Abstract
We have characterized the organization, complexity, and expression of the porcine (Sus scrofa domestica) immunoglobulin lambda (IGL) light chain locus, which accounts for about half of antibody light chain usage in swine, yet is nearly totally unknown. Twenty-two IGL variable (IGLV) genes were identified that belong to seven subgroups. Nine genes appear to be functional. Eight possess stop codons, frameshifts, or both, and one is missing the V-EXON. Two additional genes are missing an essential cysteine residue and are classified as ORF (open reading frame). The IGLV genes are organized in two distinct clusters, a constant (C)-proximal cluster dominated by genes similar to the human IGLV3 subgroup, and a C-distal cluster dominated by genes most similar to the human IGLV8 and IGLV5 subgroups. Phylogenetic analysis reveals that the porcine IGLV8 subgroup genes have recently expanded, suggesting a particularly effective role in immunity to porcine-specific pathogens. Moreover, expression of IGLV genes is nearly exclusively restricted to the IGLV3 and IGLV8 genes. The constant locus comprises three tandem cassettes comprised of a joining (IGLJ) gene and a constant (IGLC) gene, whereas a fourth downstream IGLJ gene has no corresponding associated IGLC gene. Comparison of individual BACs generated from the same individual revealed polymorphisms in IGLC2 and several IGLV genes, indicating that allelic variation in IGLV further expands the porcine antibody light chain repertoire.
Keywords: Sus scrofa, porcine, light chain, lambda, IMGT, gene rearrangement
Introduction
Adaptive humoral immunity is mediated by a diverse array of genes which recombine during B cell development to generate a vast repertoire of antigen-binding immunoglobulin (IG) proteins. Antigen binding is carried out using the variable domains of both a heavy chain and a light chain. The antibody variable domain is encoded by a variable (V) gene, a diversity (D) gene (heavy chain-only) and a joining (J) gene which rearrange via recognition and cleavage of flanking recombination signal (RS) sequences by the RAG1 and RAG2 complex and subsequent double strand break repair whereby the intervening sequence is excised from the genome (McBlane et al. 1995; Kim and Oettinger 2000). Three complementarity determining regions (CDR) within the variable domain of each heavy and light chain exhibit a large degree of sequence variability. These CDR provide antibodies with an incredibly diverse antigen-binding repertoire (Wu and Kabat 1970; Lefranc and Lefranc 2001).
A large body of knowledge concerning the pig (Sus scrofa domestica) IG heavy (IGH) locus has been accumulated. Its germline organization on chromosome 7q25–q26 (Yerle et al. 1997) of the constant (IGHC) genes and the 15 C-proximal V (IGHV) genes have been mapped, and its expression has been characterized (Eguchi-Ogawa et al. 2010). However, the two light chain loci, kappa (IGK) and lambda (IGL), on chromosomes 3q12–q14 and 14q16–q21, respectively (Yerle et al. 1997), are only partially characterized in the vicinity of their constant genes, and no information is available on the organization of their variable genes (Butler et al. 2006). A search of GenBank and IMGT/LIGM-DB (Giudicelli et al. 2006) reveals the presence of approximately 100 cDNA sequences for the porcine kappa locus and only two partial lambda cDNA sequences, thus highlighting the lack of information on swine light chain loci. Here, we interrogated available porcine genome sequence (Schook et al. 2005; Humphray et al. 2007; Archibald et al. 2010), as well as additional sequence generated by our lab and available in GenBank and IMGT/LIGM-DB to completely characterize the organization, complexity, and expression of the porcine light chain lambda (IGL) locus. Our findings show that the locus organization is typical of mammalian species, but is substantially modified so that expression is dominated by V genes that have undergone a recent expansion.
Materials and methods
Identification and sequencing of bacterial artificial chromosomes
The Sus scrofa genome build 9 was queried to identify bacterial artificial chromosomes (BACs) containing IGLV sequences using the Basic Local Alignment Search Tool (BLAST) within the Ensembl database (Altschul et al. 1990; Hubbard et al. 2002). Nucleotide sequences in the region of interest for each BAC were downloaded from GenBank for further analysis. The CHORI (Children’s Hospital Oakland Research Institute)-242 BAC library used was derived from a single Duroc sow (http://bacpac.chori.org/porcine242.htm). Four BACs were identified that span the IGL locus (CH242-141B5, CH242-524K4, CH242-158O5, CH242-288F14) and two additional BAC clones (CH242-298L14 and CH242-82N3) were found to overlap and extend upstream of the locus (Fig. 1a). Of these BACs, three represent completely sequenced contigs (CH242-158O5, CH242-298L14, and CH242-82N3) and the remaining three are only partially sequenced. CH242-288F14 is fragmented into five contigs; however, its content almost completely overlaps CH242-158O5 and CH242-298L14. Additionally, CH242-141B5 is fragmented into seven contigs and CH242-524K4 is fragmented into 19 contigs (not shown). Due to a number of gaps in the overlap between these two BACs, it was uncertain if the entire IGL locus was represented. Thus, the two BAC clones were re-sequenced after overnight growth and BAC DNA purification using the Qiagen Plasmid Midi Prep with Qiagen-tip 500 columns. Purified DNA was submitted to the University of Minnesota Biomedical Genomics Center for library preparation and paired-end sequencing using the Illumina GAIIx platform.
