Abstract
Kink turns (k-turns) are widespread elements in RNA that mediate tertiary contacts by kinking the helical axis. We have found that the ability of k-turns to undergo ion-induced folding is conferred by a single base pair that follows the conserved A·G pairs, that is, the 3b·3n position. A Watson–Crick pair leads to an inability to fold in metal ions alone, while 3n=G or 3b=C (but not both) permits folding. Crystallographic study reveals two hydrated metal ions coordinated to O6 of G3n and G2n of Kt-7. Removal of either atom impairs Mg2+-induced folding in solution. While SAM-I riboswitches have 3b·3n sequences that would predispose them to ion-induced folding, U4 snRNA are strongly biased to an inability to such folding. Thus riboswitch sequences allow folding to occur independently of protein binding, while U4 should remain unfolded until bound by protein. The empirical rules deduced for k-turn folding have strong predictive value.
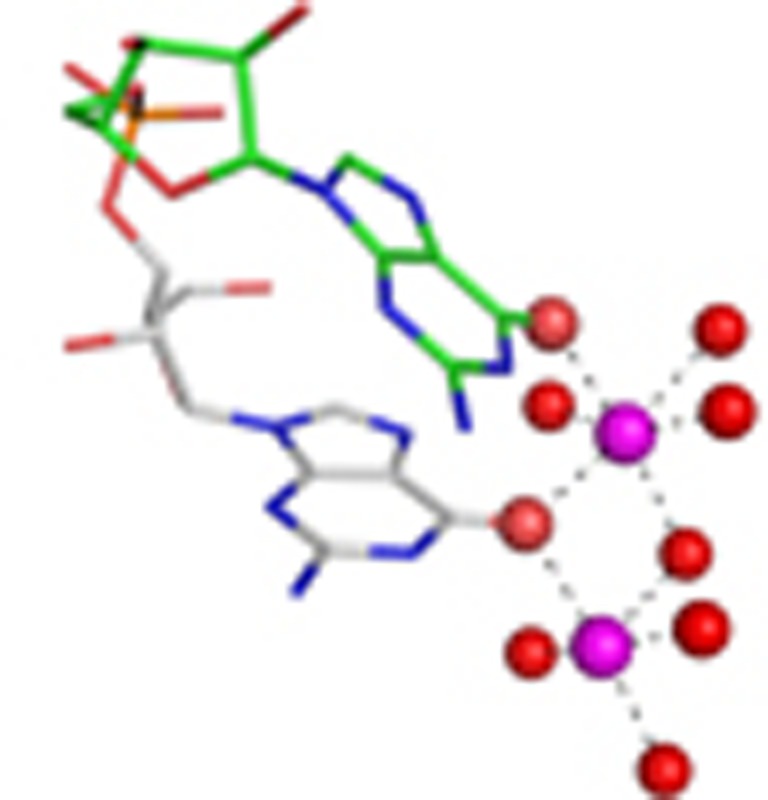 The k-turn is a widespread RNA element that adopts a kinked structure that mediates tertiary contacts and frequently binds specific proteins. Here, McPhee et al. show that the ability of a given k-turn to fold in the presence of metal ions alone—or to otherwise require protein binding—is attributable to a specific base pair.
The k-turn is a widespread RNA element that adopts a kinked structure that mediates tertiary contacts and frequently binds specific proteins. Here, McPhee et al. show that the ability of a given k-turn to fold in the presence of metal ions alone—or to otherwise require protein binding—is attributable to a specific base pair.
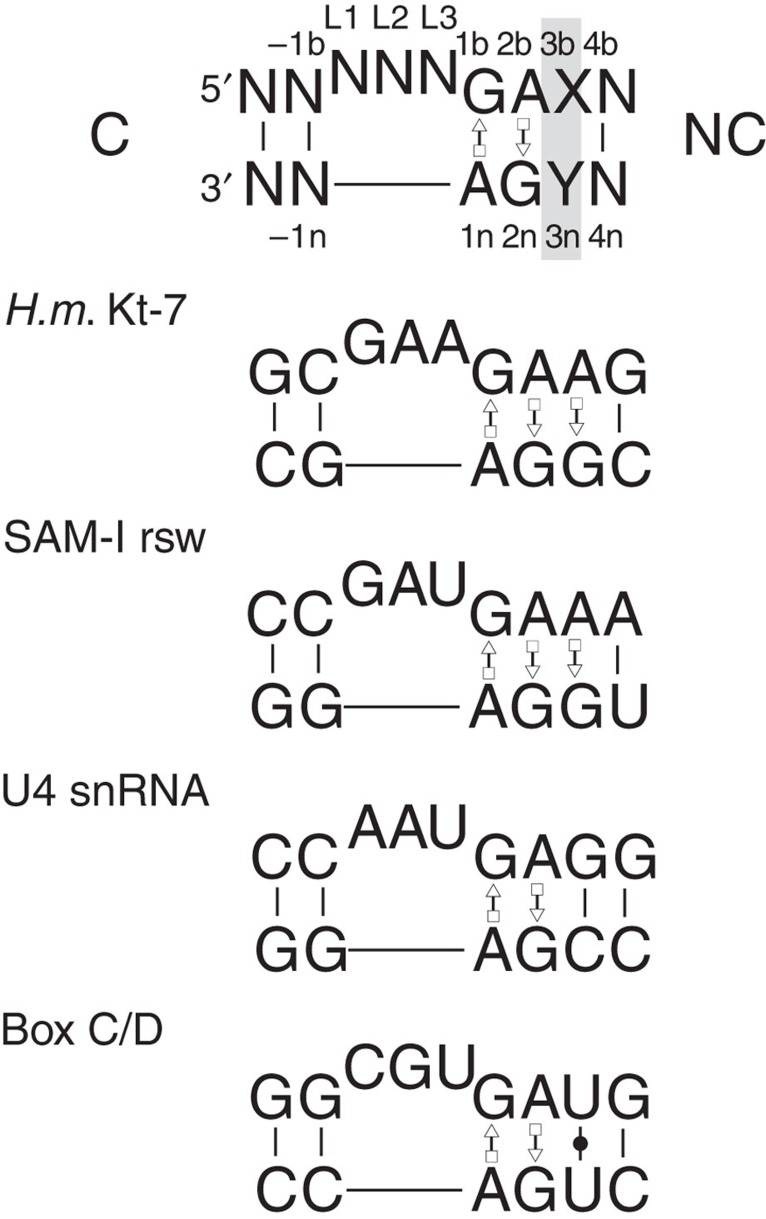
Kink turns (k-turns) are ubiquitous sequences that generate a tight kink within an RNA helix1, mediating tertiary interactions in the folding of large assemblies such as the ribosome, and often serving as the target for specific binding proteins. Because of this, k-turns have a key role in the assembly of ribosomes, the spliceosome2 and box C/D3 and H/ACA4,5 snoRNPs, as well as seven distinct riboswitch species6. The standard k-turn comprises a duplex interrupted by a three-nucleotide bulge followed by G·A and A·G base pairs (Fig. 1), and the adenine nucleobases make key cross-strand hydrogen bonds that stabilize the kinked conformation7,8,9.
Figure 1. The sequences of some standard k-turns.
The top sequence shows a standard k-turn with the standard nomenclature for nucleotide positions indicated8 and the 3b·3n position highlighted. Below are shown the sequences of the k-turns of H. marismortui ribosomal Kt-7, the SAM-I riboswitch, human U4 snRNA and box C/D snoRNA.
