Abstract
MicroRNAs (miRNAs) are small, non-coding RNAs that control protein expression. Aberrant miRNA expression has been linked to various human diseases, and thus miRNAs have been explored as diagnostic markers and therapeutic targets. Although it is challenging to target RNA with small molecules in general, there have been successful campaigns that have identified small molecule modulators of miRNA function by targeting various pathways. For example, small molecules that modulate transcription and target nuclease processing sites in miRNA precursors have been identified. Herein, we describe challenges in developing chemical probes that target miRNAs and highlight aspects of miRNA cellular biology elucidated by using small molecule chemical probes. We expect that this area will expand dramatically in the near future as strides are made to understand small molecule recognition of RNA from a fundamental perspective.
Introduction
RNA is essential for many cellular process, from translation [1] to gene regulation [2] to the production of metabolites [3] and viral replication and propagation [4,5]. Aberrant RNA function or expression is also causative of disease. Yet, small molecules that target RNA have been limited mainly to antibacterials that target the ribosome. These compounds serve as invaluable therapeutics and chemical probes that have elucidated the intricacies of translation [6]. There is clear potential for many RNAs, both human and viral, to be targeted with small molecules yet such investigations have been only sparsely reported. One key to advance this area is a fundamental understanding of selective small molecule-RNA recognition events.
Approaches have been developed to identify and design lead small molecules for both protein and DNA [7–9]. Rules have been established to target the DNA minor groove. The eponymous “Dervan Rules” have allowed for the facile design of small molecules that read out the hydrogen bond donor and acceptor patterns displayed by base pairs [10]. Transition state mimicry approaches can facilitate design of enzyme inhibitors; small molecule screening can be used to identify leads for other types of proteins. Substrate mimicry has been applied to RNA; riboswitches can be targeted with small molecules that mimic the metabolite that they bind.
RNA, however, is generally considered to be “undruggable” with small molecules [11,12]. This perception is due to a variety of factors, including: (i) there are limited data on the molecular recognition of RNA by small molecules that elicit a biological response; (ii) little is known about chemotypes that impart selective recognition of and affinity for RNA; and (iii) perhaps a false perception that all RNA secondary structures are redundant within the transcriptome, making RNA-selective targeting difficult if not impossible.
In this review, we describe the development of RNA-directed chemical probes and lead therapeutics that target microRNAs (miRNAs). Since their initial discovery by Ambrose and Ruvkin in Caenorhabditis elegans [13], miRNAs have been discovered in many kingdoms of life. Because of their essential roles, the development of chemical probes that selectively target a miRNA could be extraordinarily powerful. Coupled with RNA-seq, complex cellular networks could be mapped out by boosting or inhibiting miRNA activity.
MiRNA discovery and biogenesis
Subsequent to their discovery, thousands of miRNAs have been identified and annotated in an online database, miRBase [14]. As of July 2014, miRBase contains 30,424 miRNAs from 206 species [14]. Signature miRNA expression profiles have been associated with nearly every cellular process from development to human disease. Thus, there is a large interest in understanding their exact roles in cellular biology, to develop miRNA expression patterns as diagnostics, and to drug miRNAs as therapeutic targets.
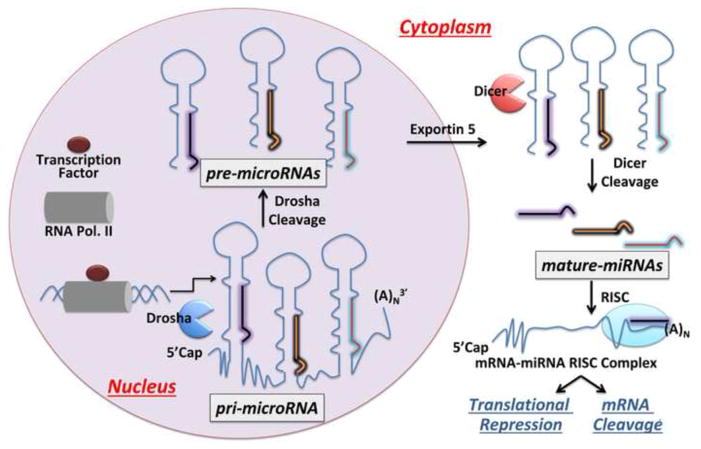
After their initial discovery, intense studies were undertaken to understand miRNA synthesis and biogenesis. MiRNAs are transcribed as precursors mainly by RNA polymerase II [15], however some viral miRNAs are transcribed by RNA polymerase III [16]. These precursor miRNAs fold into stem-loop structures encoding the mature miRNA that are capped with 7-methylguanosine at the 5′ end and polyadenylated at the 3′ end [17]. Often multiple miRNA precursors are transcribed on a single transcript, or a primary miRNA (pri-miRNA), that can be several kilobases in length [18]. Precursor miRNAs (including pri-miRNAs) are cleaved in the nucleus by the microprocessor complex between the nuclease Drosha and DiGeorge Syndrome Critical Region 8 protein (DGCR8) [19], affording pre-miRNA(s).