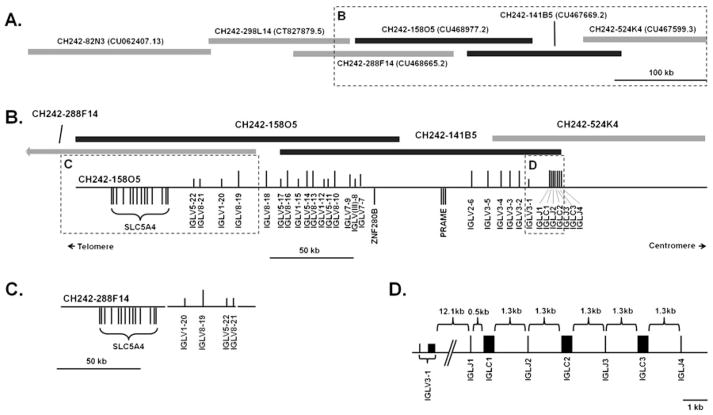
Fig. 1.
Organization of the porcine (Sus scrofa domestica) immunoglobulin lambda (IGL) locus. a BACs used to determine locus organization. Grey and black bars represent allelic variation. b IGL locus gene organization. Genes are displayed along a scaffold line as vertical bars representing functional genes (long bars), pseudogenes (short bars), or ORF (intermediate bars). c Rearranged gene order in IGLV1-19 to IGVL1-22 in BAC CH242-288F14. d Organization of the IGL J–C locus. The non-Ig genes ZNF280B, PRAME, and SLC5A4 are shown for genomic context
Characterization of the porcine IG lambda locus
Approximately 20 million high-quality reads were sorted by molecular tag to differentiate samples and assembled using a combination of the software programs ABySS and Velvet (Simpson et al. 2009; Zerbino and Birney 2008). Generated contigs were assembled against the existing BAC sequences from GenBank using Sequencher 4.10.1 (Gene Codes Corporation). The insert of CH242-141B5 retained a 651-bp gap in a highly repetitive region of the IGLC locus and CH242-524K4 retained several gaps, primarily in the IGLC locus and further downstream. The entire J–C region was re-sequenced using primer walking, PCR, and chain-termination sequencing to resolve the gaps. It included using primers specific for each IGLJ paired with either a reverse IGLJ-specific primer or with a conserved IGLC primer for PCR amplification and chain-termination sequencing. The re-sequenced portion encompassed the entire constant region and the first six IGLV genes. All IGLV genes identified by next generation re-sequencing were present on previously shotgun-sequenced contigs. The complete locus was manually annotated and interrogated for immunoglobulin features such as RS (i.e., heptamers and nonamers), promoters (i.e., octamers), and gene structure using the annotation software Artemis (Rutherford et al. 2000).
The sequences of CH242-141B5, CH242-524K4, CH242-158O5, CH242-288F14, CH242-298L14, and CH242-82N3 were acquired from GenBank (accession numbers: CU467669, CU467599, CU468977, CU468665, CT827879, and CU062407, respectively) and assessed for IGLV, IGLJ, and IGLC genes using BLAST. Phylogenetic analyses were performed in CLC Sequence Viewer (CLC Bio) and Dendroscope (Huson et al. 2007) using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) with 1,000 bootstrap iterations. Genes were annotated according to IMGT®, the international ImMunoGeneTics information system® (Lefranc et al. 2009) (http://www.imgt.org). Translated amino acid sequences of the IGLV genes were compared and CDR and framework (FR) boundaries were annotated according to IMGT unique numbering for V region and V domain (Lefranc et al. 2003). IGLC gene translations were annotated according to IMGT unique numbering for C domain (Lefranc et al. 2005). Expression of germline IGLV genes was deduced using 116 BLAST hits from 398,837 porcine expressed sequence tags (ESTs) obtained from GenBank and deposited at: http://pigest.ku.dk/index.html (Gorodkin et al. 2007) using an E-value threshold of 10−12 and ≥98% identity.
Nomenclature
The porcine IGLV genes are named according to IMGT nomenclature (Lefranc and Lefranc 2001; Lefranc 2007, 2008), using human V gene subgroup nomenclature to maintain consistency with porcine heavy chain, light chain, and cattle lambda light chain nomenclature (Eguchi-Ogawa et al. 2010; Butler et al. 2005, 2006; Pasman et al. 2010). The porcine IGLV, IGLJ and IGLC genes were submitted to IMGT/GENE-DB (Giudicelli et al. 2005). BLAST at NCBI was used to organize IGLV genes into subgroups using a 75% identity threshold according to IMGT criteria (Lefranc and Lefranc 2001; Lefranc 2007, 2008). Genes were deemed pseudogenes if they contained a truncation, stop codon, frameshift, or a defective initiation codon. Additionally, genes were described as ORF (open reading frame) if they were missing one (or more) key amino acids (1st-CYS 23, CONSERVED-TRP 41, hydrophobic 89, and 2nd-CYS 104). RS were deemed non-canonical if the heptamer was anything other than “ CACAGTG” for V-HEPTAMER (or “CACTGTG” for J-HEPTAMER) or if the nonamer contained at least two nucleotides which were each present in less than 10% of all RS described by Ramsden et al. (1994).