k-turn-containing RNA exists in a two-state equilibrium between the kinked conformation and a relatively extended structure, and is strongly biased towards the extended structure in the absence of some specific process promoting folding10. Several factors can drive the equilibrium towards the kinked structure of the k-turn. These include tertiary contacts11 and protein binding12,13,14. The L7Ae family15 is a particularly important class of proteins that selectively bind k-turns, that includes the human 15.5 kDa protein. L7Ae-related proteins are bound to k-turns in the ribosome16, U4 snRNA17 and box C/D18,19. Metal ions are a third factor—some but not all, k-turns will fold upon addition of metal ions. Both divalent and monovalent ions can induce k-turn folding, but much higher concentrations of the latter are required. Half-complete folding of Kt-7 occurs with [Mg2+]1/2=90 μM or [Na+]1/2=30 mM (ref. 10).
However k-turns differ markedly in their ability to fold in response to metal ions. Some will fold intrinsically in the presence of physiological concentrations of metal ions, while others require stabilization by other means, such as the binding of proteins. These contrasting folding properties must be very important in the ordered assembly and function of their RNA species. In this work we have set out to discover the molecular basis of these differences. We have discovered the key determinant of ability to undergo ion-induced folding resides in a single base pair that follows the conserved A·G pairs (3b·3n). The most readily folded sequence has A·G in this position, and we find that the O6 of the guanine directly coordinates metal ions. Analysis of many sequences of the k-turns of the SAM-I riboswitch and U4 snRNA reveals a strong correlation between the folding ability conferred by their sequence, and their biological function. The deduced sequence rules have strong predictive value, and can be applied to many natural RNA sequences such as those of the ribosome.
Results
Ion-induced folding determined by a key sequence element
The extensively-studied Kt-7 of the Haloarcula marismortui ribosome, and the k-turn of the SAM-I riboswitch, both fold into the characteristic kinked structure on addition of metal ions alone (Supplementary Figs 1 and 2; refs 10,11). However, in marked contrast the k-turns of the archaeal box C/D (Supplementary Fig. 3) and the human U4 snRNA (Supplementary Fig. 4) do not fold upon addition of metal ions. Both box C/D and U4 k-turns fold on binding of L7Ae protein, and indeed co-crystal structures of both show that these k-turns are folded2,3, so each is intrinsically capable of adopting the k-turn conformation, yet metal ions alone fail to achieve folding. These biologically important k-turns divide into two classes on the basis of their ability to be folded by metal ions, evidently a result of their sequence. As the G·A and A·G pairs are strongly conserved at the 1b·1n and 2b·2n positions (the nomenclature8 is shown in Fig. 1), the important difference must lie elsewhere, and our suspicion turned to the 3b·3n position that follows the conserved G·A pairs.