When a pre-miRNA is liberated, it is translocated to the cytoplasm by Exportin-5 (EXP5) and cleaved by the nuclease Dicer, which forms a complex with the Trans Activating Response RNA-binding protein (TRBP), into an miRNA duplex of 21–25 nucleotides in each strand [20]. The duplex, which contains both the active miRNA and a passenger strand, is loaded onto argonaute (AGO) protein to form the RNA-induced silencing complex (RISC) [20]. In most cases, the passenger strand is removed; however, in some instances both strands are involved in silencing [21,22]. The RISC complex binds to 3′ untranslated regions (UTRs) of mRNAs and either induces cleavage of the mRNA (full miRNA-mRNA complementarity) or translationally represses it (partial miRNA-mRNA duplex complementarity) [23].
Once a mature miRNA is produced, a variety of factors control the downstream mRNA it targets (single or multiple messages). The concentration of the miRNA plays a significant role in mediating gene regulation as well as the stability of the miRNA-mRNA duplex, with higher affinity duplexes active at lower concentrations. For instance, if the concentration of a miRNA is lower than the affinity of miRNA-mRNA complex, then its effect on the mRNA is generally considered negligible [24], as validated by Mullakandov and coworkers [25].
Aberrant miRNAs expression is associated with disease
In general, miRNA expression is tightly controlled. Both up- and down-regulation of miRNA expression can be directly attributed to disease [26,27]. Seed-targeting oligonucleotides and antagomirs that knock out miRNA activity and reverse phenotype convincingly show that miRNAs can be causative of disease [28,29].
The mechanisms by which miRNA expression is altered in disease are varied (Figure 1). Often these effects are at the transcriptional level in which there is an altered binding of transcription factors to gene promoters. Tumor protein p53 (p53), myelocytomatosis viral oncogene homolog (MYC) and Twist-related protein 1 (TWIST) have been shown to affect miRNA expression [30–33]. Regulation of transcription factors leads to double negative feedback loop mechanisms in turn controlling miRNA expression [34]. Further, miRNA expression can be regulated at the post-transcriptional level and aberrant regulation is associated with cancer [27]. In principle, each mechanism could be targeted for therapeutic applications (Figure 1). Below, we highlight specific interrogation of these pathways with small molecules to serve as lead therapeutics and chemical probes of miRNA function.
Figure 1.
Biogenesis of microRNAs. MiRNAs are transcribed by RNA Polymerase II into primary miRNA (pri-miRNA) transcripts that are 5′ capped and 3′ poly(A) tailed. The transcribed pri-miRNA is then processed into precursor miRNA (pre-miRNA) by Drosha and exported to the cytoplasm by Exportin-5. Dicer processes the exported pre-miRNA into the mature miRNA. The mature miRNA forms a complex with AGO2 to form the miRNA-mRNA RISC complex, which can either transitionally repress the mRNA or initiate its cleavage.
Targeting miRNA transcription factors
It is possible to modulate the expression of miRNAs using small molecules that either activate or repress their transcription (Figure 1). Deiters and co-workers inhibited transcription of miR-21 [35]. MiR-21 is frequently up-regulated in many cancers [36], resulting in down-regulation of pro-apoptotic proteins. Transcriptional inhibitors were identified by completing a small molecule screen in which a 3′ UTR complementary to miR-21 was inserted into a luciferase mRNA reporter [35]. This study identified a diazobenzene 1, which was optimized to afford 2, as miR-21 transcriptional inhibitors (Figure 2A). 2 inhibits expression of miR-21 in several cancer cell lines, not transcription globally, suggesting selective modulation.