Results
Organization of the porcine immunoglobulin lambda locus
The porcine IGL locus spans approximately 229 kb on chromosome 14 (Fig. 1). The IGLV locus is organized in two distinct clusters containing 22 IGLV genes, fewer as compared to humans and cattle, which have three clusters containing a total of 52 and 25 IGLV genes, respectively (Frippiat et al. 1995; Pasman et al. 2010). The C-proximal cluster contains six IGLV genes, of which one is a pseudogene. The C-distal cluster, approximately 65.1 kb upstream, contains 16 IGLV genes, of which eight are pseudogenes. Two of the pseudogenes possess premature stop codons (IGLV3-1 and IGLV8-21), four have mutated start sites (IGLV(III)-8, IGLV1-15, IGLV1-20, and IGLV5-22), five are frameshifted (IGLV(III)-8, IGLV5-11, IGLV5-17, IGLV1-20, and IGLV5-22), and one (IGLV1-12) is missing the V-EXON (Table 1). In addition, multiple codons are deleted from both IGLV3-1 and IGLV5-11. Two additional genes are classified as ORF (IGLV7-7 and IGLV7-9) since they lack the highly conserved cysteine residue at IMGT position 104 (2nd-CYS), which is critical for intrachain disulfide bond formation with the cysteine at position 23 (1st-CYS) and are therefore unlikely to be functional (Tables 1 and 2) (Lefranc et al. 2003; Bergman and Kuehl 1979). IGLV8-16 contains an insertion within CDR3, creating a premature stop codon at the 3′ end of the gene. However, due to exonuclease and terminal deoxynucleotidyl transferase (TdT) activity in the V–J junction during recombination, it is plausible for this stop codon to be altered and generate a functional V region. The intervening region between the IGLV clusters contains the genes PRAME and ZNF280B, syntenic with cattle (Pasman et al. 2010).
Table 1.
Porcine (Sus scrofa domestica) IGLV subgroups, gene and allele names (allele functionality, clone names and accession numbers are provided)
| IMGT subgroup | IMGT gene name | IMGT allele name | Fct | Clone names | Clone accession numbers |
|---|---|---|---|---|---|
| IGLV1 | IGLV1-12 | IGLV1-12*01 | Pa | CH242-158O5 | CU468977.2 |
| CH242-141B5 | CU467669.2 | ||||
| IGLV1-15 | IGLV1-15*01 | Pb | CH242-158O5 | CU468977.2 | |
| CH242-141B5 | CU467669.2 | ||||
| IGLV1-20 | IGLV1-20*01 | Pc | CH242-158O5 | CU468977.2 | |
| IGLV1-20*02 | Pc | CH242-141B5 | CU467669.2 | ||
| IGLV2 | IGLV2-6 | IGLV2-6*01 | F | CH242-141B5 | CU467669.2 |
| IGLV3 | IGLV3-1 | IGLV3-1*01 | Pd | CH242-141B5 | CU467669.2 |
| IGLV3-1*02 | Pd | CH242-524K4 | CU467599.3 | ||
| IGLV3-2 | IGLV3-2*01 | F | CH242-141B5 | CU467669.2 | |
| IGLV3-2*02 | F | CH242-524K4 | CU467599.3 | ||
| IGLV3-3 | IGLV3-3*01 | F | CH242-141B5 | CU467669.2 | |
| IGLV3-3*02 | F | CH242-524K4 | CU467599.3 | ||
| IGLV3-4 | IGLV3-4*01 | F | CH242-141B5 | CU467669.2 | |
| IGLV3-4*02 | F | CH242-524K4 | CU467599.3 | ||
| IGLV3-5 | IGLV3-5*01 | F | CH242-141B5 | CU467669.2 | |
| IGLV5 | IGLV5-11 | IGLV5-11*01 | Pe | CH242-158O5 | CU468977.2 |
| CH242-141B5 | CU467669.2 | ||||
| IGLV5-14 | IGLV5-14*01 | F | CH242-158O5 | CU468977.2 | |
| CH242-141B5 | CU467669.2 | ||||
| IGLV5-17 | IGLV5-17*01 | Pf | CH242-158O5 | CU468977.2 | |
| CH242-141B5 | CU467669.2 | ||||
| IGLV5-22 | IGLV5-22*01 | Pg | CH242-158O5 | CU468977.2 | |
| IGLV5-22*02 | Pg | CH242-288F14 | CU468665.2 | ||
| IGLV7 | IGLV7-7 | IGLV7-7*01 | ORFh | CH242-158O5 | CU468977.2 |
| CH242-141B5 | CU467669.2 | ||||
| IGLV7-9 | IGLV7-9*01 | ORFi | CH242-158O5 | CU468977.2 | |
| CH242-141B5 | CU467669.2 | ||||
| IGLV8 | IGLV8-10 | IGLV8-10*01 | F | CH242-158O5 | CU468977.2 |
| CH242-141B5 | CU467669.2 | ||||
| IGLV8-13 | IGLV8-13*01 | F | CH242-158O5 | CU468977.2 | |
| CH242-141B5 | CU467669.2 | ||||
| IGLV8-16 | IGLV8-16*01 | F | CH242-158O5 | CU468977.2 | |
| CH242-141B5 | CU467669.2 | ||||
| IGLV8-18 | IGLV8-18*01 | F | CH242-158O5 | CU468977.2 | |
| IGLV8-19 | IGLV8-19*01 | F | CH242-158O5 | CU468977.2 | |
| IGLV8-19*02 | F | CH242-288F14 | CU468665.2 | ||
| IGLV8-21 | IGLV8-21*01 | Pj | CH242-158O5 | CU468977.2 | |
| IGLV8-21*02 | Pj | CH242-288F14 | CU468665.2 | ||
| IGLV(III) | IGLV(III)-8 | IGLV(III)-8*01 | Pk | CH242-158O5 | CU468977.2 |
Fct functionality, F functional, P pseudogene, ORF open reading frame
V-EXON is missing
INIT-CODON replaced by Val (ATG>GTG)
INIT-CODON replaced by Arg (ATG>CGG); frameshift at position 98; 2nd-CYS replaced by Val; STOP-CODON at pos. 105
STOP-CODON in L-PART1, codons 80–87 are deleted
14 nt deletion and frameshift from codons 2–6, frameshift at position 45
Frameshift at position 57
L-PART1 and V-EXON are fused, INIT-CODON is replaced by Leu (ATG>CTG), frameshift in FR3-IMGT
2nd-CYS replaced by Tyr
2nd-CYS replaced by Ser
STOP-CODON in L-PART1
INIT-CODON replaced by Lys (ATG>AAG); frameshift in FR1-IMGT
Table 2.