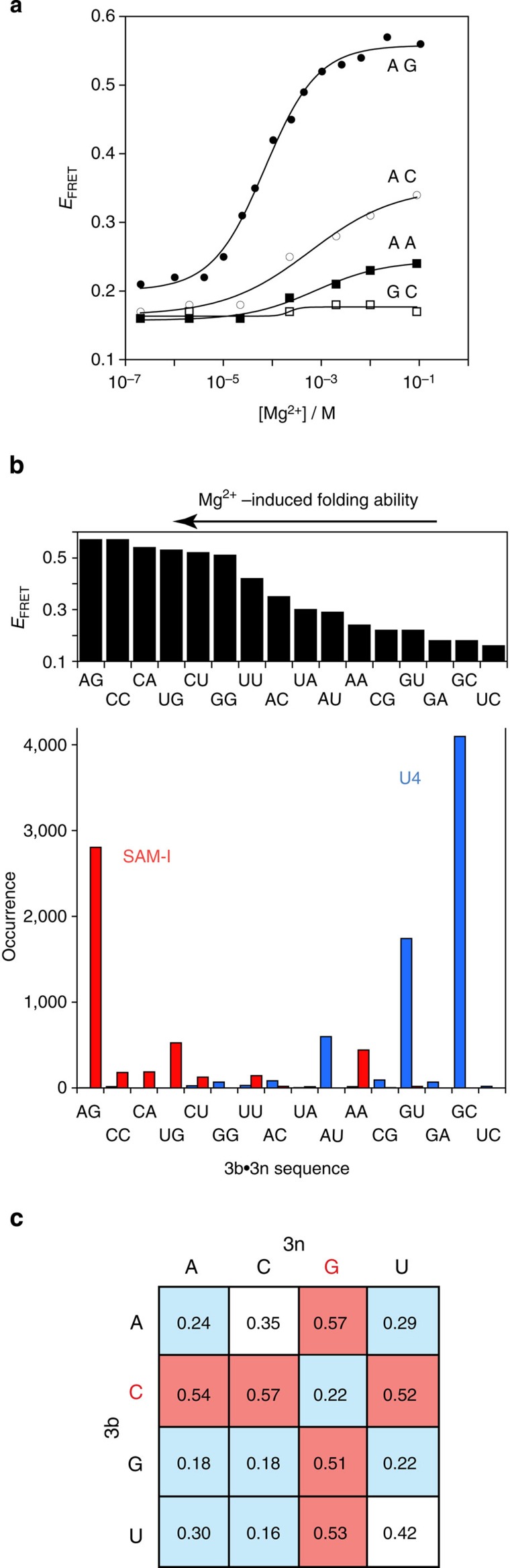
We took a short RNA duplex with a central Kt-7 sequence and fluorophores at both 5′ termini, enabling us to follow folding into the kinked conformation by the increase in FRET efficiency (EFRET) as the end-to-end distance shortens. The experiment was performed in a background of 90 mM Tris-borate (pH 8.3), with Mg2+ titrated as the only cation present. For the natural Kt-7 sequence EFRET increased from ~0.2 to 0.56 on addition of Mg2+ ions, with a value of [Mg2+]1/2=70 μM. The analysis was repeated for the 15 species in which the 3b·3n position was replaced by each combination of the four nucleotides (Fig. 2a, Supplementary Table 1). A range of folding abilities were found from full folding (for example, natural Kt-7; 3b·3n=A·G) to those exhibiting a complete inability to fold under these conditions (for example, 3b·3n=G-C). Yet even the 3b·3n=G-C sequence underwent folding upon addition of L7Ae protein, so it is not intrinsically unable to adopt the k-turn structure.
Figure 2. The ion-induced folding of k-turns as a function of sequence, and a correlation with biological function.
(a) Plot of EFRET (measured in the ensemble) as a function of Mg2+ ion concentration for Kt-7 (3b·3n=A·G) and three representative 3b·3n variants of contrasting folding ability. The data (points) have been fitted (lines) to a two-state model for ion-induced folding. (b) Histograms of EFRET versus 3b·3n sequence (upper) and distribution of SAM and U4 3b·3n sequences (lower) in natural sequences found in the Rfam database20 ranked (L to R) by the ability to fold in Mg2+ ions. (c) An array showing EFRET as function of 3b (vertical) and 3n (horizontal) sequence for all Kt-7 variants. Sequences that are readily folded by Mg2+ ions (ΔEFRET≥0.5) are shaded red, while those poorly folded (ΔEFRET≤0.3) are shaded blue. The ascending diagonal contains the Watson–Crick pairs.
3b·3n sequences correlate with biological function
Thus the 3b·3n sequence acts as a key discriminator, conferring ion-dependent folding properties. Is this reflected in the distribution of k-turn sequences as a function of biological role? We examined the distribution of 3b·3n sequences in two important functional RNA species, comparing several thousand SAM-I and U4 k-turn sequences downloaded from the Rfam database20. These two were chosen because of their contrasting environments. The SAM-I k-turn mediates a key tertiary contact6,11 to create a ligand-binding pocket in a riboswitch not known to bind a protein, while the U4 snRNA k-turn binds the 15.5 kDa protein during spliceosome assembly17. The results are plotted as a histogram in Fig. 2b, showing the occurrence of the 3b·3n sequences for the two species ranked horizontally by the folding ability of Kt-7 with the same 3b·3n sequence. It is apparent that the two species cluster at opposite ends of the folding spectrum. The SAM-I k-turn sequences are strongly biased towards an ability to fold in Mg2+ ions, with 60% having 3b·3n=A·G, which is the best folding sequence, and just 0.1% being C-G or G-C. By contrast, 97% of the U4 k-turn sequences are predicted to be unable to fold in Mg2+ ions, with a very strong bias to 3b·3n=G-C or G-U, and less than 0.03% being A·G. Interestingly, modification of the human U4 k-turn sequence by conversion of 3b·3n from G-C to A·G conferred an ability to fold in response to addition of Mg2+ ions (Supplementary Fig. 4).
