Abstract
The mammary epithelium is thought to be stabilized by cell-cell adhesion mediated mainly by E-cadherin (E-cad). Here, we show that another cadherin, retinal cadherin (R-cad), is critical for maintenance of the epithelial phenotype. R-cad is expressed in nontransformed mammary epithelium but absent from tumorigenic cell lines. In vivo, R-cad was prominently expressed in the epithelium of both ducts and lobules. In human breast cancer, R-cad was down-regulated with tumor progression, with high expression in ductal carcinoma in situ and reduced expression in invasive duct carcinomas. By comparison, E-cad expression persisted in invasive breast tumors and cell lines where R-cad was lost. Consistent with these findings, R-cad knockdown in normal mammary epithelium stimulated invasiveness and disrupted formation of acini despite continued E-cad expression. Conversely, R-cad overexpression in aggressive cell lines induced glandular morphogenesis and inhibited invasiveness, tumor formation, and lung colonization. R-cad also suppressed the matrix metalloproteinase 1 (MMP1), MMP2, and cyclooxygenase 2 gene expression associated with pulmonary metastasis. The data suggest that R-cad is an adhesion molecule of the mammary epithelium, which acts as a critical regulator of the normal phenotype. As a result, R-cad loss contributes to epithelial suppression and metastatic progression.
Introduction
Tumor invasion and metastasis are partly regulated by a switch from epithelial to mesenchymal cadherins in the transformed epithelium. E-cadherin (E-cad) down-regulation in epithelial carcinomas is associated with malignant progression (1–6). In contrast, N-cadherin (N-cad), normally expressed in neural and mesenchymal tissues, is up-regulated in invasive carcinomas, a change that is indicative of the epithelial-to-mesenchymal transition (EMT; refs. 7–14). Retinal cadherin (R-cad), another classic cadherin, has recently been linked to epithelial carcinomas (15, 16), although earlier studies showed R-cad to function in normal brain, retina (17, 18), muscle, pancreas, gastrointestinal tract, and kidney development (19–21).
Two studies have suggested that R-cad has a positive effect on tumor progression. Ectopic expression of R-cad in BT-20 breast tumor cells induced lamellipodia and motility via Rho GTPase activation (15). R-cad was also shown to compete with E-cad for p120, thus stimulating E-cad endocytosis and cell motility (16). Another study of rabdomyosarcomas showed a transforming effect of R-cad on myoblasts due to Rac1 activation and impairment of cell cycle arrest (22). These studies suggested an oncogenic effect of R-cad. Conversely, another study implicated R-cad in suppression of malignancy—expression was found to be down-regulated in gastrointestinal tumors due to DNA methylation (23).
To address these discrepancies, we sought to determine if R-cad expression is characteristic of normal mammary epithelium and whether it is up-regulated or down-regulated with malignancy. Indeed, we found that R-cad is expressed in nontransformed mammary cell lines and is down-regulated in breast carcinoma cell lines. In contrast, E-cad, also present in normal cells, remains expressed in most epithelial tumor cell lines. Clinical specimens showed similar relationships. R-cad was highly expressed in normal duct and lobules, and in ductal carcinoma in situ (DCIS) but significantly reduced in invasive duct carcinomas (IDC). Normal E-cad expression, however, persisted in IDC. Manipulation of R-cad expression in culture showed strong effects on cell properties. Small interfering RNA (siRNA) knockdown of R-cad in normal MCF10A breast cells disrupted acinus formation and stimulated an invasive phenotype. Conversely, exogenous expression of R-cad in invasive cell lines induced acini and suppressed invasion. Furthermore, R-cad suppressed tumor formation and lung colonization in vivo and was associated with repression of the matrix metalloproteinase 1 (MMP1), MMP2, cyclooxygenase 2 (Cox2) gene signature characteristic of pulmonary metastasis (24). Thus, R-cad is a critical epithelial cadherin that exerts potent suppression of breast cancer progression.
Materials and Methods
Cell lines
HMEC, 184A1, 184B5, and AB5 normal epithelial cell lines were obtained from Dr. Stampfer (Lawrence Berkeley Laboratories) and cultured in mammary epithelial growth media, including BPE, hydrocortisone, human epidermal growth factor (EGF), insulin, and antibiotics (Cambrex). MCF10A and AB5 cell lines were grown in DMEM/F12 with 5% horse serum, 20 ng/mL EGF, 0.5 µg/mL hydrocortisone, 100 ng/mL cholera toxin, 10 µg/mL insulin, and 1% antibiotics (Sigma). All other breast cancer cell lines were from American Type Culture Collection and grown in DMEM with 10% fetal bovine serum and 1% antibiotics. The 3475 metastatic subline was from Dr. Massague (Memorial Sloan Ketterring Cancer Center). The cadherin-11 (cad-11)/R-cad null MDA-MB-231 cell line (231v) was derived by limiting dilution and was able to grow tumors in severe combined immunodeficient mice fat pads.
Antibodies
A monoclonal R-cad antibody (25) was kindly provided by ICOS Corporation. This antibody was raised against a glutathione S-transferase fusion protein of the human R-cad encompassing the extracellular cadherin repeats EC1 to EC3 (25). Monoclonal E-cad, N-cad, Cad-11, and ZO-1 antibodies were obtained from BD Biosciences. Monoclonal anti-Rac1 was obtained from Cell Signaling.
R-cad infection
A cDNA clone of mouse R-cad, obtained from Dr. Masatoshi Takeichi (RIKEN Center for Developmental Biology), was subcloned into the pLNCX-EGFP vector (Clonetech) for retroviral production. Viral suspensions were produced in Phoenix A cells and used to infect cells. Highly expressing cells were isolated by EGFP fluorescence-activated cell sorting and selected in 0.4 mg/mL G418. The 6× myc-tagged R-cad construct was subcloned into a pCXN2 vector driven by the β-actin promoter.
R-cad knockdown by siRNA
MCF10A cell suspensions were plated at 2 × 105 cells/mL in six-well dishes in growth medium without antibiotics. Three R-cad siRNA sequences and a nontargeting siRNA sequence were obtained from Ambion. The siRNA that caused the most effective knockdown (ID 146004) was used (CCACGUUUUCAGCUGUGGAtt and UCCACAGCUGAAAACGUGGtcc). We also used a pool of three R-cad siRNAs from Santa Cruz (CAACAGCAUGGACUUCAAA, GGAAGGAAAUCUCAACUAU, and CAACACGUCCAUCAUCAAA). SiRNA was transfected at 30 nmol/L into cells using the siPORT Neo FX transfection agent (Ambion). At 48 h later, 1 × 105 cells were applied onto Matrigel-coated transwells for invasion assays.
Immunoblotting
Proteins were extracted in 1% Triton-X-100 lysis buffer including protease inhibitors. Protein (30 µg) was loaded on 7.5% SDS-polyacrylamide gels and transferred to Immobilon membranes. Blots were probed with indicated antibodies and developed by chemiluminescence.
Immunofluorescence
Cells were plated on collagen-coated coverslips, fixed in 3.7% paraformaldehyde for 15 min, permeabilized for 2 min in 0.1% Triton X-100, blocked in PBS containing 2% bovine serum albumin, incubated with 2 µg/mL of primary antibody for 1 h at room temperature, washed and incubated for 1 h with 1:5,000 FITC-conjugated or TRITC-conjugated secondary antibodies, and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Images were captured using a Zeiss Axioskop 2 microscope.