IMGT protein display of in-frame porcine (Sus scrofa domestica) IGLV genes
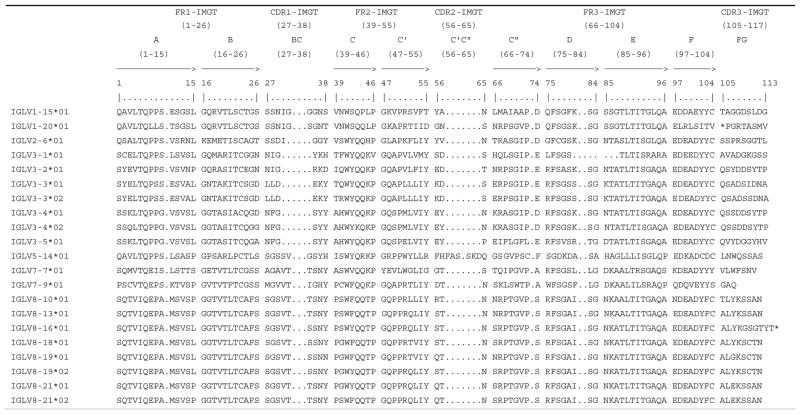
|
The protein display is shown using IMGT header (IMGT Repertoire, http://www.imgt.org) and IMGT unique numbering for V region and V domain (Lefranc et al. 2003)
The presence of additional IGLV clusters farther upstream was ruled out by analyzing contiguous, overlapping sequence represented by the fully sequenced BACs CH242-298L14 and CH242-82N3 (Fig. 1a). These BACs extend approximately 445 kb upstream from the most upstream IGLV gene, IGLV5-22. Flanking the IGL locus upstream from IGLV5-22 on these BACs are the genes SLC5A4, SLC5A1, YWHAH, and DEPCD5 (ordered from IGL-proximal to distal), syntenic with the cattle upstream flanking genes (UCSC Genome Browser, assembly: bos_taurus_UMD_3.1/bosTau6). This effectively rules out the possibility of additional upstream IGLV clusters and makes the germline porcine IGL locus more compact than that of cattle.
In contrast to the mouse IGL locus which is organized in tandem V–J–C cassettes (Sanchez et al. 1991), the organization of the porcine IGL locus is similar to most other mammals having tandem J–C cassettes downstream of the IGLV genes (Fig. 1d). Approximately 91 kb of sequence on CH242-524K4 lies downstream of IGLJ4, the most 3′ IGLJ gene. However, no corresponding IGLC gene was identified, based on both chain-termination and next generation sequencing. Likewise, PCR failed to amplify a product when using an IGLJ4-specific forward primer when paired with either of two conserved constant region reverse primers, despite positive results with the other IGLJ-specific primers (data not shown). The canonical amino acid motif W/F-G-X-G was present in the IGLJ genes save IGLJ1, which also has a non-canonical RS, indicating that swine only possess two functional J–C cassettes: IGLJ2–IGLC2 and IGLJ3–IGLC3 (Table 3).
Table 3.
Porcine (Sus scrofa domestica) IGLJ nucleotide (nt) and amino acid (AA) sequences and associated J-RS
| IGLV gene | Fct | J-NONAMER | J-SPACER | J-HEPTAMER | J-REGION nt and AA sequences |
|---|---|---|---|---|---|
| IGLJ1 | P | GGGTTTTGT | TCGAGCCTCAGT | CAGCGTG |
|
| IGLJ2 | F | GGTTTATGT | TTGAGGCTGTAT | CACTGTG |
|
| IGLJ3 | F | GGTTTATGT | TTGAGGCTGTAT | CACTGTG |
|
| IGLJ4 | F | GGTTTATGT | TTGAGGCTGTAT | CACTGTG |
|
Fct functionality, F functional, P pseudogene
Phylogenetic analysis of IGLV genes
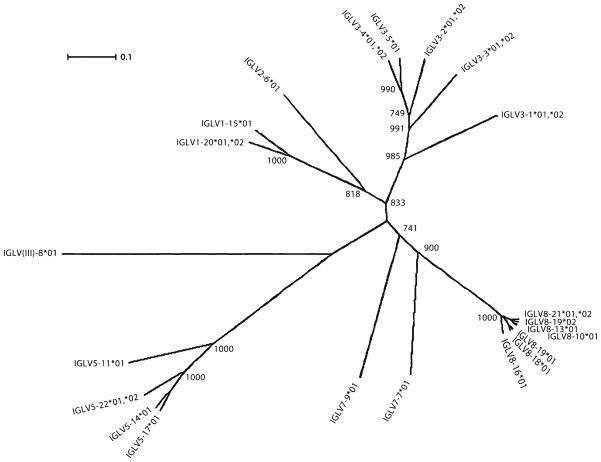
The C-proximal IGLV cluster contains five genes (four functional and one pseudogene) all belonging to either the IGLV3 or IGLV2 subgroups. The second cluster is comprised of 16 genes, six of which belong to the IGLV8 subgroup, four belong to IGLV5, three belong to IGLV1, two belong to IGLV7, and one, which most closely resembles members of IGLV clan III yet is distinct from any defined subgroup. This organization differs from cattle, which possess four IGLV2 subgroup genes and four IGLV3 subgroup genes in the most C-proximal IGLV cluster and exclusively contain IGLV1 genes in their second and third IGLV clusters (Pasman et al. 2010). Interestingly, IGLV8 subgroup genes are at least 92.4% identical to each other compared to IGLV1, IGLV3, and IGLV5 genes which are only 84.5%, 74.2%, and 79.5% identical, respectively. This suggests that porcine IGLV8 genes result from a recent expansion by gene duplication (Fig. 2).