Figure 2.
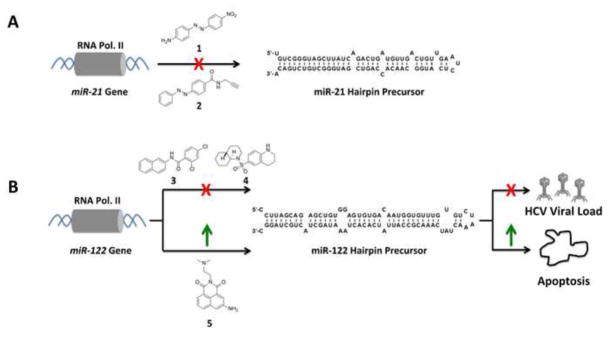
Targeting transcription of miRNAs. (A) Chemical structures of small molecules 1 and 2 that inhibit transcription of miR-21 and the secondary structure of precursor miR-21. (B) Chemical structures of small molecules 3, 4 (inhibitors) and 5 (activator) that affect the transcription of miR-122. The downstream effects of modulating miRNA-122 are also shown.
Small molecules were also discovered to modulate transcription of miR-122, a highly expressed, liver-specific miRNA whose aberrant expression is associated with hepatocellular carcinoma (HCC; down-regulated) [37] and Hepatitis C viral infections (HCV; up-regulated) [38]. Two small molecules that inhibit transcription (3 and 4, Figure 2B) and one that promotes transcription of pri-miR-122 (5, Figure 2B) were identified using a luciferase reporter system [39]. Compounds 3 and 4 reduced HCV viral load by ~50% at 10 μM. In contrast, 5, which is a transcriptional inducer of miR-122, boosts miR-122 levels in an HCC cell line and induces apoptosis at 3 μM. Thus, small molecules that target miRNA transcriptional factors could be developed into anti-cancer and anti-viral agents [39].
Targeting RNA biogenesis by inhibiting Dicer and Drosha processing
Many sites within RNAs can be bound by small molecules; however, it is likely that binding to only a percentage of these sites will have a functional consequence. Thus, exquisite selectivity would likely not be a pre-requisite for an RNA-targeting small molecule agent, provided binding to off-target sites are lower affinity and/or the sites are not functional (no biological consequence). In miRNA precursors, sites that are cleaved by the nucleases Drosha or Dicer are functional sites, the inhibition of which would decrease production of the mature miRNA and de-repress downstream mRNA targets (Figure 1).
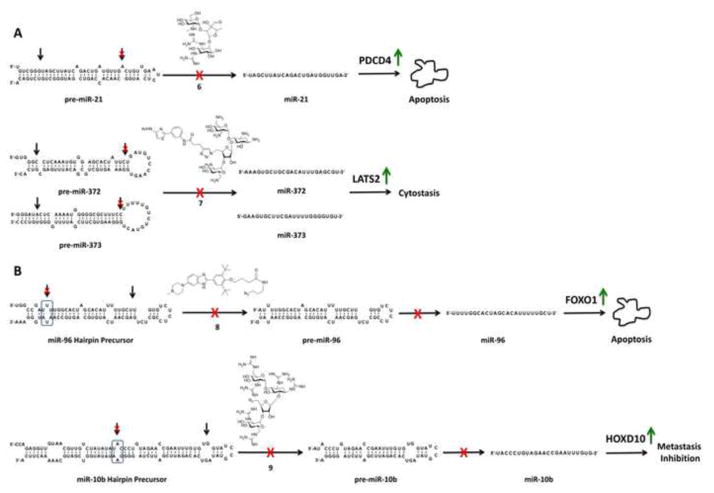
Maiti and co-workers showed that inhibition of miR-21 processing by Dicer is an effective strategy to affect miRNA biology [40]. Aminoglycoside antibiotics were screened to identify compounds that inhibit Dicer processing, which showed that streptomycin (6; Figure 3A) inhibits miR-21 maturation. In vitro experiments showed that streptomycin binds directly to pre-miR-21. Further, a luciferase reporter using the programmed cell death protein 4 (PDCD4) 3′ UTR, a known target of miR-21, showed induction of luciferase activity upon addition of streptomycin. These results were further supported by an Annexin V/propidium iodide assay, which showed induction of apoptosis in Jurkat (T cell leukemia) cells.