Empirical sequence rules for ion-induced folding
Examination of the 3b·3n sequences displayed in array form and scored by folding ability (Fig. 2c) reveals some interesting patterns. First, Watson–Crick pairs (Fig. 2c—ascending diagonal) plus GU are all poor folders, with G-C and C-G especially bad. By contrast, the presence of 3n=G (Fig. 2c—third column) or 3b=C (Fig. 2c—second row) associates with ability to fold in Mg2+ ions. However, since 3b·3n=C-G is unfolded in metal ions, the first rule takes precedent over the second. The two best-folding k-turns both have G at the 3n position, and 95% of SAM-I k-turns have G at 3n. Moreover, the k-turns of the glycine21, lysine22 and cobalamine23 riboswitches also have 3n=G.
A structural explanation of the 3n=G rule
Systematic investigation of the ion-induced folding of Kt-7 shows that the most readily folded sequences are those with either 3n=G or 3b=C. There are no high-resolution crystal structures available for 3b=C k-turns, and at the present time we cannot rationalize this effect. However, we can provide a molecular explanation for the 3n=G behaviour.
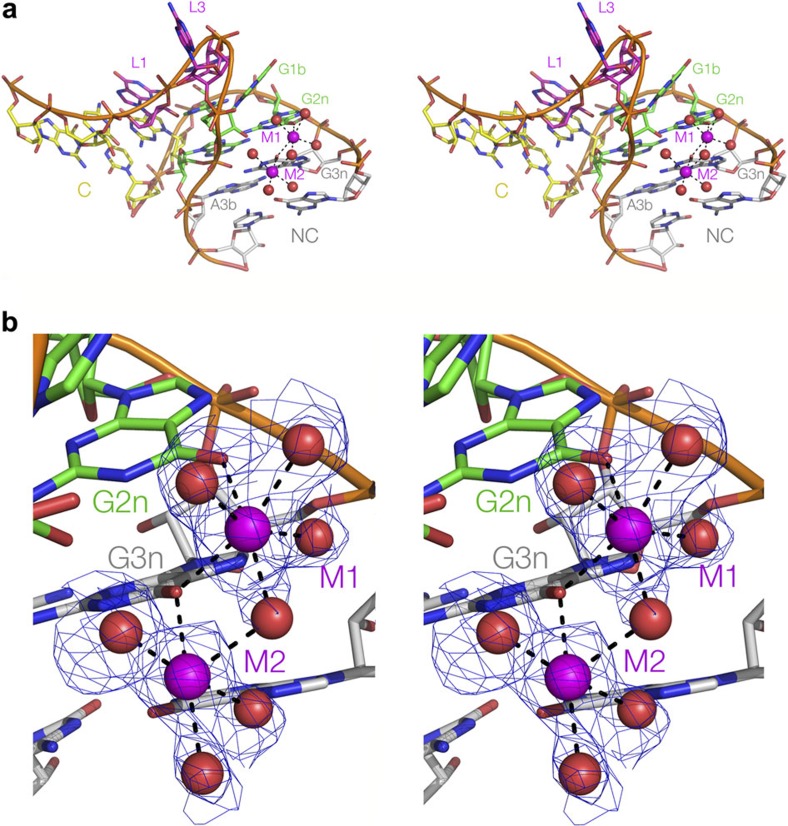
We have previously presented a crystal structure of H. marismortui Kt-7 as a protein-free duplex at 2.3 Å resolution24. We subsequently obtained crystals diffracting to 2.0 Å (Table 1), whereupon we observed two hydrated metal ions bound in the major groove of the NC helix adjacent to G2n and G3n (Fig. 3a). The electron density for the inner coordination sphere of water molecules is very clear, and both metal ions have octahedral symmetry. Thus they are most probably Mg2+ ions, although we cannot exclude the possibility that they are Na+ ions on the basis of the metal-O distances. Ion M1 has exchanged two adjacent inner-sphere water molecules with G2n and G3n O6 atoms, while G3n O6 makes an inner-sphere contact with both ions (Fig. 3b, Supplementary Fig. 5).
Table 1. Data collection and refinement statistics.
| Macromolecules | Kt-7 |
| Data collection | |
| Space group | P6322 |
| Cell dimensions | |
| a, b, c (Å) | 70.08, 70.08, 47.52 |
| α, β, γ (°) | 90.00, 90.00, 120.00 |
| Resolution (Å) | 37.41-2.00 (2.05-2.00)* |
| Rsym or Rmerge | 6.5 (132.8) |
| I/σI | 14.82 (1.4) |
| CC(1/2) | 0.999 (0.501) |
| Completeness (%) | 98.32 (99.59) |
| Redundancy | 6.3 (6.4) |
| Refinement | |
| Resolution (Å) | 37.42-2.00 (2.52-2.00) |
| No. of reflections | 4916 |
| Rwork/Rfree | 0.184/0.234 (0.259/0.291) |
| No. of atoms | |
| RNA | 417 |
| Ligand/ion | 2 |
| Water | 23 |
| B-factors | |
| RNA | 47.6 |
| Ligand/ion | 33.7 |
| Water | 44.6 |
| r.m.s deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles | 0.81 |
*Highest resolution shell is shown in parenthesis.
Figure 3. A structural basis for the differential ability of k-turn sequences to undergo ion-induced folding.