Fig. 2.
Phylogenetic analysis of porcine (Sus scrofa domestica) IGLV nucleotide sequences using UPGMA with 1,000 bootstrap iterations. Genes comprising the most highly expressed subgroup, IGLV8, are very tightly clustered, suggestive of a relatively recent expansion. Nodes are labeled with bootstrap values
Allelic variation
BACs at the 5′ and 3′ ends of the IGLV locus overlap with approximately 99% identity. All eight IGLV genes in these overlaps differ by at least a single nucleotide (Table 1). IGLV3-3, IGLV3-4, IGLV8-19, and IGLV8-21 differ by two, four, six, and eight amino acids between alleles, respectively (Table 2). A gene order polymorphism was found in the second IGLV cluster at the 5′ end of the locus, where the last four IGLV genes have a different order (Fig. 1b and c). However, it is not certain if it is real or is an artifact due to whole genome shotgun sequencing and assembly error.
The CH242-141B5 insert terminates downstream of IGLJ3 and is therefore missing IGLC3 and IGLJ4. There are no nucleotide substitutions in any of the first three IGLJ genes or their respective RS between BACs. Sequence analysis revealed, however, that IGLC2 on CH242-524K4 contains a non-synonymous SNP resulting in an A1>T (using IMGT numbering) amino acid substitution. Other than this polymorphism, all of the constant region exons are identical (Table 4).
Table 4.
MGT protein display of porcine (Sus scrofa domestica) IGLC genes
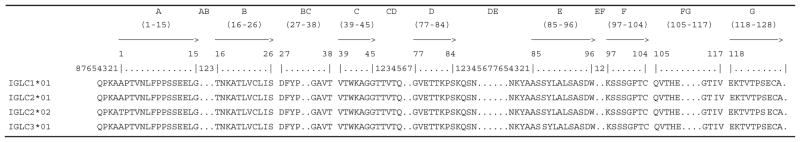
|
The protein display is shown using IMGT header (IMGT Repertoire, http://www.imgt.org) and IMGT unique numbering for C domain (Lefranc et al. 2005)
Expression of IGLV genes
Only IGLV3 and IGLV8 subgroup genes are predicted to be expressed based on functionality and RS (Table 5), in agreement with a previous report (Butler et al. 2006). BLAST analysis of a porcine EST database revealed that all functional IGLV8 subgroup genes are expressed, with IGLV8-13 and IGLV8-18 expressed most abundantly, but only two IGLV3 subgroup genes were expressed (Fig. 3). In contrast, IGLV3 family members were reported to dominate the expressed pre-immune lambda repertoire in neonatal pigs (Butler et al. 2006). Additionally, low level expression of IGLV5-14 was observed despite the presence of non-canonical heptamer and nonamer sequences (Table 5).
Table 5.
Genomic features of the porcine (Sus scrofa domestica) IGLV genes
| IGLV gene | Fct | Octamer (promoter) | nt | Ini. | L-PART1 (nt) | 5′ Splice site | Intron (nt) | 3′ Splice site | V-EXON (nt) | V-HEPTAMER | V-SPACER (nt) | V-NONAMER |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IGLV3-1*01 | P | ATTTGCAT | 96 | ATG | 46 | GT | 358 | AG | 282 | CCCAGTG | 37 | ACACAAACT |
| IGLV3-2*01 | F | ATTTGCAT | 96 | ATG | 46 | GT | 370 | AG | 299 | CACAGTG | 23 | ACACAAACT |
| IGLV3-2*02 | F | ATTTGCAT | 96 | ATG | 46 | GT | 363 | AG | 299 | CACAGTG | 17 | ACACAAACT |
| IGLV3-3*01 | F | ATTTGCAT | 106 | ATG | 46 | GT | 143 | AG | 299 | CACAGTG | 23 | ACACAAACC |
| IGLV3-4*01 | F | ATTTGCAT | 82 | ATG | 46 | GT | 151 | AG | 299 | CACAGTG | 23 | ACACAAACC |
| IGLV3-5*01 | F | ATTTGCAT | 82 | ATG | 46 | GT | 151 | AG | 299 | CACAGTG | 23 | ACACAAACT |
| IGLV3-5*02 | F | ATTTGCAT | 82 | ATG | 46 | GT | 151 | AG | 299 | CACAGTG | 23 | ACACAAACC |
| IGLV2-6*01 | F | ATTTGTAT | 101 | ATG | 46 | GT | 116 | AG | 302 | TACAGTG | 23 | ACACAAACC |
| IGLV7-7*01 | ORF | ATCTGCAT | 101 | ATG | 46 | GT | 97 | AG | 301 | CACCGTG | 23 | ACATGAGCC |