Figure 3.
Chemical structures of small molecules that inhibit Dicer and Drosha processing of precursor miRNAs and the corresponding miRNA secondary structures. (A) 6 and 7 inhibit Dicer processing. Inhibition of miR-21 processing by streptomycin up-regulates PDCD4, which induces apoptosis. Inhibition of miR-372 and -373 processing by 7 causes up-regulation of LATS2 inducing cytostatic effects. (B) 8 and 9 inhibit Drosha processing of miR-96 and miR-10b, respectively. Inhibition of miR-96 processing up-regulates FOXO1 thereby inducing apoptosis. Inhibition of miR-10b processing induces up-regulation of HOXD10, which can inhibit cell migration and metastasis. Drosha and Dicer processing sites are indicated with arrows.
Duca and co-workers identified small molecule inhibitors of the Dicer processing of pre-miR-372 and pre-miR-373, which are implicated in gastric cancer [41]. A variety of neomycin-nucleobase conjugates were screened to identify binders to the pre-miRNAs, affording 7 (Figure 3A). Compound 7 (25 μM) inhibits maturation of pre-miR-372 and pre-miR-373 and decreases production of large tumor suppressor homologue 2 (LATS2) protein, which induces cytostatic effects. Down-regulation of miR-17-5p was also observed when cells were treated with 7, which is likely due to structural similarities between the pre-miRNAs.
Recently, we described the development of rational design approach, named Inforna, to target miRNAs in a transcriptome-wide and target agnostic manner [42]. The basic premise of these studies was to leverage data about the RNA motifs that bind small molecules and cross-reference these data to the motifs found in Dicer and Drosha processing sites as reported in miRBase [14]. RNA motif-small molecule interactions were identified and annotated by using two methods developed in our laboratory, Two-Dimensional Combinatorial Screening (2DCS) [43] and Structure-Activity Relationships Through Sequencing (StARTS) [44]. In 2DCS, a small library of RNA motifs, which contains thousands of members, is probed for binding to a library of small molecules under highly stringent conditions. Thus, the optimal RNA motifs that bind small molecules are selected. StARTS is a statistical approach that predicts the fitness of RNA-small molecule interactions (based on affinity and selectivity). Taken together, Inforna integrates the results of 2DCS selections and their subsequent analysis by StARTS, thereby identifying the ideal RNA target for a given small molecule and making the approach target agnostic.
Inforna predicted lead small molecules for 26 miRNAs; ~45% of these interactions inhibited production of the desired mature miRNA in cell lines without lead optimization [42]. Most notable was the small molecule 8 that binds the Drosha site in the miR-96 hairpin precursor. MiR-96 is over-expressed in various cancers, in particular breast cancer [45,46]. Indeed, 8 inhibits maturation of miR-96 as determined by qRT-PCR of mature (90% at 40 μM), pre-, and pri-miRNA expression levels in MCF7 cells. Further, inhibition of miR-96 de-represses its downstream target, forkhead box protein O1 (FOXO1), and promotes apoptosis. siRNA knockdown of FOXO1 confirmed that 8’s mode of action was via the miR-96-FOXO1 circuit. These studies revealed other interesting aspects of miRNA biology. For example, FOXO1 is a target of miR-96, miR-182, and miR-27a [46]. However, our studies showed that inhibition of miR-96 biogenesis is sufficient to promote apoptosis [42].
Inforna predicted other sites in miRNA precursors that could bind 8 including the Dicer sites in miR-433, -301a, -320c, and 320d-1 and the Drosha site in 449c. Binding of 8 to these sites, however, is predicted to be less fit (Figure 4A) than the Drosha site in miR-96. Indeed, transcriptome-wide qRT-PCR profiling showed that 8 selectively inhibits miR-96 maturation (Figure 4B). Interestingly, miR-96, miR-182, and miR-183 are transcribed in a single transcript. Since miR-182 and miR-183 expression levels are unaffected by 8, there is no cooperativity between nuclease processing of pri-miRNAs. Furthermore, 8 is more selective than a seed-targeting LNA oligonucleotide and an antagomir that target miR-96. Although the perception is that oligonucleotides are exquisitely selective, oligonucleotides can bind to off-targets that have mismatches with high affinity.