Archival breast tumors and immunohistochemistry
Formalin-fixed paraffin-embedded tissue blocks, including five cases of normal breast tissue and 18 cases of breast cancer, were selected from the Department of Pathology Archives of Montefiore Medical Center. Four-micrometer sections were heated at 60°C for 30 min, deparaffinized, hydrated, and incubated in epitope retrieval buffer (DAKO) at 95°C for 45 min. The slides were then incubated with an antibody to R-cad (1:400 dilution) or E-cad (1:25 dilution) for 30 min at room temperature in a DAKO Autostainer. The immunoreactivity was detected using DAKO EnVision reagents.
Migration and invasion assays
Cell migration and invasion assays were performed in 24-well Boyden chambers using 8-µm transwell filters, uncoated for migration assays, or coated with 20 µg Matrigel for invasion assays, except for MCF10A cells, wherein 2 µg were used for coating, as detailed in refs. (26, 27). Invasion or migration was expressed as the number of migrated cells bound per microscopic field and averaged from at least four fields per assay in at least three experiments.
Growth in Matrigel or collagen
The embedding and growth of cells in Matrigel was performed as described (28). Cells (5 × 105/mL) were plated in eight-well chambers precoated with 100% growth factor–reduced Matrigel (12 mg/mL, Trevigen). Cells were cultured in 0.3 mL growth media/2% Matrigel, for 1 wk, fixed, and photographed. MCF10A (5 × 105/mL) were plated on collagen-coated dishes (8 mg/mL collagen). After 24 h, cells were fixed and visualized by microscopy.
RNA purification and reverse transcription-PCR
Total RNA was prepared with Trizol (Invitrogen). RNA was reverse transcribed in the presence of oligo-dT primer. Primers to amplify R-cad cDNA are 5′atgaccgatgaccgcgggcgccggcgt3′, 5′catcgttcaggccacagata3′, 5′ggactggaaggacatctgga3′, and 5′atcctcttcaccacctccat3′. PCR was performed with Taq polymerase in 50 µL volume using 30 cycles of denaturation at 92°C for 30 s, annealing at 58°C for 30 s and 72°C for 1 min.
Real-time quantitative PCR analysis
The expression of Cox2, Epiregulin (EREG), MMP1, and MMP2 in control and 3475-R-cad cells was determined by reverse transcription-PCR (RT-PCR) and real-time quantitative PCR (qPCR) using primer pairs: COX2, 5′gaacattcctaccaccagcaa3′ and 5′tgcctgacacctttcaaattc3′; EREG, 5′aactctggatcccctgaggta3′ and aatatgtggctttgaccgtga; MMP1, 5′agaattcctgcatttgcttca 3′ and 5′ttcccagcgactctagaaaca3′; and MMP2, 5′gcagatctcaggagtgacagg3 and gatggcacccatttacaccta. Total RNA was reverse transcribed, and qPCR was run on a Lightcycler Roche 480 with the LightCycler Roche 480 Master kit (Roche). PCR was performed by denaturing at 95°C for 5 min, 45 cycles of denaturation at 95°C, annealing at 60°C, and extension at 72°C for 10 s. Results were normalized to β-actin mRNA levels.
Animals
All procedures involving mice were conducted in accordance with NIH regulations concerning the use and care of experimental animals. This study was approved by the Albert Einstein College of Medicine Animal Use Committee.
Tumor formation
3475-myc or 3475-R-cadmyc 1 cells (5 × 105) were mixed with 0.1 mL PBS and 0.1 mL Matrigel and injected bilaterally into the fourth inguinal mammary fat pad of 4-wk-old athymic BALB/c mice (n = 7). Mice were monitored biweekly for tumor growth. The experiment was repeated thrice.
Lung metastasis
Cells (1 × 106) per 0.2 mL PBS were injected into the tail vein of athymic BALB/c female mice. Four weeks later, mice were sacrificed, and lungs were perfused by injection of 2 mL of 10% formalin. Paraffin blocks were sectioned at 5 µm. Four to five sets of five serial sections, at 50 µm intervals, were obtained from each lung. Analysis was performed on 3 to 10 random sections at levels separated by at least 150 µm to minimize duplicate analysis. The number of metastases per section was normalized to the area of the lung section. Data are displayed as mean number of metastases ± SE. Statistical analysis was performed using the Mann-Whitney U test.
Results
R-cad is present in nontransformed breast cells but absent from breast tumor cell lines
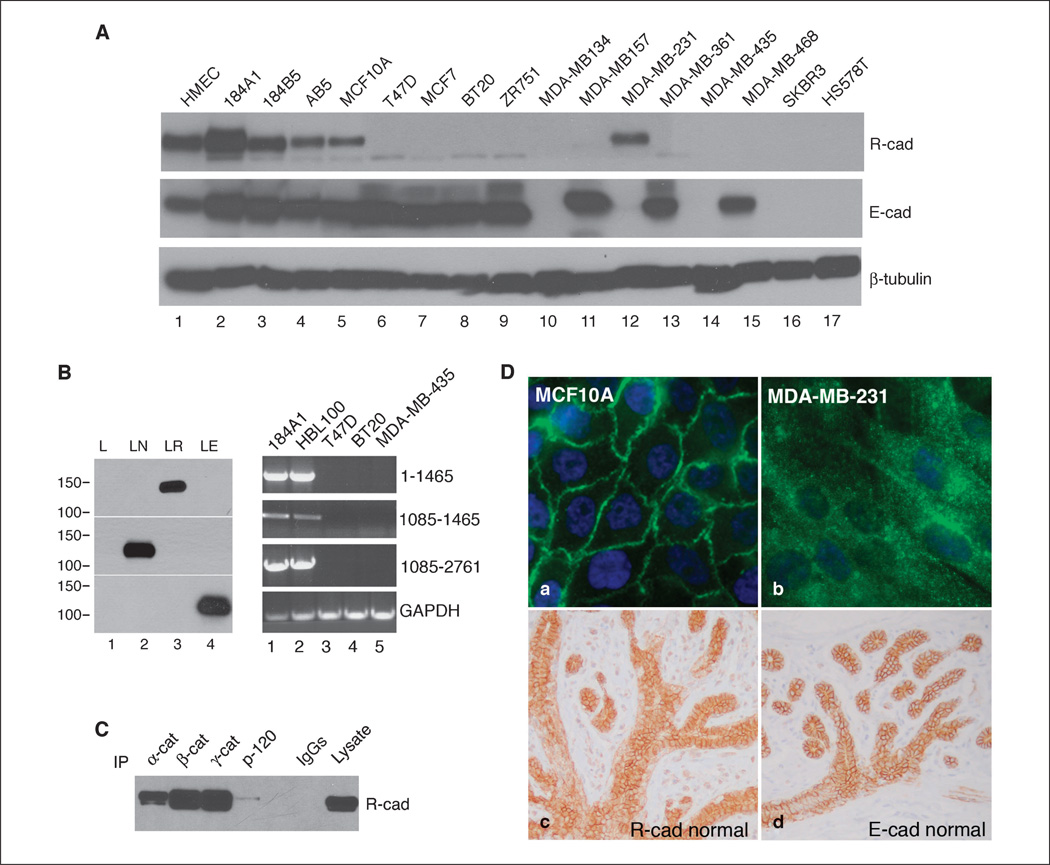
We screened a panel of human mammary cell lines for R-cad expression by Western blot analysis using an R-cad–specific monoclonal antibody (25). This antibody recognized a single ~125 kDa immunoreactive polypeptide band and did not cross-react with N-cad or E-cad (Fig. 1B). R-cad expression was detected at varying levels in all five nontransformed breast cell lines (HMEC, 184A1, 184B1, AB5, and MCF10A), but in 12 malignant cell lines, only MDA-MB-231 showed expression (Fig. 1A). In contrast, E-cad was present in all normal mammary cell lines (Fig. 1A, lanes 1–5) and seven well-differentiated breast tumor cell lines (T47D, MCF-7, BT-20, ZR751, MDA-MB-157, MDA-MB-361, and MDA-MB-468), although not in five poorly differentiated cell lines, such as MDA-MB-134, MDA-MB-435, MDA-MB-231, SKBR3, and HS578T. We confirmed R-cad expression by RT-PCR using primer sets spanning the R-cad mRNA sequence (Fig. 1B). R-cad transcripts were detected in the R-cad–positive cell lines 184A1 and HBL-100 (not shown in Fig. 1A) and absent from R-cad–negative T47D, BT-20, and MDA-MB-435 cell lines.