| IGLV(III)-8 | P | ATTTGCAT | 93 | AAG | 47 | GT | 111 | AG | 267 | CACAGTG | 21 | ACTCAAACC |
| IGLV7-9*01 | ORF | ATTTGCAT | 108 | ATG | 43 | GT | 85 | AG | 288 | CACAGTG | 23 | GACACAAAG |
| IGLV8-10*01 | F | ATTTGCAT | 106 | ATG | 46 | GT | 97 | AG | 304 | CACAGTG | 23 | ACCCAAACC |
| IGLV5-11*01 | P | ACTGGCAT | 87 | ATG | 46 | GT | 109 | TG | 303 | CACAATG | 23 | AAAAAACAT |
| IGLV1-12*01 | P | ATTTGCAT | 108 | ATG | 46 | GA | – | – | – | – | – | – |
| IGLV8-13*01 | F | ATTTGCAT | 106 | ATG | 46 | GT | 97 | AG | 304 | CACAGTG | 23 | ACCCAAACC |
| IGLV5-14*01 | F | ACTGGCAT | 88 | ATG | 46 | GT | 121 | AG | 323 | CACTGTG | 23 | AGAAGAATC |
| IGLV1-15*01 | P | ATTGGCAT | 107 | GTG | 46 | GT | 106 | AG | 310 | CACAGTG | 23 | ACCAAAACC |
| IGLV8-16*01 | F | ATTTGCAT | 106 | ATG | 46 | GT | 97 | AG | 312 | CACAGTG | 23 | ACCCAAACC |
| IGLV5-17*01 | P | ACTGGCAT | 87 | ATG | 46 | GT | 121 | AG | 302 | CACTGTA | 23 | TGGGAACGG |
| IGLV8-18*01 | F | ATTTGCAT | 106 | ATG | 46 | GT | 97 | AG | 304 | CACAGTG | 23 | ACTCAAACC |
| IGLV8-19*01 | F | ATTTGCAT | 106 | ATG | 46 | GT | 97 | AG | 304 | CACAGTG | 23 | ACCCAAACC |
| IGLV8-19*02 | F | ATTTGCAT | 105 | ATG | 46 | GT | 98 | AG | 304 | CACAGTG | 23 | ACCCAAACC |
| IGLV1-20*01 | P | ATTTGCAT | 88 | CGG | 46 | GT | 108 | AG | 309 | CACAGTG | 23 | ACCAAAACC |
| IGLV1-20*02 | P | ATTTGCAT | 91 | CGG | 46 | GT | 108 | AG | 309 | CACAGTG | 23 | ACCAAAACC |
| IGLV8-21*01 | P | ATTTGCAT | 106 | ATG | 46 | CC | 96 | AG | 304 | CACAGTG | 23 | ACCCAAACC |
| IGLV5-22*01 | P | ATTTGCAT | 103 | CTG | - | - | - | - | 354 | CACAGTG | 23 | AGAAGAATC |
IMGT standardized labels are written in capital letters
Fct functionality, F functional, P pseudogene, ORF open reading frame
Fig. 3.
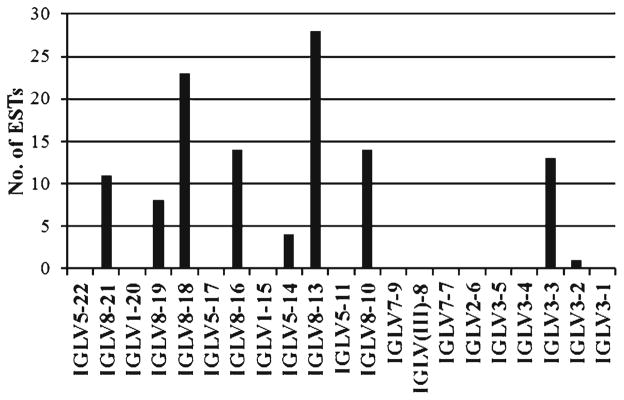
Expression of porcine (Sus scrofa domestica) IGLV genes. BLAST analysis revealed 116 hits to germline IGLV sequences from 398,837 expressed sequence tags in GenBank and deposited at http://pigest.ku.dk/index.html (Gorodkin et al. 2007). The x-axis is organized by gene order in the locus with the most 5′ (most C-distal) IGLV genes on the left. Genes belonging to the IGLV8 subgroup dominate the expression profile
A canonical octamer (ATTTGCAT) necessary for transcription was present upstream of all IGLV genes except for six (IGLV2-6, IGLV-7, IGLV5-11, IGLV5-14, IGLV1-15, and IGLV5-17). The octamer was located 106 bp upstream of the seven most highly expressed IGLV genes (six IGLV8 subgroup genes and IGLV3-3). The mean distance among all IGLV genes was 94 bp and ranged from 82 to 108 bp (Table 5), which is typical of mammals (Parslow et al. 1984).
Discussion
The porcine IGL locus organization is typical among mammalian species, containing multiple V genes followed by tandem J–C cassettes. Among described species, it is most similar in organization to, but is more compact than cattle, another member of the order Cetartiodactyla. The capacity for recombinatorial diversity is significantly limited since more than half of the V genes are pseudogenes or ORFs and one of four J genes lacks a paired C gene. The lack of IGLC4 downstream of IGLJ4 and a non-canonical heptamer and mutated W/F-G-X-G motif associated with IGLJ1 suggest that swine use only two J–C cassettes (IGLJ2–IGLC2 and IGLJ3–IGLC3). The only porcine lambda cDNA previously described is identical to IGLJ3 (Lammers et al. 1991).