(a,b) Crystal structure of Kt-7 reveals two Mg2+ ions bound in the major groove of the NC helix. The structure is shown as parallel eye stereographic images. (a) An overall view of the k-turn, with bound ions. (b) Closer view of the bound ions. The electron density for the metal and directly bound water is taken from the Fo−Fc omit map contoured at 2σ. Further maps are shown in Supplementary Figure 5. Both ions have exchanged inner-sphere water ligands to bond to guanine O6 atoms; M1 is directly bonded to O6 of both G2n and G3n, while the latter is directly bonded to both ions.
Removal of O6 from G2n or G3n impairs ion-induced folding
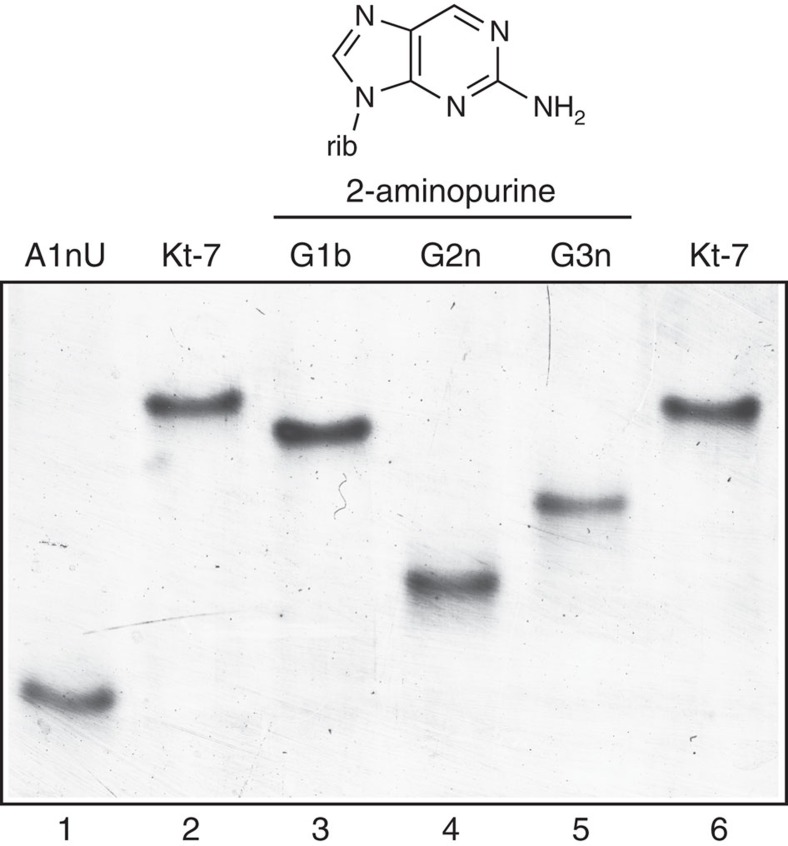
Having observed the two ions bound to G3n and G2n in the crystal, we then sought to test the importance of these interactions in the folding of Kt-7 in solution. This was examined by atomic mutagenesis whereby the participating O6 atoms were selectively removed by individual substitution of guanine by 2-aminopurine. Folding was analysed using a gel electrophoretic method10. A 65 bp RNA duplex with a central k-turn-containing RNA section was electrophoresed in 15% polyacrylamide in the presence of 90 mM Tris-borate (pH 8.3), 2 mM Mg2+. The folded structure of the unmodified k-turn results in pronounced electrophoretic retardation (Fig. 4). However, removal of either G2n or G3n O6 atoms significantly impaired the ability to fold on addition of Mg2+ ions, that is, resulted in less retarded electrophoretic mobility, whereas the corresponding modification of G1b had a minor effect. This provides a direct connection between the metal ions observed to bind to G2n and G3n O6 atoms by crystallography, and the ability of the k-turn to fold in response to the presence of Mg2+ ions. Thus binding of the divalent metal ions to guanine O6 at the 2n and 3n positions is the key determinant allowing the Kt-7 k-turn to fold unassisted by protein binding.
Figure 4. Removal of the O6 atoms from G2n and G3n impairs ion-induced folding of Kt-7.
The scheme at the top shows the structure of 2-aminopurine, showing that it differs from guanine by removal of O6. A 65 bp duplex with a central k-turn-containing RNA section was electrophoresed in 15% polyacrylamide in the presence of 2 mM Mg2+ ions. Folding of the unmodified species (tracks 2 and 6) results in a marked retardation compared with the A1nU variant (track 1). Substitution of G2n (track 4) or G3n (track 5) by 2-aminopurine lead to a marked impairment of folding (that is, increased mobility), whereas the same substitution at G1b (track 3) had a rather smaller effect.
Discussion
While all k-turns can be folded by protein binding and/or the formation of tertiary contacts, not all will fold spontaneously in the presence of metal ions, and we have found that a major determinant of this behaviour resides in the 3b·3n sequence. From a systematic analysis of 3b·3n sequence variants of Kt-7, we have formulated a set of rules that have predictive value; application of these can convert the U4 k-turn from non-ion folding into one that is fully folded, in the presence of Mg2+ ions for example. One of the rules is that 3n=G, and a high-resolution structure of Kt-7 as a free duplex RNA provides an explanation. Two hydrated, octahedrally-coordinated metal ions are directly bound to the O6 atoms of G2n and 3n. These are probably Mg2+ ions, although they could conceivably be Na+ ions, but since both ions can induce folding of Kt-7 then probably either can coordinate at this position. The binding can be directly connected with folding in solution, since selective removal of the O6 atoms leads to impairment of folding in the presence of Mg2+ as the only cation.