Figure 1.
R-cad is expressed in mammary epithelium and normal cell lines but is down-regulated in tumor cell lines. A, cell lysates from normal (lanes 1–5) and tumorigenic breast cell lines (lanes 6–17) were immunoblotted with antibodies against R-cad, E-cad, or β-tubulin. B, left, L cells transfected with control vector (lane 1), N-cad (lane 2), R-cad (lane 3), and E-cad (lane 4) were immunoblotted for R-cad, N-cad, or E-cad. Right, R-cad transcripts were examined by RT-PCR using RNA from R-cad–positive (lanes 1 and 2) and R-cad–negative (lanes 3–5) breast cell lines using three pairs of R-cad primers spanning the human R-cad mRNA sequence. C, lysates from MCF10A cells were immunoprecipitated with anti–α-catenin (lane 1), β-catenin (lane 2), γ-catenin (lane 3), and p120 (lane 4) and immunoblotted with anti–R-cad. Control IgGs were used (lane 5), and an MCF10A lysate was used as positive control (lane 6). D, MCF10A and MDA-MB-231 cells were stained with anti–R-cad followed by FITC secondary detection. Normal breast tissue was stained for R-cad (c) and E-cad (d). Note the crisp membrane staining of R-cad and E-cad in both the central ducts and surrounding lobular acini.
Consistent with a role in epithelial adhesion, R-cad strongly localized to cell-cell contacts in MCF10A cells (Fig. 1D) and coimmunoprecipitated with α-catenin, β-catenin, γ-catenin, and, to a lesser extent, p120 (Fig. 1C). Although R-cad protein was detected in a single malignant cell line, MDA-MB-231, its localization was mostly cytosolic, apparently unable to form junctions in these cells (Fig. 1D, b).
To determine whether R-cad is expressed in mammary epithelium in vivo, we performed immunohistochemical staining of normal breast tissue (Fig. 1D, c and d). R-cad showed crisp membrane localization in the mammary epithelium in both ducts and lobules, where its staining pattern was identical to that of E-cad, implying that both cadherins contribute to the adhesive structure of the mammary gland.
R-cad is down-regulated during breast tumor progression
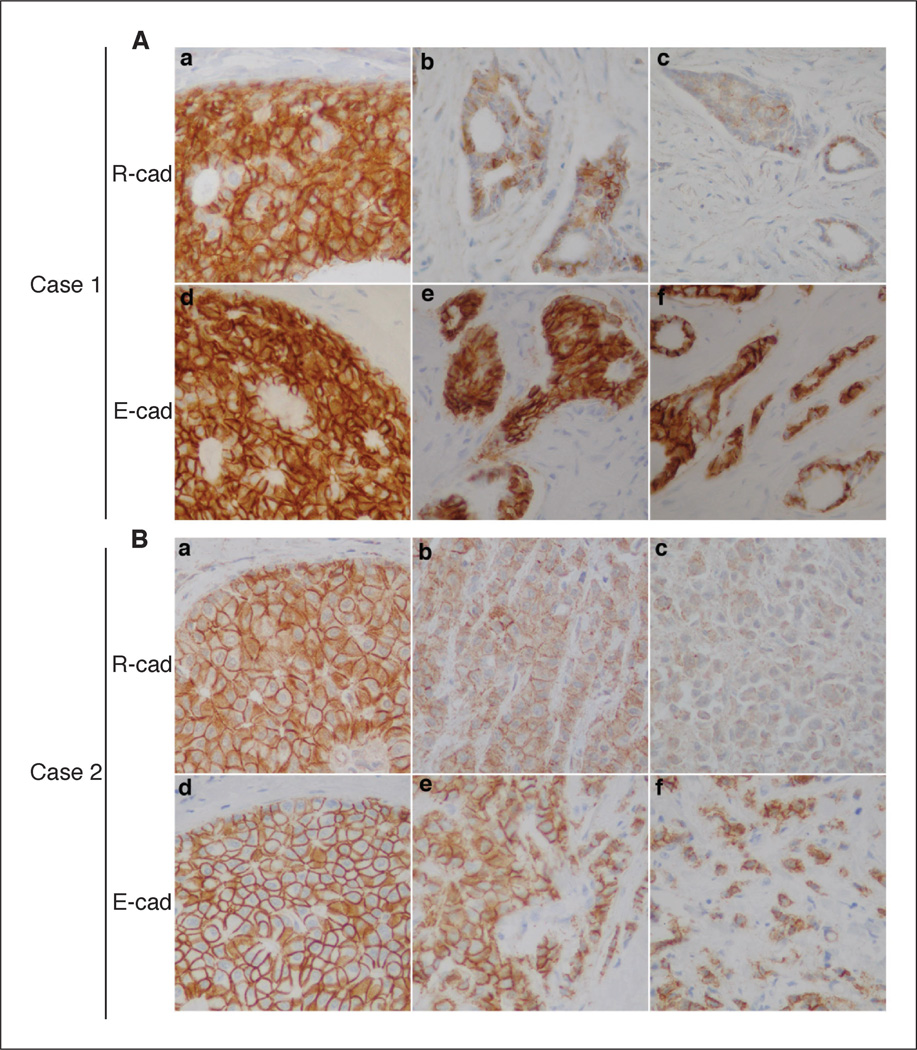
The results from the cell line panel suggested that R-cad might be down-regulated with tumor progression. We, therefore, performed immunohistochemical analysis of R-cad and E-cad in 18 archived specimens of IDC; eight also contained regions of DCIS (Fig. 2). Like E-cad, R-cad staining was strong in DCIS, but IDC that retained E-cad frequently showed reduced R-cad expression, particularly in the most anaplastic or invasive regions. Using a quantitative scoring system (Supplementary Table S1), the mean R-cad score was significantly reduced in IDCs compared with DCIS (0.39 versus 0.74; P < 0.0007). Of the 18 IDC, 10 exhibited an irregular pattern of R-cad expression with extensive areas of R-cad loss, whereas eight others had homogenous expression of R-cad throughout. One case had cytoplasmic R-cad staining, reminiscent of that seen in MDA-MB-231 cells (Supplementary Fig. S1), whereas three others had mixed membrane and cytoplasmic staining. Thus, R-cad is ubiquitously expressed in normal ducts and noninvasive tumors (DCIS) and frequently down-regulated in IDC, especially in regions of advanced progression.