Screening of EST databases indicates that the vast majority of expressed IGLV genes belong to the IGLV8 subgroup, followed by IGLV3 and IGLV5. The IGLV8 subgroup is the result of a recent expansion, presumably by gene duplication, suggesting that its products have provided a strong selective advantage. Our EST analysis differs from an earlier report which showed that the C-proximal IGLV3 is the most highly expressed IGLV subgroup in the porcine pre-immune repertoire (Butler et al. 2006). The preferential use of C-proximal IGLV2 and IGLV3 subgroup genes has been reported in humans (Frippiat et al. 1995). Similarly, C-proximal IGHV gene preference has been reported in mice (Yancopolous et al. 1984; Malynn et al. 1990), rabbits (Knight and Becker 1990), and humans (Berman et al. 1991). However, in mice, IGHV gene usage shifts upstream with animal age, possibly due to an increased presence of peripheral B cells bearing more distal genes as a result of environmental stimulation, since the number of pre-B cells possessing C-proximal rearrangements remains unchanged in the bone marrow of adult mice (Malynn et al. 1990; Jung et al. 2006). It is plausible that swine may similarly shift IGLV usage upstream with age and peripheral antigenic stimulation. In addition, several IGLV3 genes containing canonical RS were not detected in the EST database, due possibly to poor reactivity to exogenous antigen so that the B cells expressing these genes are rare, or the B cells may have been deleted due to reactivity to self-antigens.
The size of the expressed repertoire may be increased by allelic variation. Polymorphisms, comprising from two to eight amino acid differences, were identified in four of the eight IGLV genes that were present in overlapping BACs from the single animal used to construct the BAC library. Thus, allelic variation in swine V regions may significantly increase the complexity of the lambda repertoire.
The two most C-proximal human IGLV clusters are dominated by 34 IGLV genes largely belonging to IGLV2 and IGLV3. The reduced complexity of C-proximal IGLV in pigs compared to cattle suggests that this cluster has undergone contraction and expansion during the divergent evolution of these two species. The third human IGLV cluster is dominated by IGLV1, IGLV5, and IGLV7 which correspond to the swine IGLV genes of the distal cluster. IGLV8, however, only occurs as a single gene in the most distal region of the third human IGLV cluster, suggesting that significant gene order reshuffling as well as expansion and/or contraction has occurred specifically during suid evolution. Thus, the immunogenetic loci may represent ideal regions for identifying intra-species gene order and gene content polymorphisms.
Acknowledgments
We would like to thank Erin Babineau for excellent technical assistance, and Dr. Jane Loveland for generous review of the manuscript. This work was supported by a grant from the U.S. National Pork Board (10-139).
Contributor Information
John C. Schwartz, Email: schwa753@umn.edu, Department of Veterinary and Biomedical Sciences, University of Minnesota, 1971 Commonwealth Avenue, St. Paul, MN 55108, USA
Marie-Paule Lefranc, IMGT®, the international ImMunoGeneTics information system®, Laboratoire d’ImmunoGénétique Moléculaire, Institut de Génétique Humaine UPR CNRS 1142, Université Montpellier 2, Montpellier, France.
Michael P. Murtaugh, Department of Veterinary and Biomedical Sciences, University of Minnesota, 1971 Commonwealth Avenue, St. Paul, MN 55108, USA
References
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Archibald AL, Bolund L, Churcher C, Fredholm M, Groenen MAM, Harlizius B, Lee KT, Milan D, Rogers J, Rothschild MF, Uenishi H, Wang J, Schook LB the Swine Genome Sequencing Consortium. Pig genome sequence – analysis and publication strategy. BMC Genom. 2010;11:438. doi: 10.1186/1471-2164-11-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman LW, Kuehl WM. Formation of an intrachain disulfide bond on nascent immunoglobulin light chains. J Biol Chem. 1979;254:8869–8876. [PubMed] [Google Scholar]
- Berman JE, Nickerson KG, Pollock RR, Barth JE, Schuurman RK, Knowles DM, Chess L, Alt FW. VH usage in humans: biased usage of the VH6 gene in immature B lymphoid cells. Eur J Immunol. 1991;21:1311–1314. doi: 10.1002/eji.1830210532. [DOI] [PubMed] [Google Scholar]
- Butler JE, Wertz N, Sun J, Wang H, Lemke C, Chardon P, Piumi F, Wells K. The pre-immune variable kappa repertoire of swine is selectively generated from certain subfamilies of V κ2 and one Jκ gene. Vet Immunol Immunop. 2005;108:127–137. doi: 10.1016/j.vetimm.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Butler JE, Sun J, Wertz N, Sinkora M. Antibody repertoire development in swine. Dev Comp Immunol. 2006;30:199–221. doi: 10.1016/j.dci.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Eguchi-Ogawa T, Wertz N, Sun XZ, Puimi F, Uenishi H, Wells K, Chardon P, Tobin GJ, Butler JE. Antibody repertoire development in fetal and neonatal piglets: XI. The relationship of variable heavy chain gene usage and the genomic organization of the variable heavy chain locus. J Immunol. 2010;184:3734–3742. doi: 10.4049/jimmunol.0903616. [DOI] [PubMed] [Google Scholar]
- Frippiat JP, Williams SC, Tomlinson IM, Cook GP, Cherif D, Le Paslier D, Collins JE, Dunham I, Winter G, Lefranc M-P. Organization of the human immunoglobulin lambda light-chain locus on chromosome 22q11.2. Hum Mol Genet. 1995;4:983–991. doi: 10.1093/hmg/4.6.983. [DOI] [PubMed] [Google Scholar]