Figure 4.
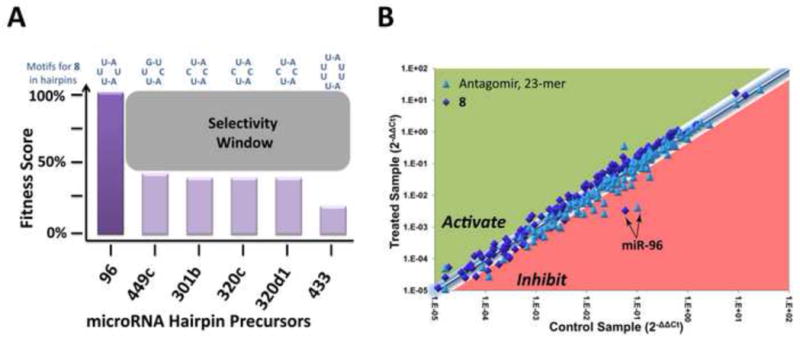
Compound 8 selectively inhibits miR-96. (A) Fitness plot for 8, which indicates optimal and sub-optimal RNA targets. Selectivity window based on fitness score predicts if a miRNA hairpin precursor can be targeted by 8. (B) Profiling the change in expression of 149 miRNAs upon treatment with 8 (diamond) and an antagomir targeting miR-96 (triangle). Only miR-96 is affected (≥2.5 fold) upon treatment with 8. However, treatment with an antagomir affects expression of 12 miRNAs (≥2.5-fold).
Inforna also identified a small molecule, guanidinylated Neomycin B (9; Figure 3B) that inhibits maturation of pri-miR-10b by binding the Drosha processing site. 9 boosts production of pri-miR-10b in cellulo and removes repression of a downstream target, HomeoboxD10 (HOXD10), which functions as a tumor suppressor [47].
Outlook & Future Directions
Since the discovery of miRNAs, a great amount of effort has been expended to define a detailed mechanism for miRNA biogenesis. Likewise, much effort will be required to untangle the intricate cellular biology of miRNAs. The examples above, however, show that miRNA expression can be altered with small molecules, providing promise to expand miRNAs from diagnostic signatures of disease to bona fide therapeutic targets. Although RNA is difficult to target selectively and afford a biological response, progress is being made on various fronts. A better understanding of RNA structure and study of RNA-small molecule recognition events could aid in the design of lead small molecules. Given the importance of miRNAs and the promise they hold, high affinity miRNA-binding scaffolds could decode miRNA function and its roles in various cellular pathways. Further, small molecule leads could be developed into therapeutic agents for disease-associated miRNAs and perhaps personalized medicines.
Highlights.
Challenges associated with selective recognition of RNA by small molecules
Review of methods to target miRNAs with small molecules
Downstream biological responses of small molecules that modulate miRNA function
Future directions in the area to develop lead RNA-directed therapeutics and chemical probes of function
Acknowledgments
This work was funded by the National Institutes of Health (R01GM097455 to MDD) and the Swiss National Science Foundation Prospective Researcher Fellowship (to BV).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 2.Bayne EH, Allshire RC. RNA-directed transcriptional gene silencing in mammals. Trends Genet. 2005;21:370–373. doi: 10.1016/j.tig.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Parsons J, Castaldi MP, Dutta S, Dibrov SM, Wyles DL, Hermann T. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat Chem Biol. 2009;5:823–825. doi: 10.1038/nchembio.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson A, Leeper TC, Athanassiou Z, Patora-Komisarska K, Karn J, Robinson JA, Varani G. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein. Proc Natl Acad Sci U S A. 2009;106:11931–11936. doi: 10.1073/pnas.0900629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yonath A, Bashan A. Ribosomal crystallography: initiation, peptide bond formation, and amino acid polymerization are hampered by antibiotics. Annu Rev Microbiol. 2004;58:233–251. doi: 10.1146/annurev.micro.58.030603.123822. [DOI] [PubMed] [Google Scholar]
- 7.Dervan P. Design of sequence-specific DNA-binding molecules. Science. 1986;232:464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen PE. Design of Sequence-Specific DNA-Binding Ligands. Chemistry – A European Journal. 1997;3:505–508. [Google Scholar]
- 9.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 10.White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature. 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 11.Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS Chem Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 12.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. Embo j. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 17.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. Rna. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirigin FF, Lindstedt K, Sellars M, Ciofani M, Low SL, Jones L, Bell F, Pauli F, Bonneau R, Myers RM, et al. Dynamic microRNA gene transcription and processing during T cell development. J Immunol. 