Analysis of the sequences of the k-turns of SAM-I and U4 snRNA shows a striking difference in the 3b·3n sequences of the two species. The k-turn sequences of the SAM-I riboswitch have been selected for predisposition to fold in metal ions alone. There is no protein that is known to bind to riboswitches in vivo, so this property is probably essential to permit folding and thereby generate a functional riboswitch. By contrast, the U4 k-turn binds the 15.5 kDa protein in vivo, and evidently U4 sequences have been selected for their inability to fold in metal ions alone. The U4 k-turn will be unfolded until the protein is bound, and can therefore only function as an RNA-protein complex to generate the U4/U6 snRNA complex in the spliceosome cycle. In the absence of protein binding, the extended form of the k-turn should be more flexible and this may be required to permit the formation of other interactions during the biogenesis of this complex and dynamic assembly. This analysis reveals a strong correlation between the folding properties of the isolated k-turn and their likely role in the cellular macromolecule. It emphasizes the key role of the 3b·3n sequence in the biological function of these k-turns.
We can apply our deduced folding sequence rules to the k-turns of the H. marismortui 50S ribosomal subunit. Kt-7, Kt-46, Kt-58 and Kt-78 (ref. 16), plus the J4,5 k-junction25 all have 3b·3n=A·G, and are therefore likely to fold unaided by protein. Interestingly, analysis of 2,716 bacterial Kt-7 sequences shows that while 3b·3n=A·G is relatively uncommon, 99.9% have either 3n=G or 3b=C. These k-turns all mediate tertiary interactions, and we envision that during rRNA folding this will assist the formation of long-range contacts before the structure becomes fixed by the binding of specific proteins. By contrast, Kt-15 is a complex k-turn with 3b·3n=C-G, and it does not undergo folding by addition of Mg2+ ions alone (unpublished data). In the ribosome it is bound by L7Ae. In vitro L7Ae binds k-turns with pM affinity13. The folding of Kt-15 and its tertiary contacts should therefore occur later than those not requiring protein binding, all of which will contribute to an ordered process for the folding of the ribosome.
In summary, we have found a strong correlation between the folding properties conferred by the 3b·3n sequence and the biological role of specific k-turns. The deduced sequence rules have predictive value and can be applied to new k-turn sequences.
Methods
RNA synthesis
Ribooligonucleotides were synthesized using t-BDMS phosphoramidite chemistry26. Fluorescein (Link Technologies) and Cy3 (GE Healthcare) were attached at 5′ termini as phosphoramidites during synthesis as required. Oligoribonucleotides were deprotected in 25% ethanol/ammonia solution at 20 °C for 3 h, and evaporated to dryness. They were redissolved in 100 μl dimethyl sulfoxide to which was added 125 μl 1 M triethylamine trihydrofluoride (Sigma-Aldrich) and incubated at 65 °C for 2.5 h to remove t-BDMS protecting groups. All oligonucleotides were purified by gel electrophoresis in polyacrylamide in the presence of 7 M urea. The full-length RNA product was visualized by ultraviolet shadowing. The band was excised and electroeluted using an Elutrap (Whatman) into 45 mM Tris-borate (pH 8.5), 5 mM EDTA buffer for 8 h at 200 V at 4 °C. The RNA was precipitated with ethanol, washed once with 70% ethanol and suspended in water.
Oligoribonucleotides containing 2-aminopurine (Glen Research) were deprotected using a 1:1 solution of 35% aqueous ammonia (Fisher Scientific) and 40% aqueous methylamine (Sigma-Aldrich) for 30 min. at 65 °C.
Fluorophore-labelled and 2-aminopurine-containing oligoribonucleotides were subjected to further purification by reversed-phase HPLC on a C18 column (ACE 10–300, Advanced Chromatography Technologies), using an acetonitrile gradient with an aqueous phase of 100 mM triethylammonium acetate (pH 7.0).
Duplex species were prepared by mixing equimolar quantities of the appropriate oligoribonucleotides and annealing them in 50 mM Tris–HCl (pH 7.5), by slow cooling from 90 to 4 °C. They were purified by electrophoresis in 12% polyacrylamide under nondenaturing conditions and recovered by electroelution, followed by ethanol precipitation.
Expression and purification of A. fulgidus L7Ae
The gene encoding full-length Archaeoglobus fulgidus L7Ae was cloned into a modified pET-Duet1 plasmid (Novagen)28 using the HindIII and EcoRI sites. The L7Ae gene was fused upstream of a hexahistidine-encoding sequence with a PreScission-cleavable linker. The hexahistidine-L7Ae fusion protein was expressed in Escherichia coli BL21-Gold (DE3) pLysS cells (Stratagene) induced with 0.2 mM IPTG at 20 °C for 12 h.
Harvested cells were resuspended in 20 mM Tris–HCl, (pH 8.0), 500 mM NaCl, 10 mM imidazole, 1 mM phenylmethylsulfonyl fluoride (buffer A) and lysed by sonication. The protein suspension was heated at 85 °C for 20 min in the presence of 10 mM MgCl2 to denature endogenous protein and this was removed by centrifugation at 18,000 r.p.m. for 30 min at 4 °C. L7Ae was loaded onto a HisTrap column (GE Healthcare), washed with 25 mM imidazole in buffer A, and the protein was eluted with 500 mM imidazole in buffer A. The six-His tag was cleaved from L7Ae by PreScission protease in 20 mM HEPES-Na (pH 7.6), 100 mM NaCl, 0.5 mM EDTA (buffer C) at 4 °C for 16 h. L7Ae was applied to a heparin column (GE Healthcare) and eluted at 250 mM NaCl in a gradient from 50 to 2,000 mM NaCl in 20 mM HEPES-Na (pH 7.6). The protein was further purified using a Superdex 200 gel filtration column in a buffer containing 5 mM Tris-HCl (pH 8.0), 100 mM NaCl.