Figure 2.
R-cad is expressed in DCIS and down-regulated in invasive ductal carcinoma of the breast. Archival breast tumors were analyzed by immunohistochemical staining for R-cad and E-cad. A and B, sections from formalin-fixed and paraffin-embedded IDCs that included areas of DCIS were immunostained with the ICOS anti–R-cad antibody. Crisp membranous staining for R-cad was detected in regions of DCIS (Aa, Ba) but diminished in regions of IDC (Ab, Bb), especially in poorly differentiated areas of the tumor (Ac, Bc). By comparison, E-cad staining was strong in all areas of these tumors.
In contrast to the abrupt down-regulation of R-cad in IDC relative to DCIS, E-cad expression persisted in the IDC of most tumors, suggesting that R-cad might be lost before E-cad during malignant progression. Consistent with these findings, we found that R-cad loss predated that of E-cad in MCF10A cells in response to transforming growth factor β, which induces EMT and cell motility. The down-regulation of epithelial R-cad and E-cad was accompanied by gain of the mesenchymal N-cad and fibronectin proteins, suggesting reduced R-cad characterizes EMT and cancer progression (Supplementary Fig. S2).
R-cad suppresses cellular invasiveness and induces morphogenesis into mammary acini
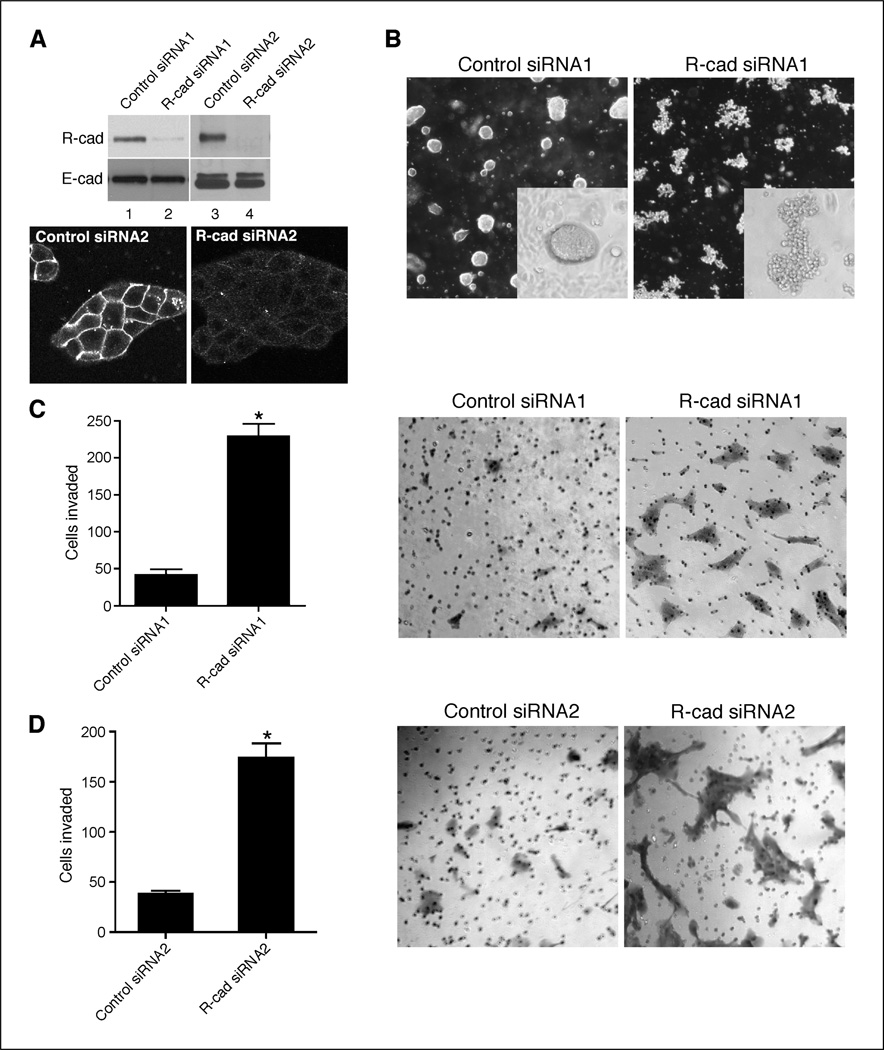
We sought to determine if R-cad is critical for maintenance of the epithelial phenotype using siRNA-mediated knockdown in normal breast cells and overexpression in invasive cells. We used two siRNAs preparations consisting of either a single oligonucleotide or a pool of three oligonucleotides. Knockdown of R-cad in MCF10A cells with either preparation led to suppression of R-cad with no effect on E-cad expression compared with control nontargeting siRNAs (Fig. 3A). R-cad loss was confirmed by immunostaining of MCF10A knockdowns with anti–R-cad (Fig. 3A). The effect of R-cad loss on MCF10A behavior was tested in Matrigel invasion and morphogenesis assays. R-cad suppression stimulated invasion through a 2-µg thin Matrigel layer (Fig. 3C and D) and disrupted spheroid formation in collagen gels (Fig. 3B). These data provide evidence that R-cad is critical for morphogenesis, thus suppressing latent invasive behavior of mammary epithelial cells.
Figure 3.
R-cad knockdown in MCF10A cells disrupts morphogenesis and stimulates invasiveness. A, MCF10A were transfected with control siRNA (lanes 1 and 3) or two different R-cad siRNA1 and siRNA2 (lanes 2 and 4) for 48 h. Cell lysates were immunoblotted with an anti–R-cad or anti–E-cad antibody. B, control cells (left) or MCF10A knockdown cells (right) were plated onto collagen gels for spheroids evaluation. Each of the illustrated cell groups tested viable by DAPI nuclear staining (not shown). C and D, control (left) or MCF10A knockdowns (right) were applied onto 2-µg Matrigel-coated transwells for 18 h. Representative migrations of cells were counted and photographed. Columns, mean of duplicate experiments; bars, SE. *, P < 0.05.