- Giudicelli V, Chaume D, Lefranc M-P. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V, Duroux P, Ginestoux C, Folch G, Jabado-Michaloud J, Chaume D, Lefranc M-P. IMGT/LIGM-DB, the IMGT® comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 2006;34:D781–D784. doi: 10.1093/nar/gkj088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodkin J, Cirera S, Hedegaard J, Gilchrist MJ, Panitz F, Jørgensen C, Scheibye-Knudsen K, Arvin T, Lumholdt S, Sawera M, Green T, Nielsen BJ, Havgaard JH, Rosenkilde C, Wang J, Li H, Li R, Liu B, Hu S, Dong W, Li W, Yu J, Wang J, Stærfeldt H, Wernersson R, Madsen LB, Thomsen B, Hornshøj H, Bujie Z, Wang X, Wang X, Bolund L, Brunak S, Yang H, Bendixen C, Fredholm M. Porcine transcriptome analysis based on 97 non-normalized cDNA libraries and assembly of 1,021,891 expressed sequence tags. Genome Biol. 2007;8:R45. doi: 10.1186/gb-2007-8-4-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Huminiecki L, Kasprzyk A, Lehvaslaiho H, Lijnzaad P, Melsopp C, Mongin E, Pettett R, Pocock M, Potter S, Rust A, Schmidt E, Searle S, Slater G, Smith J, Spooner W, Stabenau A, Stalker J, Stupka E, Ureta-Vidal A, Vastrik I, Clamp M. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphray SJ, Scott CE, Clark R, Marron B, Bender C, Camm N, Davis J, Jenks A, Noon A, Patel M, Sehra H, Yang F, Rogatcheva MB, Milan D, Chardon P, Rohrer G, Nonneman D, de Jong P, Meyers SN, Archibald A, Beever JE, Schook LB, Rogers J. A high utility map of the pig genome. Genome Biol. 2007;8:R139. doi: 10.1186/gb-2007-8-7-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinform. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Kim DR, Oettinger MA. V(D)J recombination: site-specific cleavage and repair. Mol Cells. 2000;10:367–374. [PubMed] [Google Scholar]
- Knight KL, Becker RS. Molecular basis of the allelic inheritance of rabbit immunoglobulin VH allotypes: implications for the generation of antibody diversity. Cell. 1990;60:963–970. doi: 10.1016/0092-8674(90)90344-e. [DOI] [PubMed] [Google Scholar]
- Lammers BM, Beaman KD, Kim YB. Sequence analysis of porcine immunoglobulin light chain cDNAs. Mol Immunol. 1991;28:877–880. doi: 10.1016/0161-5890(91)90051-k. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P. WHO-IUIS Nomenclature Subcommittee for Immunoglobulins and T cell receptors report. Immunogenetics. 2007;59:899–902. doi: 10.1007/s00251-007-0260-4. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P. WHO-IUIS Nomenclature Subcommittee for Immunoglobulins and T cell receptors report. Immunoglobulins and Tcell receptors report August 2007, 13th International Congress of Immunology, Rio de Janeiro, Brazil. Dev Comp Immunol. 2008;32:461–463. doi: 10.1016/j.dci.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P, Lefranc G. The immunoglobulin factsbook. Academic Press; London: 2001. pp. 1–458. [Google Scholar]
- Lefranc M-P, Pommié C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P, Pommié C, Kaas Q, Duprat E, Bosc N, Guiraudou D, Jean C, Ruiz M, Da Piédade I, Rouard M, Foulquier E, Thouvenin V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev Comp Immunol. 2005;29:185–203. doi: 10.1016/j.dci.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT®, the international ImMunoGeneTics information system®. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn BA, Yancopoulos GD, Barth JE, Bona CA, Alt FW. Biased expression of JH-proximal VH genes occurs in the newly generated repertoire of neonatal and adult mice. J Exp Med. 1990;171:843–859. doi: 10.1084/jem.171.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane JF, Van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- Parslow TG, Blair DL, Murphy WJ, Granner DK. Structure of the 5′ ends of immunoglobulin genes: a novel conserved sequence. Proc Natl Acad Sci U S A. 1984;81:2650–2654. doi: 10.1073/pnas.81.9.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman Y, Saini SS, Smith E, Kaushik AK. Organization and genomic complexity of bovine λ-light chain gene locus. Vet Immunol Immunop. 2010;135:306–313. doi: 10.1016/j.vetimm.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Baetz KA, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 1994;22:1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sanchez P, Nadel B, Cazenave PA. V lambda–J lambda rearrangements are restricted within a V–J–C recombination unit in the mouse. Eur J Immunol. 1991;21:907–911. doi: 10.1002/eji.1830210408. [DOI] [PubMed] [Google Scholar]
- Schook LB, Beever JE, Rogers J, Humphray S, Archibald A, Chardon P, Milan D, Rohrer G, Eversole K. Swine Genome Sequencing Consortium (SGSC): a strategic roadmap for sequencing the pig genome. Comp Funct Genom. 2005;6:251–255. doi: 10.1002/cfg.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Kabat EA. An analysis of the sequences of the variable regions of Bence Jones proteins and their myeloma light chains and their implications for antibody complementarity. J Exp Med. 1970;132:211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopolous GD, Desiderio SV, Paskind M, Kearney JF, Baltimore D, Alt FW. Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature. 1984;311:727–733. doi: 10.1038/311727a0. [DOI] [PubMed] [Google Scholar]
- Yerle M, Lahbib-Mansais Y, Pinton P, Robic A, Goureau A, Milan D, Gellin J. The cytogenetic map of the domestic pig (Sus scrofa domestica) Mamm Genom. 1997;8:592–607. doi: 10.1007/s003359900512. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]