The protein concentration was measured by absorbance at 280 nm using a molar extinction coefficient of 5,240 M−1 cm−1 for L7Ae. The protein was concentrated to 20 mg ml−1 in buffer containing 5 mM Tris-HCl (pH 8.0), 100 mM NaCl and stored at −20 °C as aliquots.
FRET analysis of k-turn folding
FRET efficiency was measured from a series RNA duplex species terminally 5′-labelled with fluorescein and Cy3, containing central k-turn sequences and variants.
Absorption spectra were measured in 90 mM Tris-borate (pH 8.3) in 2 μl volumes using a Thermo Scientific NanoDrop 2000c spectrophotometer. Spectra were deconvoluted using a corresponding RNA species labelled only with Cy3, and fluorophore absorption ratios calculated using a MATLAB program. Fluorescence spectra were recorded in 90 mM Tris-borate (pH 8.3) at 4 °C using an SLM-Aminco 8,100 fluorimeter. Spectra were corrected for lamp fluctuations and instrumental variations, and polarization artifacts were avoided by setting excitation and emission polarizers crossed at 54.7°. Values of FRET efficiency (EFRET) were measured using the acceptor normalization method29 implemented in MATLAB. EFRET as a function of Mg2+ ion concentration was analysed on the basis of a model in which the fraction of folded molecules corresponds to a simple two-state model for ion-induced folding, that is,
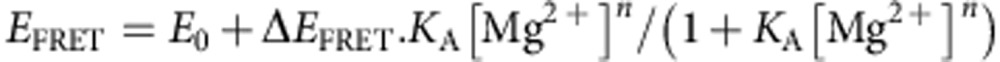 |
where E0 is the FRET efficiency of the RNA in the absence of added metal ions, ΔEFRET is the increase in FRET efficiency at saturating metal ion concentration, [Mg2+] is the prevailing Mg2+ ion concentration, KA is the apparent association constant for metal ion binding and n is a Hill coefficient. Data were fitted to this equation by nonlinear regression. The metal ion concentration at which the transition is half complete is given by [Mg2+]1/2=(1/KA)1/n.
The same RNA oligonucleotides as used in the Mg2+-induced folding were used for the L7Ae binding experiments, and FRET was measured and analysed using the same approach. L7Ae was added from a stock solution to a solution of 2 nM solution of RNA.
Gel electrophoretic analysis of k-turn folding
RNA species were electrophoresed in 13% polyacrylamide (29:1, acrylamide:bis) gels in 90 mM Tris.borate (pH 8.3) plus 2 mM Mg2+ ions. Electrophoresis was performed at 120 V at 4 °C for at least 72 h, with recirculation of the buffer at >1 litre h−1. Gels were stained using SYBR Gold (Life Technologies), washed in MilliQ water and visualized on a Typhoon FLA 9500 (GE Healthcare).
The sequences used for the electrophoretic experiments were (written 5′ to 3′):
Kt-7 upper strand:
5′- CGCAAGCGACAGGAACCTCGCCAGUCAGUGGCGAAGAACCAU GUCAGGGGACTGTCAAGTTGAACAGG -3′
Kt-7 lower strand:
5′- CCTGTTCAACTTGACAGTCCCCUGACAUGGGGAGCCACUGA CUGGCGAGGTTCCTGTCGCTTGCG -3′
The DNA sections of these oligonucleotides are shown underlined. Modified nucleotides were introduced into the RNA sections as required.
Crystal structure determination and refinement
The crystallized construct had the sequence (written 5′ to 3′):
5′- GGCGAAGAACCGGGGAGCC -3′
This self-complementary sequence forms the structure shown in Supplementary Fig. 6, containing two Kt-7 motifs.
A solution of 1 mM RNA in 5 mM Tris–HCl (pH 8.0) and 100 mM NaCl was heated to 95 °C for 1 min. The solution was slow cooled to 20 °C and MgCl2 was added to a final concentration of 10 mM. The hanging-drop vapour diffusion method was used for crystallization. A volume of 1.0 μl of RNA was mixed 1:1 with well solution comprising 3.5 M Na formate, 0.1 M Na acetate (pH 4.6) at 20 °C. Crystals (approximate dimensions 150 × 20 × 20 μm3) with space group P6322 grew in a few days. Crystals were briefly washed in well solution supplemented with 30% glycerol. The crystals were flash frozen by mounting in nylon loops and plunging into liquid nitrogen. A 2.0 Å resolution data set was collected on beamline I03 of the Diamond Light Source (Harwell, UK). The resolution cutoff for the data was determined by examining both CC1/2 and difference map of the magnesium ions, as described previously30,31. The structure was determined by molecular replacement. H. marismortui Kt-7 (PDB 4C40) was used as the search model using the program PHASER32. The remaining ligands and waters were added to the model on the basis of inspection of electron density difference maps.
Structural models were built in Coot33 and RCrane34. The structure was refined with Refmac5 (ref. 35) from the CCP4 suite of programs36 and Phenix refine37. Model geometry and the fit to electron-density maps were monitored with MOLPROBITY38 and the validation tools in COOT.