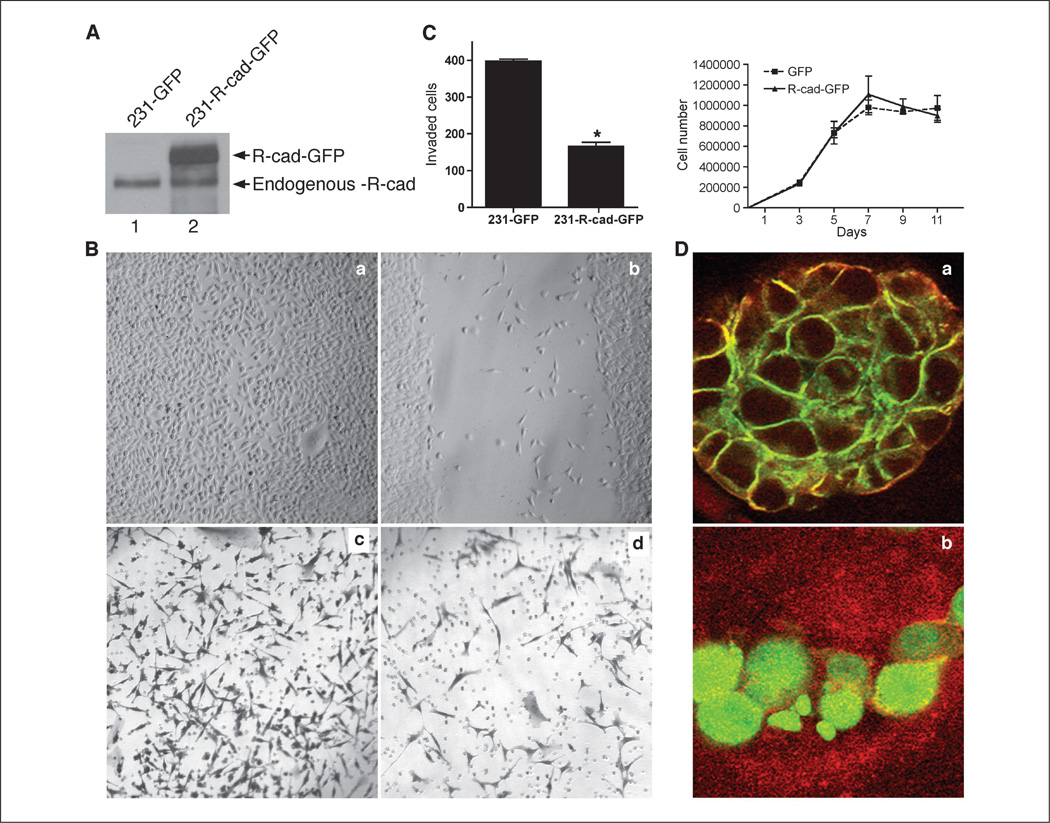
We next tested whether R-cad overexpression suppresses migration or invasion of MDA-MB-231 cells, which are highly invasive in vitro and tumorigenic in vivo. Although R-cad is endogenously expressed in these cells, the localization is mostly cytosolic and does not restrict cellular invasiveness, because R-cad knockdown did not affect this process (Supplementary Fig. S3). However, transduction of the cells with a green fluorescent protein (GFP)–tagged R-cad (R-cad-GFP) retroviral construct resulted in membrane localization of R-cad (Fig. 4A and D). Thus, the higher level of expression in transfected cells allows normal processing and localization. The mechanism of abnormal cytoplasmic localization in MDA-MB-231 cells was not explored further but could represent a nonmembrane isoform, mutated R-cad, or a binding interaction that becomes saturated with overexpression. We next measured whether R-cad overexpression causes changes in cellular behavior using wound healing and Matrigel invasion assays (Fig. 4B and C). R-cad-GFP delayed the migration of MDA-MB-231 cells into the wound compared with GFP controls, which closed the wound by 24 hours. Moreover, R-cad-GFP inhibited Matrigel-invasion as shown by the low number of invading cells relative to controls. The reduced migration and invasion induced by R-cad were not due to inhibition of cell proliferation, because growth analysis over a period of 11 days showed that R-cad-GFP expressing MDA-MB-231 cells grew at similar rates as controls (Fig. 4C). R-cad, therefore, inhibits invasion but does not suppress growth.
Figure 4.
R-cad overexpression in the MDA-MB-231 cell line suppresses migration and invasion and induces morphogenesis. A, lysates of MDA-231 expressing GFP (lane 1) or R-cad-GFP (lane 2) were immunoblotted with antibody to R-cad. A 125-kDa endogenous R-cad protein was detected in both cell lines (lanes 1 and 2) and a 140-kDda R-cad-GFP fusion protein was detected in infected cells (lane 2). B, GFP (a) or R-cad-GFP (b) expressing cells were grown to confluence on 35-mm Petri dishes and wounded using a sterile pipette. A representative from triplicate experiments. Invasiveness of the cells was tested in Bowden chambers. GFP (c) or MDA-MB-231/R-cad-GFP cells (d) were applied onto 20 Ag Matrigel-coated transwell filters for 24 h. C, left, the number of invading cells was counted in each well. Mean ± SE from five experiments. P < 0.05 (Mann-Whitney test). Right, GFP and R-cad-GFP expressing cells were plated at 1 × 105 in six-well plates and counted at 0, 3, 5, 7, 9, and 11 d postplating. Mean ± SE from three experiments. D, MDA-231/Rcad-GFP cells (a) and GFP cells (b) were plated on Matrigel for 7 d, stained with an anti–ZO-1 antibody followed by TRITC detection, and imaged by confocal microscopy.
Given the suppressive effect of R-cad on motility and invasion, we also examined whether R-cad reverses the mesenchymal morphology of MDA-MB-231 cells in three-dimensional Matrigel cultures (Fig. 4D). Interestingly, whereas GFP controls formed an undifferentiated morphology in Matrigel, R-cad-GFP–expressing cells formed acinus-like structures (Fig. 4D). Moreover, R-cad-GFP was targeted to the cell surface and induced a polarized morphology with relocation of ZO-1 from the cytoplasm to the cell cortex, so that all acini showed colocalization of these two markers at the cell surface. Thus, R-cad induces differentiation and morphogenesis of carcinoma cells into structures resembling mammary acini.
Because of the cytoplasmic R-cad expression of MDA-MB-231, we also evaluated the effects of R-cad expression on the highly invasive breast carcinoma cell line, HS578T, which is R-cad negative. In these cells, low levels of R-cad expression dramatically suppressed invasion. In addition, R-cad changed the morphology of colonies in Matrigel from loose stellate cell groups into tight rounded acinus-like groupings (Supplementary Fig. S4A–C). These effects were entirely consistent with the observations in MDA-MB-231 and occurred despite the continued expression of N-cad in HS578T.
Suppression of invasion by R-cad does not depend on competition of endogenous cadherins
Competition between R-cad and E-cad for p120 was reported to cause E-cad endocytosis, thus reducing cell-cell adhesion (16). We, therefore, tested whether R-cad expression in MDA-MB-231 cells alters endogenous expression of the proinvasive cad-11, because loss of this cadherin might attenuate invasion (29, 30). Cad-11, α-cat, β-cat, and p120 were analyzed in both sparse and confluent cultures. As expected, increases in cell-cell adhesion caused by R-cad led to increases in α-cat, β-cat, and p120 levels (Supplementary Fig. S5A) and localization of these molecules to cell-cell contacts due to stabilization by R-cad (Supplementary Fig. S5B). Indeed, R-cad expressing cells had reduced cad-11 expression relative to controls, and the effect was apparent at low and high cell densities (Supplementary Fig. S5A, lanes 1–6). However, R-cad expression into an MDA-MB-231 variant cell line devoid of cad-11 and R-cad (231v) suppressed invasion and induced acini (Supplementary Fig. S6A–C), suggesting cad-11 modulation is not a pivotal pathway for invasive suppression by R-cad.
We next tested whether R-cad activates Rac1, which induces lamellipodia that are necessary for establishment of epithelial cell-cell contacts (31). R-cad enhanced Rac activity in MDA-MB-231v cells, but only in cells grown in Matrigel (three dimensional) and not on the surface of plastic dishes (two dimensional; Supplementary Fig. S6D). Thus, Rac1 activation by R-cad likely supports adhesion and morphogenesis.
