogeneity as assessed by MATH analysis was associated with decreased survival among HNSCC patients. With NGS expected to soon play a significant role in clinical oncology, MATH should provide a simple way to incorporate intra-tumor heterogeneity into clinical trial design, treatment selection, and the care of the HNSCC patient.
Acknowledgments
I wish to thank Dr. Ed Mroz for helpful comments and suggestions on an earlier draft of this review.
References
- 1.Burrell RA, Swanton C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol. 2014;8(6):1095–1111. doi: 10.1016/j.molonc.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedard PL, et al. Tumour heterogeneity in the clinic. Nature. 2013;501(7467):355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park SY, et al. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120(2):636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russnes HG, et al. Insight into the heterogeneity of breast cancer through next-generation sequencing. J Clin Invest. 2011;121(10):3810–3818. doi: 10.1172/JCI57088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolff AC, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: american Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138(2):241–256. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allison KH, Dintzis SM, Schmidt RA. Frequency of HER2 heterogeneity by fluorescence in situ hybridization according to CAP expert panel recommendations: time for a new look at how to report heterogeneity. Am J Clin Pathol. 2011;136(6):864–871. doi: 10.1309/AJCPXTZSKBRIP07W. [DOI] [PubMed] [Google Scholar]
- 8.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461(7265):809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 9.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148(5):886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crockford A, et al. Implications of intratumour heterogeneity for treatment stratification. J Pathol. 2014;232(2):264–273. doi: 10.1002/path.4270. [DOI] [PubMed] [Google Scholar]
- 12.Lim JS, Lee SC. Understanding intra-tumour heterogeneity—the next holy grail of cancer therapeutics? Ann Acad Med Singap. 2014;43(2):72–73. [PubMed] [Google Scholar]
- 13.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49(3):211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mroz EA, et al. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer. 2013;119(16):3034–3042. doi: 10.1002/cncr.28150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonilla-Velez J, et al. Impact of human papillomavirus on oropharyngeal cancer biology and response to therapy: implications for treatment. Otolaryngol Clin North Am. 2013;46(4):521–543. doi: 10.1016/j.otc.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renovanz M, Kim EL. Intratumoral heterogeneity, its contribution to therapy resistance and methodological caveats to assessment. Front Oncol. 2014;4:142. doi: 10.3389/fonc.2014.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shackleton M, et al. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138(5):822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Polyak K. Tumor heterogeneity confounds and illuminates: a case for Darwinian tumor evolution. Nat Med. 2014;20(4):344–346. doi: 10.1038/nm.3518. [DOI] [PubMed] [Google Scholar]
- 20.Andre N, Carre M, Pasquier E. Metronomics: towards personalized chemotherapy? Nat Rev Clin Oncol. 2014;11(7):413–431. doi: 10.1038/nrclinonc.2014.89. [DOI] [PubMed] [Google Scholar]
- 21.Gatenby RA, et al. Adaptive therapy. Cancer Res. 2009;69(11):4894–4903. doi: 10.1158/0008-5472.CAN-08-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marabelle A, et al. Intratumoral immunization: a new paradigm for cancer therapy. Clin Cancer Res. 2014;20(7):1747–1756. doi: 10.1158/1078-0432.CCR-13-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30(7):679–692. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]