Author contributions
S.A.M. performed spectroscopy, biochemistry, bioinformatics and synthesis; L.H. performed crystallography; S.A.M., L.H. and D.M.J.L. devised experiments, analysed the data and wrote the paper.
Additional information
How to cite this article: McPhee, S. A. et al. A critical base pair in k-turns that confers folding characteristics and correlates with biological function. Nat. Commun. 5:5127 doi: 10.1038/ncomms6127 (2014).
Accession codes: Atomic coordinates and structure factor amplitudes have been deposited with the PDB with accession code 4CS1.
Supplementary Material
Supplementary Figures 1-6 and Supplementary Tables 1
Acknowledgments
We thank Diamond Light Source for beam time (proposal mx7705 and 8268), and the staff of beamlines I02, I03, I04 and I24, Dr Tim Wilson for discussion, and Cancer Research UK and the Wellcome Trust for financial support.
References
- Klein D. J., Schmeing T. M., Moore P. B. & Steitz T. A. The kink-turn: a new RNA secondary structure motif. EMBO J. 20, 4214–4221 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic I., Nottrott S., Hartmuth K., Luhrmann R. & Ficner R. Crystal structure of the spliceosomal 15.5 kD protein bound to a U4 snRNA fragment. Molec. Cell 6, 1331–1342 (2000). [DOI] [PubMed] [Google Scholar]
- Moore T., Zhang Y., Fenley M. O. & Li H. Molecular basis of box C/D RNA-protein Interactions; Cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure 12, 807–818 (2004). [DOI] [PubMed] [Google Scholar]
- Hamma T. & Ferré-D'Amaré A. R. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure 12, 893–903 (2004). [DOI] [PubMed] [Google Scholar]
- Li L. & Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443, 302–307 (2006). [DOI] [PubMed] [Google Scholar]
- Montange R. K. & Batey R. T. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441, 1172–1175 (2006). [DOI] [PubMed] [Google Scholar]
- Lescoute A., Leontis N. B., Massire C. & Westhof E. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 33, 2395–2409 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. & Lilley D. M. J. The role of specific 2'-hydroxyl groups in the stabilization of the folded conformation of kink-turn RNA. RNA 13, 200–210 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daldrop P. & Lilley D. M. J. The plasticity of a structural motif in RNA: structural polymorphism of a kink turn as a function of its environment. RNA 19, 357–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody T. A., Melcher S. E., Norman D. G. & Lilley D. M. J. The kink-turn motif in RNA is dimorphic, and metal ion dependent. RNA 10, 254–264 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder K. T., Daldrop P. & Lilley D. M. J. RNA tertiary interactions in a riboswitch stabilize the structure of a kink turn. Structure 19, 1233–1240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B., Melcher S. E., Wilson T. J., Norman D. G. & Lilley D. M. J. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA 11, 1192–1200 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B. & Lilley D. M. J. The importance of G.A hydrogen bonding in the metal ion- and protein-induced folding of a kink turn RNA. J. Molec. Biol. 381, 431–442 (2008). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. Single-molecule observation of the induction of k-turn RNA structure on binding L7Ae protein. Biophys. J. 103, 2541–2548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Bork P. & Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 22, 2166–2167 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N., Nissen P., Hansen J., Moore P. B. & Steitz T. A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000). [DOI] [PubMed] [Google Scholar]
- Nottrott S. et al. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5' stem-loop of U4 snRNA. EMBO J. 18, 6119–6133 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J. F., Tran E. J. & Maxwell E. S. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 30, 931–941 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N. J., Dickmanns A. & Lührmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22, 8342–8352 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S. W. et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 41, D226–D232 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird N. J. & Ferre-D'Amare A. R. Modulation of quaternary structure and enhancement of ligand binding by the K-turn of tandem glycine riboswitches. RNA 19, 167–176 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin S. & Lafontaine D. A. A loop loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 13, 1256–1267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peselis A. & Serganov A. Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch. Nat. Struct. Molec. Biol. 19, 1182–1184 (2012). [DOI] [PubMed] [Google Scholar]
- Huang L. & Lilley D. M. J. The molecular recognition of kink turn structure by the L7Ae class of proteins. RNA 19, 1703–1710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Daldrop P., Huang L. & Lilley D. M. J. The k-junction motif in RNA structure. Nucleic Acids Res. 42, 5322–5331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaucage S. L. & Caruthers M. H. Deoxynucleoside phosphoramidites-a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 22, 1859–1862 (1981). [Google Scholar]
- Wilson T. J., Zhao Z.-Y., Maxwell K., Kontogiannis L. & Lilley D. M. J. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry 40, 2291–2302 (2001). [DOI] [PubMed] [Google Scholar]
- Huang L., Yin P., Zhu X., Zhang Y. & Ye K. Crystal structure and centromere binding of the plasmid segregation protein ParB from pCXC100. Nucleic Acids Res. 39, 2954–2968 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg R. M. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 211, 353–388 (1992). [DOI] [PubMed] [Google Scholar]
- Karplus P. A. & Diederichs K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Rajashankar K. & Dauter Z. Weak data do not make a free lunch, only a cheap meal. Acta Crystallogr. D Biol. Crystallogr. 70, 253–260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating K. S. & Pyle A. M. RCrane: semi-automated RNA model building. Acta Crystallogr. D Biol. Crystallogr. 68, 985–995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-6 and Supplementary Tables 1