2012;188:3257–3267. doi: 10.4049/jimmunol.1103175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu HY, Yan Z, Xu Y, Hu H, Menzel C, Zhou YH, Chen W, Khaitovich P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 2009;10:413. doi: 10.1186/1471-2164-10-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 24.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 25.Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 27.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 28.Swarbrick A, Woods SL, Shaw A, Balakrishnan A, Phua Y, Nguyen A, Chanthery Y, Lim L, Ashton LJ, Judson RL, et al. miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat Med. 2010;16:1134–1140. doi: 10.1038/nm.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 30.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 34.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 35•.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47:7482–7484. doi: 10.1002/anie.200801555. This was the first demonstration utilizing small molecule for modulating microRNA transcription. Small molecule transcriptional inhibitors of miR-21, a microRNA upregulated in cancer were identified. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8:706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 38.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 39•.Young DD, Connelly CM, Grohmann C, Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc. 2010;132:7976–7981. doi: 10.1021/ja910275u. It was shown that a small molecule could be used for both up and down regulating the transcription of a microRNA. miR-122 is upregulated in HCV and downregulated in HCC. Small molecule modulators were identified for enhancing and repressing miR-122 transcriptionally. This was the first report that identifies activators of microRNA transcription. [DOI] [PubMed] [Google Scholar]
- 40•.Bose D, Jayaraj G, Suryawanshi H, Agarwala P, Pore SK, Banerjee R, Maiti S. The tuberculosis drug streptomycin as a potential cancer therapeutic: inhibition of miR-21 function by directly targeting its precursor. Angew Chem Int Ed Engl. 2012;51:1019–1023. doi: 10.1002/anie.201106455. This study demonstrates that small molecules could interfere with dicer processing of precursor microRNAs. Streptomycin was shown to bind the Dicer processing site which prevents the processing of pre miR-21 to mature miR-21. [DOI] [PubMed] [Google Scholar]
- 41•.Vo DD, Staedel C, Zehnacker L, Benhida R, Darfeuille F, Duca M. Targeting the production of oncogenic microRNAs with multimodal synthetic small molecules. ACS Chem Biol. 2014;9:711–721. doi: 10.1021/cb400668h. Another example of small molecules inhibiting Dicer processing of precursor microRNAs. Neomycin-nucleobase conjugates were shown to inhibit the Dicer processing of pre miR-372 and pre-miR-373. [DOI] [PubMed] [Google Scholar]
- 42••.Velagapudi SP, Gallo SM, Disney MD. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat Chem Biol. 2014;10:291–297. doi: 10.1038/nchembio.1452. An approach termed inforna was developed to design lead small molecules targeting RNA-transcriptome-wide in a target agnostic manner. inforna allowed to identify small molecule inhibitors of microRNA hairpin precursors with a hit rate of 45%. It was the first demonstration of small molecule inhibitors of microRNA to be atleast as selective as an antagomir. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Disney MD, Labuda LP, Paul DJ, Poplawski SG, Pushechnikov A, Tran T, Velagapudi SP, Wu M, Childs-Disney JL. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J Am Chem Soc. 2008;130:11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- 44.Velagapudi SP, Seedhouse SJ, Disney MD. Structure-activity relationships through sequencing (StARTS) defines optimal and suboptimal RNA motif targets for small molecules. Angew Chem Int Ed Engl. 2010;49:3816–3818. doi: 10.1002/anie.200907257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu C, Li J, Wang X, Song L. Unregulated miR-96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS One. 2010;5:e15797. doi: 10.1371/journal.pone.0015797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velagapudi SP, Disney MD. Two-dimensional combinatorial screening enables the bottom-up design of a microRNA-10b inhibitor. Chem Commun (Camb) 2014;50:3027–3029. doi: 10.1039/c3cc00173c. [DOI] [PMC free article] [PubMed] [Google Scholar]