R-cad suppresses breast cancer progression in vivo
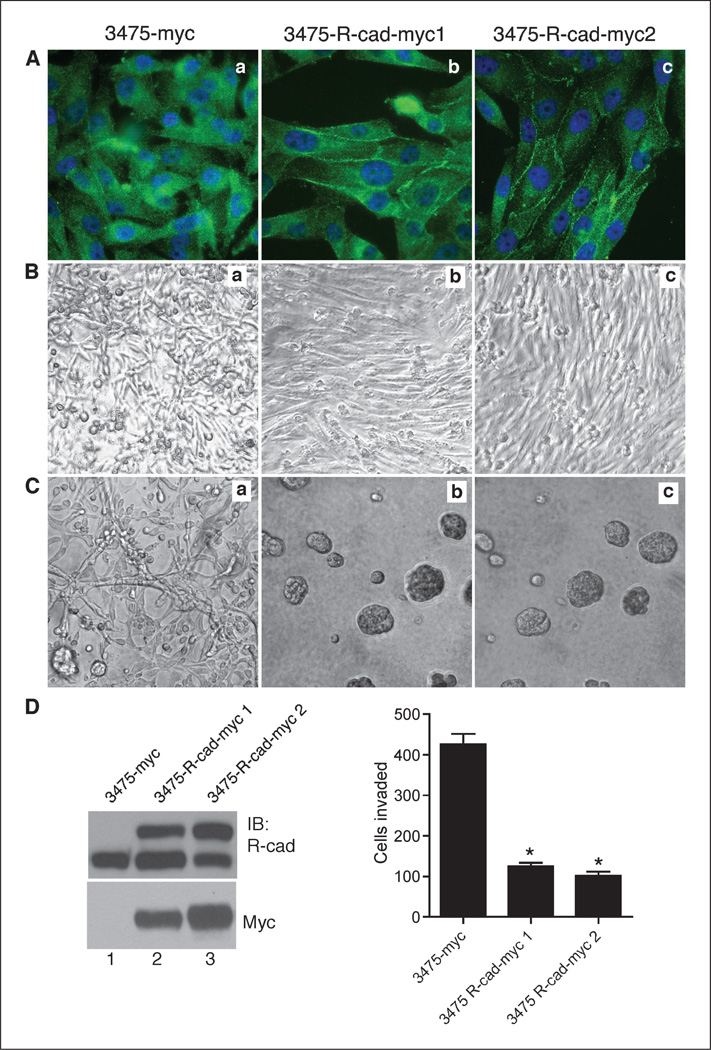
Because R-cad induced morphogenesis and suppressed invasiveness, we sought to determine whether R-cad also blocks tumor progression in vivo. We used the 3475 subline of MDA-MB-231 cells because of their extensive metastasis to lungs in vivo (32). 3475 cells were transfected with myc-tagged R-cad, and highly expressing cells were obtained from independent transfections by neomycin selection. R-cad-myc was localized at cell-cell contacts compared with the diffusely distributed R-cad of control cells (Fig. 5A). As in MDA-MB-231 cells, the transfected cells expressed endogenous 125-kDa R-cad and additional 140-kDa fusion protein (Fig. 5D), but the total expression of the two proteins was less than twice the level of the endogenous protein. Thus, exogenous expression of R-cad remained within the observed range of native expression in mammary cell lines.
Figure 5.
R-cad expression in MDA-MB-231 cells induces morphogenesis and suppresses invasion. A, the 3475 metastatic subline of MDA-231 was transfected with R-cad myc or myc vector and selected for R-cad expression by neomycin. Control cells (Aa) or R-cad-myc cells isolated from two independent transfections (Ab and Ac) were stained with anti–R-cad antibody followed by FITC-conjugated antibody. Note junctional localization of R-cad in 3475-R-cad cells (Ab, Ac) versus cytosolic endogenous R-cad in control cells (Aa). B and C, control (a) or R-cad-myc (b, c) cells were grown to confluence in two-dimensional (plastic dishes) or in three-dimensional (Matrigel) cultures for 7 d. D, left, cell extracts were immunoblotted with anti–R-cad or anti-Myc antibody. Right, 3475-R-cad cells were compared with controls in Matrigel invasion. Values of invading cells from five independent experiments. Columns, mean; bars, SE. *, P < 0.05.
R-cad caused striking changes in cellular morphology in two-dimensional and three-dimensional cultures. The expression of R-cad caused orderly monolayer growth that contrasts with the disorganized multilayer growth of the control cells (Fig. 5B). Consistently, 3475-R-cad cells formed acini in Matrigel, whereas the control cells grew as networks (Fig. 5C). Finally, R-cad expression greatly reduced invasivity in Matrigel relative to controls (Fig. 5D).
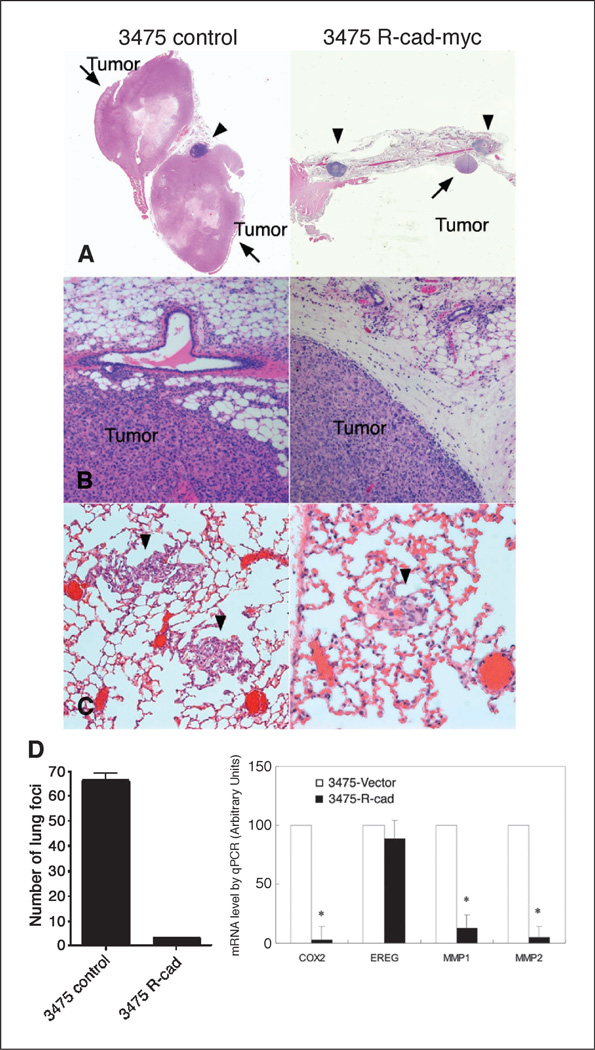
We next tested whether R-cad could inhibit tumor formation or lung metastasis (Fig. 6). Mammary fat pad implantation into athymic BALB/c female mice showed striking differences (Fig. 6A and B). Control 3475 tumor cells produced large primary tumors, up to 2 cm in diameter at 8 to 12 weeks postinjection. In contrast, 3475-R-cad cells did not produce any palpable tumors at that time. However, histologic examination growth of mammary fat pads after 6 months led to identified small tumor nodules. These data contrasted with those in vitro, in which control and R-cad–positive 3475 cells grew at comparable rates in culture, suggesting that there are in vivo constraints that do not exist in vitro (not illustrated, but see Fig. 4C). Thus, R-cad inhibition of cell migration or invasion restricts tumor growth in vivo, but not in vitro. Moreover, whereas control tumors greatly infiltrated the surrounding connective tissue and expanded into the fatty tissue (Fig. 6B, left), R-cad expressing tumors were confined to the edge of the mammary tissue with a sharp demarcation from the surrounding loose connective tissue, suggesting a defect in migration or invasion (Fig. 6B, right). We, thus, speculate that by restricting mobility, R-cad prohibits access to vascularized tumor areas that provide oxygen and nutrients for growth.
Figure 6.
R-cad suppresses tumor growth and lung colonization and inhibits the lung metastasis signature. A, control 3475 (left) or 3475-R-cad myc 1 cells (right) were injected bilaterally at 0.5 × 106 mixed 1:1 with Matrigel into the mammary fat pads of athymic BALB/c females. At 8 to 12 wk later, 3475 control cells grew into large tumors whereas 3475-R-cad cells did not produce any tumors at that time. However, after 180 wk, 3475-R-cad cells grew into small nodules in the fat pad. Arrows point to tumor nodules, and arrowheads point to lymph nodes. B, 3475-myc tumors invaded readily the surrounding connective and fat tissue (left), whereas 3475-R-cad tumors had sharp demarcated margins (right). C and D, 1.0 × 106 cells of control 3475-myc or 3475-R-cad myc 1 cells were injected into the tail vein of athymic BALB/c females. At 4 wk later, mice were sacrificed, and the number of lung foci, as in C, was quantified as mean ± SE; *, P < 0.05. D, right, RNA from control and 3475-R-cadmyc1 cells was reverse transcribed into cDNA. The level of mRNA for Cox-2, EREG, MMP1, and MMP2 were determined by qPCR and normalized to levels of β-actin mRNA. The mRNA levels for each of these genes from three experiments are displayed as mean ± SE. P < 0.05.
The next experiments examined whether R-cad limits the ability of 3475 metastatic cells to colonize the lungs after tail vein injection (33, 34). Suspensions of control or 3475-R-cad cells were injected into the tail vein of athymic BALB/c mice. Four weeks later, lungs were processed and the number of pulmonary foci was determined. While control 3475 cells produced ~65 foci per lung section, R-cad expressing cells generated an average of only three metastatic nodules per lung section (Fig. 6C and D).
Lung metastasis by 3475 cells has been associated with expression of a gene signature that includes MMP1, MMP2, COX2, and EREG (24). This signature was associated with vascular remodeling, intravasation, extravasation, and, hence, metastasis. Real-time qPCR showed that R-cad dramatically suppressed the expression of MMP1, MMP2, and COX2, but not EREG (Fig. 6D), suggesting that R-cad targets these critical regulators of metastatic seeding and growth.
Discussion
The classic cadherins play a critical role in tissue morphogenesis, including cell recognition and sorting, boundary formation and maintenance, coordinated cell movement, and the induction and maintenance of cell and tissue polarity (35). Aberrations in any of these features can lead to disruption of tissue integrity and onset of metastatic tumors.
Cell adhesion in the mammary epithelium is mediated in large part by E-cad and is necessary for epithelial maintenance and apico-basal polarity. Its expression restricts cell locomotion and proliferation. E-cad has been linked to epithelial suppression, and its deregulation in carcinomas correlates with tumor progression. Its expression is frequently repressed by gene methylation in gastric and breast carcinomas (36). Recently, the gene for R-cad or CDH4 was also found to be methylated in 78% and 95% of colorectal and gastrointestinal tumors, suggesting that it fulfills a previously unsuspected tumor-suppressive function (23). Our data further substantiate a suppressive function. We found that R-cad is expressed in normal mammary epithelium and in noninvasive tumors, such as DCIS, but significantly reduced in IDCs. Moreover, some tumors had heterogeneous expression, and R-cad was lowest in areas of poor differentiation. In contrast E-cad was uniform during neoplastic progression.
R-cad expression in normal epithelium was previously unsuspected. However, we found that R-cad is as abundant as E-cad in mammary epithelium, suggesting it plays an important role in the maintenance of the glandular architecture. Moreover, knockdown of R-cad in MCF10A mammary cells stimulated invasiveness and disrupted acini formation despite E-cad expression. Conversely, expression of R-cad in the highly aggressive MDA-MB-231 cell line induced acini and suppressed invasion. These results suggest that R-cad is a gatekeeper of the epithelial phenotype that acts as an innate suppressor of invasion. Other studies found that R-cad knockout in vivo suppressed mesenchymal-to-epithelial transition in the kidney, whereas E-cad ablation did not (19). Moreover, R-cad expression in E-cad null embryonic stem cell could rescue formation of epithelia, muscle, and gastrointestinal tract (20). All of these findings suggest that R-cad has a fundamental role in stabilizing epithelial adhesion during morphogenesis.
In contrast to our data, however, a study reported that R-cad transfection into BT-20 breast tumor cells increased motility due to Rac1 and Cdc42 activation (15). Another study showed that overexpression of R-cad in myoblasts stimulated a transformed phenotype due to Rac1 activation (22). The differences from our study may indicate that Rac1 activation by R-cad may have opposite effects on the cellular phenotype, dependent on the cell context. This is consistent with our observation that Rac1 activation by R-cad occurs in three-dimensional, but not in two-dimensional cultures. Thus, in a more natural organoid context, Rac1 supports R-cad adhesion and morphogenesis. This is consistent with the known role of Rac1 in epithelial cell-cell contact formation (31). Our studies also showed that R-cad moderately competes with other endogenous cadherins. Other studies have emphasized this effect. R-cad expression in myoblasts was accompanied with down-regulation of N-cad and M-cad (22) and in BT20 cells line with loss of E-cad and P-cad (16). Our data, however, suggest that competition of endogenous cadherins is not the critical mechanism whereby R-cad suppresses malignancy because R-cad expression in a cad-11 null MDA-MB-231 cell line still induced acini and suppressed invasion.
Interestingly, R-cad expression in the MDA-MB-231 cell line was mostly cytosolic, although ectopically expressed R-cad was localized to cell-cell contacts. This implies some property of the endogenous R-cad is different. Further studies might show a novel isoform or dominant mutation. These possibilities are particularly intriguing because the cytoplasmic form might act to quench “invasive cadherins,” such as N-cad. Indeed, R-cad was found to heterodimerize with N-cad (37), thus possibly neutralizing its invasion-promoting function.
R-cad effects in vivo were further supportive of a tumor suppressive function. R-cad overexpression in the 3475 metastatic subline suppressed tumor formation and metastasis in vivo. One possible explanation is that the reduced migration and infiltration limits access of tumor cells to vascularized areas of the tumor. Another possibility is that R-cad prevents “self-seeding,” a process whereby a tumor grows by fusion of propagating colonies that arise from local invasion (38). Thus, the findings support the notion that migration and infiltration can contribute significantly to primary tumor growth. Moreover, R-cad suppressed metastatic colonization and repressed three of the four genes associated with the lung metastasis signature. Thus, the effects of R-cad, therefore, go well beyond a simple change in adhesion.
In summary, these findings show that R-cad is a newly identified mammary epithelial cadherin that exerts a profound effect on the breast tumor phenotype. R-cad promotes morphogenesis, inhibits invasion and metastasis, and represses key genes that govern metastatic virulence.
Supplementary Material
Acknowledgments
Grant support: National Cancer Institute grant 1R01 CA90872, Susan G. Komen Foundation grant BCTR0503930, and Breast Cancer Research Foundation (R.B. Hazan). J. Hulit is supported by the Susan G. Komen postdoctoral fellowship grant PDF 0503607.
We thank Drs. Martha Stampfer, Stuart Aaronson, and David Colman for providing cell lines and Dr. Masatoshi Takeichi for R-cad cDNA.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fraga MF, Herranz M, Espada J, et al. A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer Res. 2004;64:5527–5534. doi: 10.1158/0008-5472.CAN-03-4061. [DOI] [PubMed] [Google Scholar]
- 3.Bracke M, Roy FV, Mareel M. The E-cadherin/catenin complex in invasion and metastasis. In: Gunthert U, Birchmeier W, editors. Attempts to Understand Metastasis Formation I Metastasis-related molecules. Berlin: Springer; 1996. pp. 123–161. [Google Scholar]
- 4.Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem Sci. 1999;24:73–76. doi: 10.1016/s0968-0004(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 5.Vleminckx K, Vakaet L, Jr, Mareel M, Fiers W, van Roy F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66:107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 6.Oka H, Shiozaki H, Kobayashi K, et al. Expression of E-cadherin cell adhesion molecules in human breast cancer tissues and its relationship to metastasis. Cancer Res. 1993;53:1696–1701. [PubMed] [Google Scholar]
- 7.Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- 8.Cavallaro U, Schaffhauser B, Christofori G. Cadherins and the tumour progression: is it all in a switch? Cancer Lett. 2002;176:123–128. doi: 10.1016/s0304-3835(01)00759-5. [DOI] [PubMed] [Google Scholar]
- 9.Nagi C, Guttman M, Jaffer S, et al. N-cadherin expression in breast cancer: correlation with an aggressive histologic variant–invasive micropapillary carcinoma. Breast Cancer Res Treat. 2005;94:225–235. doi: 10.1007/s10549-005-7727-5. [DOI] [PubMed] [Google Scholar]
- 10.Islam S, Carey TE, Wolf GT, Wheelock MJ, Johnson KR. Expression of N-cadherin by human squamous carcinoma cells induces a scattered fibroblastic phenotype with disrupted cell-cell adhesion. J Cell Biol. 1996;135:1643–1654. doi: 10.1083/jcb.135.6.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Hulit J, Suyama K, Chung S, et al. N-cadherin signaling potentiates mammary tumor metastasis via enhanced ERK activation. Cancer Res. 2007;67:3106–3116. doi: 10.1158/0008-5472.CAN-06-3401. [DOI] [PubMed] [Google Scholar]
- 13.Agiostratidou G, Hulit J, Phillips GR, Hazan RB. Differential cadherin expression: potential markers for epithelial to mesenchymal transformation during tumor progression. J Mammary Gland Biol Neoplasia. 2007;12:127–133. doi: 10.1007/s10911-007-9044-6. [DOI] [PubMed] [Google Scholar]
- 14.Hazan RB, Phillips GR, Qiao RF, Norton L, Aaronson SA. Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J Cell Biol. 2000;148:779–790. doi: 10.1083/jcb.148.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson E, Theisen CS, Johnson KR, Wheelock MJ. R-cadherin influences cell motility via Rho family GTPases. J Biol Chem. 2004;279:31041–31049. doi: 10.1074/jbc.M400024200. [DOI] [PubMed] [Google Scholar]
- 16.Maeda M, Johnson E, Mandal SH, et al. Expression of inappropriate cadherins by epithelial tumor cells promotes endocytosis and degradation of E-cadherin via competition for p120(ctn) Oncogene. 2006;25:4595–4604. doi: 10.1038/sj.onc.1209396. [DOI] [PubMed] [Google Scholar]
- 17.Treubert-Zimmermann U, Heyers D, Redies C. Targeting axons to specific fiber tracts in vivo by altering cadherin expression. J Neurosci. 2002;22:7617–7626. doi: 10.1523/JNEUROSCI.22-17-07617.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews GL, Mastick GS. R-cadherin is a Pax6-regulated, growth-promoting cue for pioneer axons. J Neurosci. 2003;23:9873–9880. doi: 10.1523/JNEUROSCI.23-30-09873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahl U, Sjodin A, Larue L, et al. Genetic dissection of cadherin function during nephrogenesis. Mol Cell Biol. 2002;22:1474–1487. doi: 10.1128/mcb.22.5.1474-1487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenberg P, Esni F, Sjodin A, et al. A potential role of R-cadherin in striated muscle formation. Dev Biol. 1997;187:55–70. doi: 10.1006/dbio.1997.8602. [DOI] [PubMed] [Google Scholar]
- 21.Sjodin A, Dahl U, Semb H. Mouse R-cadherin: expression during the organogenesis of pancreas and gastrointestinal tract. Exp Cell Res. 1995;221:413–425. doi: 10.1006/excr.1995.1392. [DOI] [PubMed] [Google Scholar]
- 22.Kucharczak J, Charrasse S, Comunale F, et al. R-Cadherin expression inhibits myogenesis and induces myoblast transformation via Rac1 GTPase. Cancer Res. 2008;68:6559–6568. doi: 10.1158/0008-5472.CAN-08-0196. [DOI] [PubMed] [Google Scholar]
- 23.Miotto E, Sabbioni S, Veronese A, et al. Frequent aberrant methylation of the CDH4 gene promoter in human colorectal and gastric cancer. Cancer Res. 2004;64:8156–8159. doi: 10.1158/0008-5472.CAN-04-3000. [DOI] [PubMed] [Google Scholar]
- 24.Gupta GP, Nguyen DX, Chiang AC, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 25.Cheng SL, Lecanda F, Davidson MK, et al. Human osteoblasts express a repertoire of cadherins, which are critical for BMP-2-induced osteogenic differentiation. J Bone Miner Res. 1998;13:633–644. doi: 10.1359/jbmr.1998.13.4.633. [DOI] [PubMed] [Google Scholar]
- 26.Suyama K, Shapiro I, Guttman M, Hazan RB. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell. 2002;2:301–314. doi: 10.1016/s1535-6108(02)00150-2. [DOI] [PubMed] [Google Scholar]
- 27.Albini A, Iwamoto Y, Kleinman HK, et al. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47:3239–3245. [PubMed] [Google Scholar]
- 28.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 29.Feltes CM, Kudo A, Blaschuk O, Byers SW. An alternatively spliced cadherin-11 enhances human breast cancer cell invasion. Cancer Res. 2002;62:6688–6697. [PubMed] [Google Scholar]
- 30.Pishvaian MJ, Feltes CM, Thompson P, Bussemakers MJ, Schalken JA, Byers SW. Cadherin-11 is expressed in invasive breast cancer cell lines. Cancer Res. 1999;59:947–952. [PubMed] [Google Scholar]
- 31.Ehrlich JS, Hansen MD, Nelson WJ. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell-cell adhesion. Dev Cell. 2002;3:259–270. doi: 10.1016/s1534-5807(02)00216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyckoff JB, Jones JG, Condeelis JS, Segall JE. A critical step in metastasis: in vivo analysis of intravasation at the primary tumor. Cancer Res. 2000;60:2504–2511. [PubMed] [Google Scholar]
- 34.Koop S, Schmidt EE, MacDonald IC, et al. Independence of metastatic ability and extravasation: metastatic ras-transformed and control fibroblasts extravasate equally well. Proc Natl Acad Sci U S A. 1996;93:11080–11084. doi: 10.1073/pnas.93.20.11080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 36.Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci U S A. 1995;92:7416–7419. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shan WS, Tanaka H, Phillips GR, et al. Functional cis - heterodimers of N- and R-cadherins. J Cell Biol. 2000;148:579–590. doi: 10.1083/jcb.148.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norton L, Massague J. Is cancer a disease of self-seeding? Nat Med. 2006;12:875–878. doi: 10.1038/nm0806-875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.